Abstract
Current standard cancer therapies (chemotherapy and radiation) often cause serious adverse off-target effects. Drug design strategies are therefore being developed that will more precisely target cancer cells for destruction while leaving surrounding normal cells relatively unaffected. Telomerase, widely expressed in most human cancers but almost undetectable in normal somatic cells, provides an exciting drug target. This review focuses on recent pharmacogenomic approaches to telomerase inhibition. Antisense oligonucleotides, RNA interference, ribozymes, mutant expression, and the exploitation of differential telomerase expression as a strategy for targeted oncolysis are discussed here in the context of cancer therapeutics. Reports of synergism between telomerase inhibitors and traditional cancer therapeutic agents are also analyzed.
Keywords: Telomerase inhibition, hTR, hTERT, antisense, RNAi, ribozyme, dominant-negative hTERT, mutant-template telomerase RNA
INTRODUCTION
Drug design in the area of cancer therapeutics is developing a trend toward more precise mechanisms of cancer cell destruction thereby minimizing adverse off-target effects incurred during the course of standard cancer treatment (nausea, vomiting, hair loss, fatigue, organ toxicity, etc). The key to selectively targeting cancer cells is to exploit some basic difference these cells have developed compared to their normal precursors. One such difference is the activity of the enzyme telomerase.
Telomeres, Telomerase, and Cancer
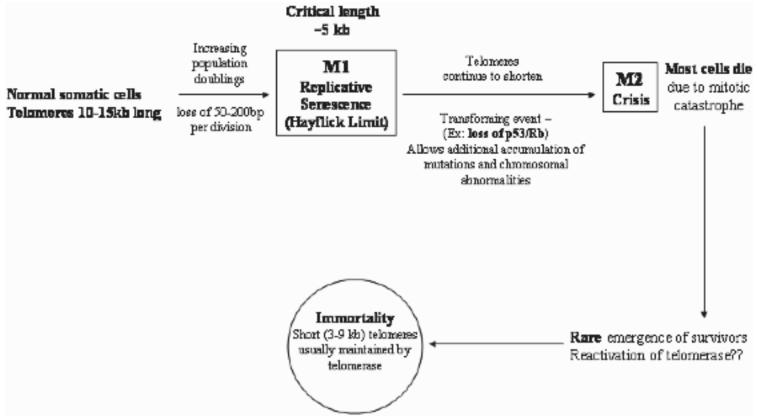
Telomerase is a ribonucleoprotein that synthesizes telomeres. Telomeres consist of tandem oligonucleotide repeats (5'-TTAGGG-3' in humans) that cap the ends of eukaryotic chromosomes. Normal human somatic cells contain up to 10-15 kilobases of telomeric repeats [1, 2]. Telomeres appear to have a dual function. First, telomeres may serve to protect the ends of chromosomes from damage and prevent the cell from recognizing the ends as double-strand breaks that might lead to adverse recombination. Second, telomeres have been proposed to act as a ‘mitotic clock’ that counts the number of divisions a cell has undergone and its capacity for continued division [3, 4]. Leonard Hayflick observed years ago that normal cells have a finite replicative capacity in vitro after which they remain metabolically active but cease to proliferate [5]. This period of growth arrest is referred to as M1 (mortality 1) or cellular senescence. A direct relationship between telomere length and cellular senescence has been established [6]. Because of the end-replication problem [7, 8], 50-200 bp of telomeric DNA is lost with every round of replication. The non-coding telomeric repeats provide a buffer that prevents the loss of genetic information during each cycle of replication. When the telomeres have eroded to a critical minimum length (∼5 kb), cellular senescence is triggered. Cellular senescence might be bypassed by repression of tumor suppressor genes, activation of oncogenes, or other mutations [9]. By escaping senescence, rare cells continue to divide and their telomeres continue to shorten until they reach a critical stage (M2 or crisis). At this point, chromosomal instability arises due to end-to-end fusions and/or chromosome breakage. DNA damage checkpoints are activated along with apoptosis. Unless the cell develops a mechanism through which to stabilize telomere length, it will not survive. Cells that escape crisis and become immortalized generally achieve telomeric stability through the reactivation of telomerase (Fig. 1).
Fig. 1.
Telomeres erode in normal somatic cells with every population doubling due to the virtual absence of telomerase. Reactivation of telomerase appears to play a key role in the development of cancer.
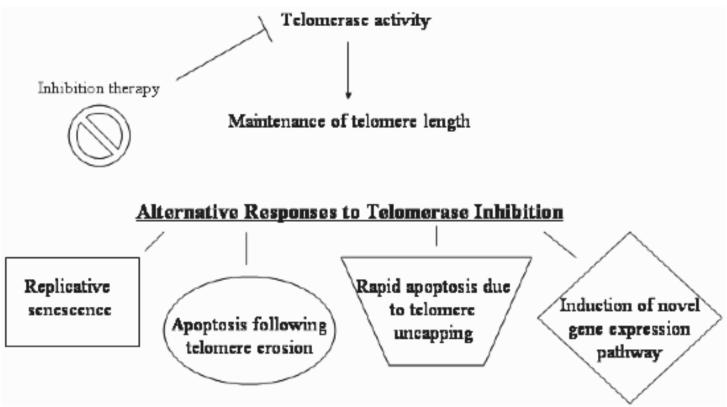
Telomerase is a ribonucleoprotein that acts to elongate telomeres in cells that possess its activity. This enzyme is expressed during embryonic development, loses its expression during differentiation of somatic cells, and is almost undetectable in most normal human somatic cells [10]. By contrast, telomerase is expressed in ∼85% of human cancers [11]. There are a few types of cells that normally express telomerase including germ line cells, stem cells, hematopoietic cells, cells lining the intestine, and other rapidly proliferating cells. The widespread expression of telomerase in a variety of human cancers, while being almost undetectable in most normal cells, makes it a very attractive drug target. Normal somatic cells are thought to harbor enough telomeric DNA reserve to withstand telomere-based therapeutics, and the few normal cells which express telomerase should also have enough reserve to withstand treatment with telomerase inhibitors. It has been shown that cancer cells often maintain much shorter telomeres than normal cells (3-9 kb compared to 10-15 kb) [12-15]. Additionally, the rapid proliferative nature of cancer cells leads to steady telomere erosion in the absence of telomerase. Telomerase-based therapeutics should therefore impact tumor cells before having any appreciable effect on telomerase-positive normal cells. Potential outcomes of telomerase-based therapeutics are illustrated in Fig. 2.
Fig. 2.
Diagram illustrating the possible outcomes of telomerase inhibition. Inhibition of telomerase prevents the maintenance of telomere length in telomerase-positive cells. As a result, telomerase may shorten, leading to eventual replicative senescence or apoptosis. Telomerase inhibition may also cause rapid cell death without telomere shortening and the induction of a novel gene expression pathway (discussed later in review).
Telomerase contains an RNA component, hTR, and a catalytic reverse transcriptase component, hTERT. While hTR is ubiquitously expressed in normal cells, it is the presence of hTERT that confers telomerase activity [6]. Without expression of hTERT there is no telomerase activity and consequently elongation of telomeres does not occur. Several different approaches to telomerase inhibition are currently in laboratory use with new ones continually being sought as a result of the growing interest in selective telomerase inhibition as a strategy for rational pharmaceutical design. Small molecules such as AZT (azidothymidine, a non-specific reverse transcriptase inhibitor) [16], chemicals such as retinoids [17], tamoxifen [18], or EGCG (epigallocatechin gallate) [19], and molecules which interfere with telomere structure (i.e., G-quadruplex stabilizers) [20, 21] have been shown to be effective in vitro inhibitors of telomerase transcription or function. While these compounds may be effective in vitro, there is legitimate concern surrounding their specificity. AZT has been shown to inhibit cell growth and telomerase activity of breast cancer cells in vitro [16]. However, AZT is not specific for telomerase, being most recognized for its use in managing HIV infection. Retinoids, which are vitamin A analogues, are able to induce telomere shortening, cell growth arrest, and cell death in acute promyelocytic leukemia (APL) cells [17]. High levels of some retinoids, however, can cause toxicity in vivo. The need is apparent, therefore, to develop specific molecular-based therapies that are both effective and exhibit an acceptable side effect profile.
The RNA component hTR and the reverse transcriptase hTERT are not the only components of telomerase required to maintain telomere length and structure. There are also a number of proteins associated with telomeric DNA that have been explored as possible drug targets. While their potential as drug targets will not be covered in this review, some basic understanding of these proteins is helpful. Although quite a number of proteins play a role in maintaining telomere structure and length, some of the more notable ones include the human telomeric proteins TRF1 and TRF2 and POT1. (For an extensive review of these and other telomeric proteins, see [22]). Briefly, TRF1 and TRF2 are telomeric repeat binding factors. TRF1 binds to the telomeric DNA duplex and acts to regulate telomere length through a negative feedback loop. The number of TRF1 proteins present on the end of a chromosome correlates with the length of the telomere [22]. TRF2 helps to form the t-loop structure at the end of the telomere [23], and likely helps to physically prevent telomerase from acting on telomere ends [22]. TRF2 plays a key role in the protection of chromosome ends (see review [24]). POT1 (protection of telomeres) is a single-stranded telomeric DNA binding protein that can associate with TRF1 and may regulate the amount or the frequency of telomere elongation [22].
The focus of this review will be on inhibitors of either the hTR or hTERT components of telomerase and therapies currently being developed that exploit the unique nature of telomerase expression.
TARGETING TELOMERASE ENZYME COMPONENTS
Antisense and Related Oligonucleotides
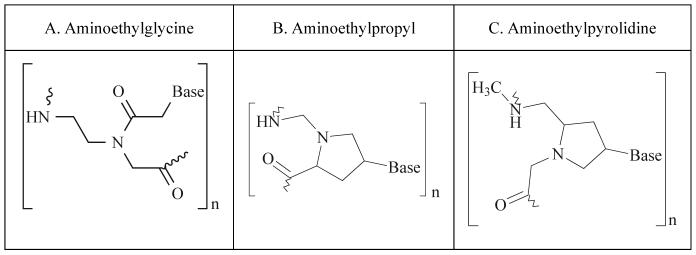
One of the oldest and most commonly used classes of telomerase inhibitory agents is antisense DNA oligonucleotides. The use of antisense molecules to block the translation of mRNA into a functional protein has been commonly used since the 1990s. In the classical sense, antisense technology utilizes nucleotides with sequence complementarity to sense RNAs. These oligonucleotides may be designed to occupy the ribosome binding site, preventing the ribosome from binding the target mRNA (Fig. 3). Antisense oligos targeted to sequences downstream of the ribosome binding site anywhere in the coding sequence will prevent ribosomal translocation, halting translation and producing a non-functional or truncated protein [25]. There have been numerous modifications of antisense molecules reflecting efforts to enhance their cellular up-take, potency, and half-life (Fig. 4). Some modifications help to recruit the activity of RNase H, which degrades the RNA strand of an RNA-DNA duplex. After the degradation of the mRNA component of the duplex, the antisense DNA molecule is released and becomes free to bind other target mRNA molecules. Phosphorothioate (PS) linkages slow the degradation of the antisense molecules in the cells and enhance their half-life [26]. Peptide nucleic acids (PNA) are molecules in which the antisense bases are connected to various peptide backbones (Fig. 5). These modifications have been found to improve the half-life of antisense oligomers and enhance hybridization properties [27]. 2'-O-Methyl- and 2'-methoxyethyl-modified RNAs increase the affinity of antisense molecules for their specific targets [25]. Several of the various types of antisense molecules have made their way into clinical trials. One, Vitravene, has gained FDA approval for the treatment of CMV-induced retinitis [28].
Fig. 3.
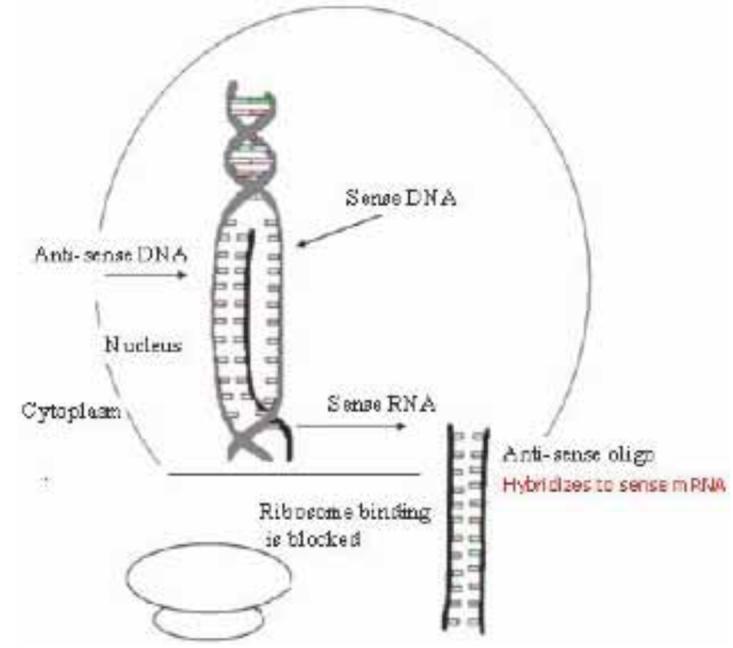
Illustation of antisense mechanism of action in blocking protein expression.
Fig 4.
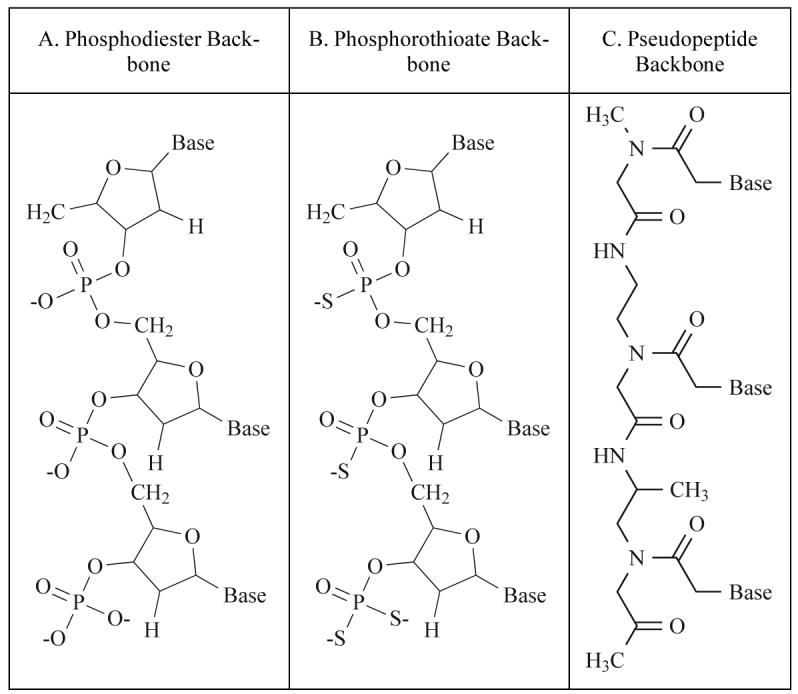
Examples of modifications made to traditional antisense oligonucleotides (A).
Fig 5.
Three variations of peptide nucleic acids (PNAs).
The use of antisense technology in telomerase inhibition is not new. In fact the first report of successful inhibition of telomerase activity involved the use of antisense oligonucleotides against hTR 10 years ago [29]. Feng et al. reported in 1995 that antisense oligonucleotides complementary to sequences within or near the human telomeric template RNA resulted in suppression of telomerase activity while antisense oligonucleotides against targets that were more distant from the telomeric template failed to inhibit the action of the ribonucleoprotein. As a result, HeLa cells that were transfected with the antisense-hTR expression construct underwent crisis after 23 to 26 population doublings and exhibited a loss of telomeric DNA repeats. In contrast, telomerase-negative foreskin cells transfected with the construct expressing antisense-hTR did not enter crisis during the same time period. This study was the first to demonstrate the therapeutic potential of antisense oligonucleotide expression as a treatment for human cancers. Building on the findings of Feng et al., others have used the anti-hTR expression vector to inhibit telomerase activity in other cancer cell lines. Human gastric cancer cells transfected with a vector expressing antisense-hTR demonstrated shortening of telomere length and an elevated level of apoptosis, suggesting that the antisense-mediated telomere shortening in gastric cancer cells acts to induce apoptosis [30]. Even if apoptosis is not induced by introduction of the antisense-hTR expression vector, the construct may still be effective in reducing the aggressiveness of cancer cells (lowering invasive capacity and tumorigenicity). This was found to be the case with various malignant glioma cell lines [31]. Upon introduction of the antisense-hTR construct, apoptosis occurred in only some of the cell populations after 30 population doublings. Other cell populations avoided apoptosis, but appeared to differentiate and diverge in morphology from their parental cells, demonstrating that telomerase inhibition can trigger distinctly different desirable results: apoptosis or differentiation. Although not a direct effecter of tumor cell death, differentiation may be a viable therapeutic outcome as it tempers the effect of the malignant phenotype.
Delivery of telomerase-inhibiting agents as a component of cancer therapeutics presents the need for stable expression of the inhibitor in vivo. An increasingly common approach to stable expression of specific genomic therapeutics is the use of modified retroviral, lentiviral, or adenoviral delivery systems. Successful delivery and expression of antisense RNA by replication-deficient retrovirus in HeLa cells and human kidney carcinoma cells has been shown to be effective in inhibiting telomerase activity by at least 75% in vitro [32]. Additionally, a hybrid adenovirus/adeno-associated virus has been used to express antisense-hTR in MCF-7 breast cancer cells [33]. The stable expression of antisense hTR in these cells resulted in significant suppression of telomerase activity and progressive telomere shortening for 30 population doublings along with induction of apoptosis, reduction of cell proliferation, and reduction of colony formation as demonstrated by soft agar assay.
While the preponderance of literature appears to report the use of antisense molecules targeting the RNA component of telomerase (hTR), antisense-mediated hTERT inhibition has also been successfully achieved. Antitumor action was observed when antisense oligodeoxynucleotides (mostly 20-mers) against various regions of the hTERT mRNA were introduced into human bladder cancer cells by transfection [34]. The amount of hTERT transcript was reduced, cell viability was significantly impaired, and G1 arrest was induced. Interestingly, when an antisense expression vector coding for antisense RNA against hTERT was transfected into human breast cancer cells, decreased telomerase activity was observed as well as significant apoptosis [35]. However, these phenomena were observed 24 hours post-transfection and were not accompanied by significant shortening of telomeric DNA. These findings support other observations of telomerase inhibition rapidly inducing apoptosis independent of telomere erosion [36, 37], and are significant because they address the traditional concern that telomerase inhibition would incur a substantial lag time between the onset of telomerase inhibition and the sufficient erosion of telomeres as to cause growth arrest of cancer cells. The simultaneous inhibition of hTR and hTERT has been found to inhibit telomerase activity synergistically [38], suggesting another strategy for antisense oligonucleotide therapy.
Improvements in antisense technology have led to improvements in introduction of the molecules into cells, stability, lengthening of half-life, and specificity of target binding. Modifications of traditional antisense oligonucleotides used in telomerase inhibition include 2'-5'-oligoadenylate (2-5A) linkages [39, 40], 2'-O-methyl-RNA [41], phosphorothioate-modified oligodeoxynucleotides (PS-ODN) [34, 42, 43], peptide nucleic acids (PNA) [44-46], and locked nucleic acids (LNA) [47].
One remaining group of compounds that warrants discussion includes the oligonucleotide N3'-P5' phosphoramidates and N3'-P5' thio-phosphoramidates. This class of oligonucleotides has attracted attention lately as one of its derivatives, GRN163L, is well on its way to becoming the first telomerase inhibitor to be available for cancer treatment. In the last five years, oligonucleotide phosphoramidates were designed with sequence complementarity to either the template region of hTR or a region ∼100 nucleotides downstream of the template region [48-50]. The N3'-P5' phosphoramidates (NP oligonucleotides) and the N3'-P5' thio-phosphoramidates (NPS oligonucleotides) form sequence-specific duplexes with target RNA. N3'-P5' thio-phosphoramidates were designed to combine the RNA binding affinity and sequence specificity of phosphoramidates with the abilitiy of the phosphorothioate oligonucleotides to interact with proteins (i.e., hTERT) [51]. Both NP and NPS oligonucleotides inhibited telomerase activity, causing telomere shortening, senescence, and eventual apoptosis; however, NP oligonucleotides were inefficient without the use of a lipid carrier to facilitate their introduction into the cells. NPS oligonucleotides were significantly more potent telomerase inhibitors than their parent molecules, even without the use of lipid carriers. NP deoxyoligonucleotides exhibited IC50 values in the 0.5-1 μM range compared to NPS deoxyoligonucleotides, which had IC50 values in the 0.5-5 nM range (both with cellular uptake enhancers) [49].
The GRN163 13-mer has emerged as an exciting N3'-P5' thio-phosphoramidate therapeutic candidate, resulting from the optimization of NP and NPS inhibitory strategies [51]. Early testing in cell-free assays demonstrated IC50 values as low as 26-44 pM [51]. Significantly, GRN163 was shown to inhibit telomerase activity in a variety of cancer cell lines in vitro 24-72 hours after treatment (using the cell-based TRAP assay Telomeric Repeat Amplification Protocol); the survival of normal human cells, WI-38 and BJ fibroblasts, was not affected, even with treatments of up to 100 μM GRN163 for 72 hours [51]. Treatment with GRN163 has been shown to induce telomere shortening, growth arrest, and cell death in human multiple myeloma [52, 53] and non-Hodgkin lymphoma [53] cell lines, as well as tumor growth suppression in a prostate cancer xenograft model [54].
A second generation oligonucleotide, GRN163L, has recently been shown to be more potent than its GRN163 predecessor, causing more rapid shortening of telomeres and cell growth inhibition and having an average of sevenfold lower IC50 values in various cell lines tested [55]. The difference in GRN163L is a lipid modification of the first generation oligonucleotide. Challenges in therapeutic oligonucleotide delivery have often made it necessary to use lipid-based transfection reagents or carriers for in vitro experimentation. Lipid modification of the oligonucleotide in this case has apparently eliminated the requirement for an extraneous lipid carrier. Manufacturing of GRN163L has been ongoing in order to supply enough of the drug for toxicity and pharmacokinetic studies in animals and Phase I clinical studies, making it potentially the first telomerase inhibitor for cancer therapeutics1. The uses of antisense technology in the inhibition of telomerase are summarized in Table 1.
Table 1.
Antisense Oligonucleotide Effects in Telomerase Inhibition
| Target | Cells Tested* | Effect of treatment | Efficiency of inhibiton | Ref. |
|---|---|---|---|---|
| hTR | HeLa (cervical) | Loss of telomeric DNA; crisis after 23-26 PD | Reduction of activity | [29] |
| hTR | SGC7901 (gastric) | Telomere length shortening; increased apoptosis | Significant reduction of activity |
[30] |
| hTR | U251-MG (malignant glioma) |
Apoptosis only in some populations after 30 PD; differentiation of some populations |
Complete ablation of activity |
[31] |
| hTR | HeLa (cervical) A498 (kidney) |
Reduction in cell viability; appearance of giant, senescent-like cells; reduction in growth rate |
≥75% inhibition of telomerase activity | [32] |
| hTR | MCF-7 (breast) | Progressive telomere shortening for 30 PD; induction of apoptosis; reduction of proliferation and colony formation |
significant suppression of telomerase activity |
[33] |
| hTERT | EJ28, 5637, J28, HT1197 (bladder) |
Reduction of hTERT mRNA (≤88%); impairment of cell viability; G1 arrest |
≤60% inhibition of activity | [34] |
| hTERT | PMC42 (breast) | Decreased telomerase activity; significant apoptosis; Rapid effect: 24 hours post-transfection No significant telomere shortening |
Up to 50% reduction of activity |
[35] |
| hTR and hTERT simultaneously |
SW480 (colon) |
Significant inhibition of proliferation by 24 hours Decrease in telomerase activity by 48 hours continuing to decrease through 72 hours Significant increase in apoptosis |
combination treatment 0.2 mmol/L for 72 hours; 80% inhibition of activity |
[38] |
| hTR (2-5A) |
U251-MG (malignant glioma) |
Significant decrease in cell viability after 5 days Significant impairment of tumor growth in nude mice |
hTR undetectable after treatment |
[39] |
| hTR (2-5A) |
PC3, DU145 (prostate) |
Significant decrease in cell viability within 6 days Significant suppression of tumor growth in nude mice |
Cell viability reduced to 9-18% within 6 days of treatment |
[40] |
| hTR (2′-O-Me) |
DU145, LNCaP (prostate) |
Telomere shortening; halt of cell proliferation after delay (in relation to telomere shortening); 90% reduction in colony forming ability in LNCaP cells |
>75% inhibition of activity up to >85% inhibition in DU145 |
[91] |
| hTERT (PS) |
EJ28 (bladder) |
Maximum decrease in hTERT mRNA 12 hours after treatment; immediate and continued reduction of cell viability with successive transfections of constructs |
>60% inhibition of activity; up to 88% reduction in hTERT mRNA |
[34] |
| hTR (PS) |
HL-60 (leukemia) |
Efficient but not selective; non-complementary constructs had nearly same effect as complementary; more efficient than PNA |
IC50 of 0.5 and 0.6 nM | [42] |
| hTR (PS) |
MKN-28, SGC7901, MKN-45 (gastric) |
After 96 hours, significant growth inhibition in poor and moderately differentiated cell lines but not in well-differentiated Apoptosis in poor and moderately differentiated lines |
Inhibition of activity at 5 mmol/L; complete inhibition at 10 mmol/L |
[43] |
| hTERT (PNA) |
DU145 (prostate) U2OS (osteogenic sarcoma) |
Efficient introduction of naked PNA against hTERT using photochemical internalization approach; effect most prominent 6 hours after treatment becoming less pronounced 24-48 hours after treatment |
Telomerase activity reduced to 8.4 ± 0.79% of control |
[46] |
| single stranded G-rich overhang (PNA) |
AT-SV1 †(Ataxia telangiectasia) |
No inhibition of telomerase activity; decrease in colony sizes; slight decrease in median telomere length Synergistic effect with PNA blocking telomerase activity |
Virtual elimination of colony formation when combined with telomerase inhibitor |
[44] |
| hTR (PNA) |
AT-SV1 †(Ataxia telangiectasia) |
Inhibition of telomerase activity; proliferation arrest after 5 to 30 generations; median telomere length shortened by 377 bp; reduction in colony size |
62% reduction of telomerase activity with 10 mM treatment |
[45] |
| hTR (LNA) |
DU145 (prostate) |
Inhibition of activity up to 40 hours post-transfection High-affinity binding and selectivity |
Some LNAs resulted in >80% inhibition of telomerase activity |
[47] |
| hTR (NP and NPS) |
various | Telomerase inhibition by both NP and NPS resulting in reduction of telomere length NP: most effective targets within template region NPS: may use PS group interacting with hTERT to stabilize secondary structure |
NP: sub-nM IC50 NPS: ∼50 pM IC50 |
[48] |
| hTR (NP and NPS) |
HME50-5E †(spontaneously immortalized breast epithelial cells) |
NP inefficient without lipid carrier; NPS efficient with or without carrier; 0.5 mM NPS caused telomere shortening, senescence by day 100, massive apoptosis by day 115 Addition of -thio group significantly increased potency |
NP: IC50 ∼0.5-1 mM with lipid NPS: IC50 0.5-5 nM with lipid |
[49] |
| hTR (GRN163) |
various | Inhibition of telomerase activity within 24-72 hours after treatment WI-38 and BJ fibroblast survival not affected by up to 100 mM treatment for 72 hours Induction of telomere shortening, growth arrest, cell death, and tumor growth suppression in nude mice |
IC50 of 26-44 pM | [51- 54] |
| (GRN163L) | various | Sevenfold lower IC50 in tested cell lines; more rapid telomeric attrition and growth inhibition; lipid modification eliminates need for carrier In early clinical trials |
[55] |
Antisense modifications: 2-5A = 2′-5′-oligoadenylate; 2′-O-Me = 2′-O-methyl-RNA; PS = phosphorothioate; PNA = peptide nucleic acid; LNA = locked nucleic acid; NP = oligonucleotide N3′-P5′ phosphoramidates; NPS = N3′-P5′ thio-phosphoramidates.
All listed are cancer cell lines unless otherwise specified (†)
RNA Interference
Since the RNA interference (RNAi) phenomenon was first described in 1998 [56], RNAi-mediated gene knockdown has become the latest must-have tool for analysis of gene function and is emerging as a potential treatment strategy for a variety of human diseases, including cancer. Briefly, RNAi involves the use of short double-stranded RNA molecules to activate a natural pathway which results in the degradation of the homologous target mRNA.
Clearly the essential components of telomerase, hTR and hTERT, present themselves as attractive targets for RNAi therapeutics. There have been attempts made to knock down expression of hTR and hTERT by transfection of small interfering RNAs (siRNAs) and stable expression of siRNA constructs in vitro using plasmids or viral expression vectors. Kosciolek et al. first inhibited telomerase using RNAi in 2003 [57]. This group used 21-nucleotide siRNAs against either hTR or hTERT. The region of hTERT that was targeted by this group was the same one used by Hahn et al. in their dominant negative mutation [58]. Human colon carcinoma cells were transfected with siRNA against either hTR or hTERT. Telomerase activity was inhibited in a dose-dependent manner, with the siRNA against hTR proving more effective than the siRNA against hTERT. The maximum effect observed in that cell line was a 75% decrease in telomerase activity in cells transfected with siRNA against hTR compared to a 65% decrease in activity in those cells treated with siRNA against hTERT. Perhaps this difference may be attributed to target selection for each component. It is also possible that direct RNAi of the RNA subunit of telomerase is more easily achieved than indirect RNAi of the protein component, hTERT. Similar results were observed when the siRNAs were used in HeLa cervical carcinoma cells as well as other types of carcinoma cells and sarcoma cells of mesodermal origin. Simultaneous treatment of HeLa cells with siRNAs against both hTR and hTERT was not found to result in greater inhibition of telomerase activity than each siRNA separately. As expected, the effects of direct siRNA transfection were transient in nature. A DNA construct expressing a hairpin structure targeting hTR was used to transfect HeLa cells to assess long-term effects [57]. At 75 days after transfection, 4 out of 5 transfected clones exhibited reduced telomeric DNA content (45%) compared to control cells. The apparent effectiveness of RNA interference directed against telomerase came as somewhat of a surprise, as RNAi is usually considered as being restricted to the cytoplasm while telomerase is regarded as being generally confined to the nucleus. While telomerase may be manufactured in the cytoplasm, its site of function is the nucleus. However, it does appear that telomerase remains in the cytoplasm at least long enough for RNA interference to cause hTR or hTERT mRNA degradation with respectable efficiency.
Soon after the initial publication of results demonstrating the ability of RNAi to diminish telomerase activity, results were published that proved the usefulness of retroviral-mediated RNA interference against hTERT [10]. The retroviral vector pMKO.1-puro [59] was used to express a short hairpin RNA (shRNA) in HeLa cells, causing suppression of hTERT mRNA and protein expression and abrogation of telomerase enzymatic function. Normal human fibroblasts (BJ and WI-38) were shown by this group to have transient telomerase activity during S phase of the cell cycle; introduction of shRNA against hTERT in these cells caused complete disruption of this transient activity, slowed their proliferation, and delayed their entry into S phase (cells accumulated in the G2/M phase). These cells continued to proliferate, although more slowly than control cells. After extended culture, morphological changes and senescence-associated β-galactosidase staining indicated that fibroblasts infected with hTERT retroviral shRNA constructs entered replicative senescence more quickly than uninfected control cells. Members of the same group recently expanded their findings on retroviral delivery of hTERT shRNA in cervical cancer cells [60]. Cells with compromised hTERT expression had impaired telomerase activity, shortened telomeres and shortened telomeric 3' overhangs (necessary for formation of the t-loop) with increasing population doublings. Even after a relatively short time (5 population doublings), infected cells demonstrated a decreased proliferative rate. Low-passage cells also had impaired colony-forming ability and tumorigenicity in mice. Finally, treatment with shRNA against hTERT appeared to significantly enhance sensitivity of cells to treatment with conventional therapeutic agents that are known to induce DNA double-strand breaks (topoisomerase inhibitors, bleomycin, and radiation).
Another example of RNA interference against hTERT has been demonstrated in Barrett's adenocarcinoma cells [61]. Two regions of hTERT were targeted using two siRNA duplexes; a mixture of these siRNAs was used to transfect the esophageal adenocarcinoma cell line SEG-1. The siRNA treatment was extremely efficient, causing a reduction in telomerase protein expression to only 5% of cells treated with control siRNAs. Telomerase activity was observed to be inhibited by day 1 and was essentially completely ablated by day 3. Greater than 80% of cells treated with the siRNAs for 3 weeks underwent apoptosis. Treatment also led to significant telomeric attrition, with more than 50% of treated cells having a complete loss of detectable telomeres. Gene expression profiling of treated cells revealed increased levels of p73, p63, and E2F1 as well as cell cycle arrest genes p21, p16, and GADD45. There was also elevated expression of several genes involved in apoptosis, including FasL, Fas, caspases 8, 7, and 3, and CARD 9.
Interesting results have been obtained using lentiviral delivery of shRNA against hTR [62]. With an infection efficiency of >95%, eliminating the need for antibiotic selection and consequential delay in assessing results, the effects of hTR inhibition were able to be analyzed almost immediately after infection. The hairpin RNA expressed by the lentiviral vector efficiently knocked down endogenous levels of hTR in vitro and caused rapid cell growth inhibition and apoptosis independent of cellular p53 status and telomere length, and without bulk telomere shortening. The same group found similar results when employing lentiviral delivery of mutant-template telomerase RNA (to be discussed in more detail later in this review). When co-expressed in vitro, the shRNA and mutant-template hTR synergistically killed cancer cells. Further experimentation with the lentiviral expression of shRNA against hTR has brought forth evidence that the aforementioned hTR knockdown in cancer cells actually induces global changes in gene expression through a novel response pathway, causing the suppression of genes likely involved in other facets of cancer progression, including angiogenesis and metastasis [63]. This exciting finding has implications for expanding not only the current knowledge base concerning the nature of telomerase inhibition and its resulting phenotypes (telomere uncapping, changes in gene expression, senescence, and apoptosis), but also potential therapeutic insights. Table 2 presents a summary of the reports of RNA interference against telomerase.
Table 2.
RNA Interference for Telomerase Inhibition
| Target | Cells Used* | Results of Treatment | Efficiency | Ref. |
|---|---|---|---|---|
| hTR | MCF-7 (breast) HCT116 (colon) |
Rapid effect with no bulk telomere shortening; p53- independent growth inhibition and apoptosis Lentiviral expression - >95% infection efficiency eliminates need for drug selection |
∼90% hTR content reduction 4 days after nfection | [62] |
| hTR | HCT116 (colon) LOX (melanoma) |
No telomere uncapping or DNA damage response; novel change in gene expression (decreased expression of cell cycle progression genes and genes involved in tumor growth, angiogenesis, and metastasis) Inhibition of cancer cell growth without any bulk telomere shortening; no resemblance of senescence |
∼96% reduction in telomerase activity | [63] |
| hTERT | HeLa (cervical) †BJ fibroblasts |
Retroviral shRNA in HeLa: decreased hTERT mRNA and protein expression BJ: shRNA slowed cell proliferation; cells entered senescence more quickly than control |
HeLa: abolished enzymatic function BJ: complete inhibition of transient telomerase activity |
[10] |
| hTERT | HeLa (cervical) | >90% reduction in hTERT mRNA; significant telomere shortening 10 PD after infection; shortened 3′- telomeric overhang; impaired colony forming and tumorigenicity in nude mice; increased sensitivity to radiation and chemotherapeutics |
>90% reduction in telomerase activity | [60] |
| hTERT | SEG-1 (Barrett's-derived esophageal adenocarcinoma) |
Inhibition of activity observed by day 1, complete by day 3 Marked reduction in median telomere length Undetectable telomeres in >50% of treated cells; telomere loss accompanied by senescence and apoptosis Increased expression of p73, p63, E2F1; increased levels of p21, p16, GADD45; elevated FasL, Fas, and caspases |
95% reduction of telomerase protein; complete inhibition of telomerase activity by day 3 | [61] |
| hTR/hTERT | various | siRNA against hTR more effective than siRNA against hTERT; increased siRNA concentrations did not increase effectiveness of telomerase inhibition; successive siRNA treatments more effective than single treatment Hairpin construct against hTR effective at least 75 days Combination treatment of siRNA against hTR and hTERT not better than either alone |
hTR siRNA: 75% inhibition of activity hTR hairpin: up to 87% reduction of activity hTERT siRNA: 65% decrease in activity |
[57] |
All listed are cancer cell lines unless otherwise specified (†).
Although RNAi technology is relatively new, advances are emerging rapidly. Two siRNA drugs intended for the treatment of age-related macular degeneration (AMD) are undergoing Phase I clinical trials, the first human clinical trials for pharmaceutical siRNA [64].
Ribozymes
Ribozymes are catalytic RNA molecules which have specific endoribonuclease activity. The ability of these molecules to cleave specific sites of target RNAs has generated considerable interest in using them as human gene therapeutic agents. The first description of a ribozyme serving as a telomerase inhibitor came in 1996. In that report [65], a hammerhead ribozyme designed to cleave at the downstream end of the template region of hTR RNA was mixed with extracts of human hepatocellular carcinoma cells. The result was inhibition of telomerase activity by up to 90%, even under reaction conditions which were sub-optimal for ribozyme activity. Later efforts to target hTR RNA with ribozymes resulted in reduction of telomerase activity in both studies [66, 67] but with slightly different outcomes. While human melanoma cells transfected with an expression vector delivering a ribozyme against hTR RNA displayed a significant increase in doubling time, a decrease in DNA synthesis, and some increased apoptosis compared to control cells, telomere shortening was not observed, even after 30 population doublings [66]. Ribozyme expression by a vector introduced into the human breast cancer cell line MCF-7 caused significant decreases in hTR expression among cell clones and dramatic decreases in telomerase activity (complete ablation in some clones, weak activity in others correlating with hTR expression) [67]. Cells bearing the ribozyme expression vector coding the ribozyme R1 (which targeted the template region of the hTR RNA) showed markedly decreased cell proliferation, morphologic changes, and signs of apoptosis. In contrast to the results caused by the ribozyme-mediated telomerase inhibition in melanoma cells [66], ribozyme-mediated telomerase inhibition in the MCF-7 cell line caused a significant shortening of telomere lengths in treated MCF-7 cells compared to control MCF-7 cells [67].
Ribozyme inhibition has also been successfully attempted against various targets on the hTERT transcript. Early attempts targeted the extreme ends of the transcript outside of the open reading frame [68]. Selecting sites at a number of locations distributed throughout the mRNA transcript, Yokoyama et al. found that 13 nucleotides downstream from the 5'-end and 59 nucleotides upstream from the poly(A) tail made the best targets for their ribozymes. In their experiments, endometrial carcinoma cells stably transfected with ribozyme-expressing plasmids maintained cell growth rates for 2 months after transfection, then slowed gradually. One clone died out completely within 4 months, and some growth-arrested cells displayed signs of apoptosis. An interesting finding was reported by a group delivering the same ribozyme into cells by two different methods [69]. Stable transfection of a plasmid encoding a ribozyme against hTERT mRNA in the region encoding the T-motif caused a reduction in mRNA levels, attenuation of telomerase activity, significantly decreased growth and evidence of apoptosis with concomitant telomere shortening in MCF-7 cells and the immortal human breast epithelial cell line HBL-100. However, upon expressing the same ribozyme via recombinant adenoviral infection in the HBL-100 cells, the group observed massive cell death through apoptosis in a matter of days before any telomere shortening could take place. These findings were expanded later by employing ovarian cancer cell lines having widely different telomere lengths [37]. Cells were transduced with adenovirus expressing the same ribozyme described previously [69]. Rapid effects were observed, with marked decreases in telomerase activity between 2 and 7 days after transduction; massive apoptosis also occurred without lag time in ovarian cancer cell lines with either long or short telomeres and without telomere shortening. Ribozyme-mediated telomerase inhibition studies are summarized in Table 3.
Table 3.
Ribozyme-Mediated Inhibition of Telomerase
| Target | Cells Used* | Results of Treatment | Efficiency | Ref. |
|---|---|---|---|---|
| hTR | HepG2, Huh-7 (liver) | First report of ribozyme inhibition of telomerase; dose-dependent telomerase inhibition | Up to 90% inhibition of telomerase activity | [65] |
| hTR | JR8, M14 (melanoma) | Significantly increased cell doubling time; decrease in DNA sythesis; morphological changes; No telomere shortening (30 PD) |
≥75% inhibition of telomerase activity |
[66] |
| hTR | MCF-7 (breast) | >50% reduction of growth rate of some clones; telomere shortening; morphological changes; visible reduction in telomerase activity by 72 hours |
Complete inhibition of activity in some clones | [67] |
| hTERT | Ishikawa, AN3CA (endometrial) | Targets outside of hTERT open reading frame (13 nt downstream from 5′-end, 59 nt upstream from poly(A) tail); cell growth rate maintained 2 months followed by slowed proliferation then death within 4 months |
Stronger inhibition using 5′-end target than 3′-end target | [68] |
| hTERT | MCF-7 (breast) HBL-100 †(immortalized breast epithelial cells) |
First use of ribozymes against hTERT; plasmid expression - decrease in hTERT mRNA and telomerase activity, decrease in growth rate, telomere shortening and apoptosis; viral expression - massive apoptosis without lag time or telomere shortening (HBL-100) |
HBL-100: 62% reduction in telomerase activity MCF-7: 46% inhibition of telomerase activity |
[69] |
| hTERT | OV-2774, OVCAR-3, OV- MZ-2A, OV-MZ-38 (ovarian) |
Adenoviral delivery of ribozyme; marked decrease in telomerase activity 2-7 days after transduction; massive apoptosis without delay (regardless of initial telomere length); no telomere shortening | >90% inhibition of telomerase activity in OV-2774 and OV- MZ-38; 75% inhibition in OVCAR-3 and OV-MZ-2A |
[37] |
All cells are cancer cell lines unless otherwise noted (†).
DISRUPTING NORMAL FUNCTION BY EXPRESSION OF MUTANTS
So far this review has focused on either ways to actively destroy hTR and hTERT transcripts or ways to prevent these normal components of the telomerase enzyme from carrying out their intended functions. Expression of mutant forms of either hTR or hTERT may be equally effective in disrupting telomerase activity by overwhelming cells with faulty versions of the primary enzyme components.
Dominant Negative hTERT
Two reports utilizing dominant negative mutations in the catalytic subunit of human telomerase made their way into the literature in 1999 [58, 70]. The first report in the literature [70] of the use of dominant negative hTERT protein mutants (DN-hTERT) for telomerase inhibition studies describes four such mutants at the following amino acid residues: 712, 868, 869, and a double mutant at residues 868 and 869. In epidermoid tumor cells with short telomeres (A431, 1-4 kb), expression of the DN-hTERT led to morphological changes and massive apoptosis within 5-10 days after induction of expression; cell division continued until death of the populations, which indicated that further telomere shortening led to eventual chromosomal damage and induction of an apoptotic response. Actual telomere length measurements were not possible due to the rapid loss of the clones. The group turned to embryonic kidney cells (HEK 293 cells) with longer telomeres in order to measure the effect of DN-hTERT expression. In HEK 293 clones expressing DN-hTERT protein, reduced telomerase activity was observed along with pronounced telomere shortening. Initial telomere lengths of the HEK 293 clones used were 10-12 kb; cell proliferation continued unabated for >109 population doublings with telomere lengths approaching 4 kb. Unfortunately, around this time the DN-hTERT expression was lost, preventing the observation of a possible apoptotic outcome. In short, the study showed that cells having short telomeres were more rapidly susceptible to the effects of telomerase inhibition than were cells with longer telomeres, perhaps because those cells with very short telomeres might require telomerase for telomere maintenance at each cell division while cells with longer telomeres do not. The senescent phenotype was not induced in cells with short telomeres, only apoptosis. These observations are important when considering the implications of telomerase inhibition as a therapeutic strategy in human cancer treatment. The average telomere length characterizing a tumor might be a factor when determining whether to use a telomerase inhibitor as a treatment either alone or in conjunction with another therapy. Additionally, the initial telomere lengths within a tumor might roughly predict the amount of time required to produce a therapeutic outcome.
A critical step in the development of telomerase inhibition as a therapeutic strategy was the description of a DN-hTERT [58] used by others [71-73] in years to follow. In this report, aspartic acid and valine at amino acid residues 710 and 711 were substituted with alanine and isoleucine, respectively, resulting in the creation of a catalytically inactive form of hTERT [58]. The expression of this DN-hTERT in several telomerase-positive human cancer cell lines produced multiple cell clones lacking detectable telomerase activity. Expression of the mutant in ovarian and breast cancer cell lines caused gradual telomere shortening as would be predicted by the rate of telomere erosion in normal, telomerase-negative cells [4]. Chromosomal metaphase spread analysis revealed a number of dicentric chromosomes and chromosomal fusions indicative of DN-hTERT -induced telomere dysfunction. Cell proliferation results in cells expressing the mutant at levels sufficient to inhibit telomerase activity showed impaired growth and eventually growth arrest and induction of apoptosis at intervals that correlated with initial telomere length. Tumorigenicity was decreased in vivo by expression of the catalytic mutant. Of therapeutic interest is the finding that the apoptotic response induced by the expression of the DN-hTERT is p53-independent [58], which is in harmony with previous findings [70]. As p53 mutations are found in at least 50% of human cancers [74], it is encouraging that there is evidence that DN-hTERT as a therapeutic telomerase inhibitor may be useful in a wide range of human malignancies, not simply those which retain p53 function. Dominant-negative hTERT studies are summarized in Table 4.
Table 4.
Dominant-Negative hTERT Expression as a Method of Telomerase Inhibition
| Cells Used* | Result of Treatment | Efficiency | Ref. |
|---|---|---|---|
| 36M (ovarian); SW613, SKBR3 (breast); LoVo (colon); HA-1 †(Chinese Hamster Ovary) |
Substituted aspartic acid with alanine (710) and valine with isoleucine (711); inhibition of activity resulted in growth arrest inversely proportional to initial telomere length then eventually morphological characteristics of crisis; elimination of tumorigenicity in nude mice |
Complete inhibition of telomerase activity |
[58] |
| A431 (skin); HEK 293 †(transformed human embryonic kidney) |
4 mutants: 712, 868, 869, 868/869; cells with short telomeres initially showed morphological changes and died 5-10 days after treatment; cells with long telomeres initially demonstrated pronounced telomere shortening over extended culture (>100 PD) |
Substantial inhibition of telomerase activity |
[70] |
| A549 (lung) | Significant telomere shortening; morphological changes and death over 30 PD; increased apoptosis when combined with some chemotherapeutics |
Complete inhibition of telomerase activity |
[71] |
| HAL-01 (leukemia) | Substantial telomere shortening; reduction of cell proliferation; morphological changes and cell death over 35 PD; enhanced apoptosis when combined with some chemotherapeutics |
Dramatic inhibition of telomerase activity |
[72] |
| K1, K2 (thyroid) | Delayed growth arrest with characteristics of senescence; elevated expression of p21WAF1 and senescence-associated β-galactosidase expression; obvious telomere erosion |
Complete inhibition of telomerase activity |
[73] |
All cells used are cancer cell lines unless otherwise noted (†).
Mutant-Template Telomerase RNA
Early theory behind this type of telomerase inhibition rested upon the highly conserved nature of the human telomeric DNA sequence. Experiments with Tetrahymena and yeast [75, 76] demonstrated that mutations in the template region of telomerase RNA caused the synthesis of mutant telomeric repeats which in turn impaired cell growth and survival. It is possible that proteins that bind telomeric DNA may not be able to bind mutant telomeric sequences and consequently may be unable to fulfill their roles in the regulation of telomere length [77]. Accordingly, early experiments in this area showed that expression of mutant-template telomerase RNA (MT-hTR) caused mutant telomerase activity and the synthesis of mutant telomeres [78]. Use of MT-hTR constructs as a possible cancer therapeutic strategy was supported by a study expressng MT-hTR in human prostate and breast cancer cell lines, in which expression decreased cellular viability and increased apoptosis despite the retention of normal telomerase RNA and stable telomere lengths [79]. Similar results were obtained in vivo with breast cancer xenografts expressing MT-hTR [79].
In the aforementioned uses of MT-hTR, only low expression of mutant template RNA was observed along with relatively slight phenotypic changes. In an effort to make delivery and expression of MT-hTR more efficient, and therefore to enable more rapid analysis of the short-term effects of expression on cancer cells, Li et al. developed a lentiviral delivery method which resulted in >95% target cell infection efficiency [62]. Rapid results were observed in human melanoma cells (LOX) and human bladder transitional epithelial carcinoma cells (UM-UC-3) infected with lentivirus expressed MT-hTR. LOX cells exhibited both rapid growth inhibition and the onset of apoptosis by day 5 post-infection, while UM-UC-3 cells were growth inhibited by day 8 and showed signs of apoptosis by day 9. Interestingly, bulk telomere shortening was not required for the onset of visible phenotypic effects, as was proven by the rapid onset of growth inhibition in LOX cells, whose telomeres are >40 kb long. In addition, the phenotypic effects were not dependent upon p53 status of the cells (see also DN-hTERT [58, 70]). These findings are summarized in Table 5.
Table 5.
Mutant-Template Telomerase RNA Expression Exerts Effects Without Telomere Shortening
| Cells Used* | Results of Treatment | Ref. |
|---|---|---|
| HT-1080 (fibrosarcoma); HEK 293 †(transformed human embryonic kidney) |
Synthesis of mutant telomeric repeats; expression of mutant for >90 PD after transfection; impaired growth; accumulation of senescent cells Low expression of MT-hTR |
[78] |
| LNCaP (prostate); MCF-7 (breast) |
Decreased viability and increased apoptosis; stable telomere lengths; reduced tumor growth rate in nude mice; even low expression of MT-hTR produced results despite retention of wild-type hTR and telomerase activity |
[79] |
| LOX (melanoma); UM- UC-3 (bladder) |
Rapid growth inhibition and apoptosis within 9 days; coexpression with siRNA sensitized cells to MT-hTR; smaller, less vascular tumors in nude mice; no bulk telomere shortening; effect independent of cellular p53 status; induction of sustained DNA damage response in telomerase-positive cancer cells >95% infection of target cells with MT-hTR construct |
[63] |
All cells used are cancer cell lines unless otherwise noted (†).
Treatment with these agents may be particularly advantageous since telomere erosion is not necessary for a phenotypic result and the time to therapeutic effect is significantly decreased compared to telomerase inhibitors that cause telomeres to gradually shorten.
UNIQUENESS OF TELOMERASE EXPRESSION AS A TARGETING STRATEGY FOR ONCOLYSIS
In addition to actively destroying telomerase enzyme components and interfering with their function, we are increasingly able to use the very nature of telomerase itself as a therapeutic weapon. The widespread expression of telomerase in human cancers [11] has already been discussed. It would be reasonable then to attempt directed cancer cell death using the hTR and hTERT enzyme components as targets for delivery.
Virus-Mediated Specific Lysis of Tumor Cells
The prospect of specifically targeting cancer cells for death while leaving surrounding healthy cells unharmed is the Holy Grail of therapeutic drug design, yet that is exactly what conditionally-replicating viruses are being designed to do in vitro and in vivo. The adenovirus TRAD (tumor- or telomerase-specific replication-competent adenovirus) has been designed to express E1A and E1B genes under the control of the hTERT promoter [80]. The result was a selective cell lysis of cancer cells (several non-small cell lung cancer and colorectal carcinoma cell lines) and no apparent cytopathic effects in normal cells (WI38 and NHLF) 7 days after adenoviral infection. Additionally, TRAD infection of human tumor xenografts in mice caused marked suppression of tumor growth as well as the induction of tumor cell necrosis. Analysis showed systemic distribution of TRAD by blood and lymph circulation in mice without adverse effect. In fact, experiments successfully demonstrated that intratumoral injection of TRAD could result in viral replication in other, distant tumors. This study has exciting implications not only for primary tumor therapy but also for treating distant metastases which may go undetected clinically for extended periods, dramatically affecting the patient's prognosis and outcome. However, although no apparent ill effects were observed in normal cells in this study, one must carefully weigh the consequences of expressing E1A and E1B (a potent combination of oncogenes) in normal human cells. Additionally, the relatively new concept of using telomerase-specific oncolytic adenoviruses seems promising but requires a deeper understanding of viral replication and associated tumor specificity (see [81]).
Selective Expression of Cytotoxic or Pro-Apoptotic Genes
The differences in hTR and hTERT expression in malignant and normal tissues also lends itself to the possibility of expressing either toxic genes (suicide genes) or genes which are important in the induction of the apoptotic process selectively in cancer cells. Several such studies have demonstrated the usefulness of the hTR or hTERT promoters in driving the expression of the diphtheria toxin A-chain gene [82], the pro-apoptotic gene Bax [83], the Fas associated protein with death domain [84], and the apoptotic signal transducers caspase-6 [85] and caspase-8 [86]. Additionally, selective expression of the herpes simplex virus thymidine kinase gene under the control of the hTERT promoter has been shown to confer sensitivity to ganciclovir treatment on cancer cells [87, 88] and the expression of bacterial nitroreductase under control of the hTR or hTERT promoters sensitizes cancer cells in vitro and in vivo to the pro-drug CB1954 [89, 90].
TELOMERASE INHIBITORS IN COMBINATION THERAPY
As reviewed here, telomerase inhibitors have been shown to be effective by themselves as potentially valuable therapeutic agents. However, their greatest use may come not as stand-alone pharmaceuticals but as part of a coordinated treatment strategy in conjunction with standard treatments including various chemotherapeutics and radiation therapy. Several of the studies cited in this review demonstrated success with their telomerase inhibitors both alone and with synergistic effects when combined with other chemotherapeutics. The method of action of the various chemotherapeutic agents include topoisomerase inhibitors [60, 69, 72], inducers of DNA damage and double-strand breaks [60, 71, 91, 92], mitotic inhibitors [71], and one agent shown to cause telomere erosion (paclitaxel) [93], [94]. Additionally, one study showed the ability of gene expression (noradrenaline transporter gene) under the control of the hTR promoter to induce the uptake of a radiopharmaceutical ([131I] MIBG) by cancer cells [95]. These combination approaches are summarized in Table 6.
Table 6.
Telomerase Inhibitors may Sensitize Cancer Cells to Standard Therapeutic Agents
| Inhibitor/Sensitizer | Standard Therapeutic Agent | Outcome |
|---|---|---|
| 2'-O-methoxyethyl RNA | Cisplatin/carboplatin | Synergistic effect |
| Antisense-hTR | Paclitaxel | Significant increase in sensitivity |
| Antisense-hTR | Cisplatin | Increase in sensitivity |
| DN-hTERT | Cisplatin/taxanes/etoposide | Increase in induction of apoptosis |
| DN-hTERT | Daunorubicin | Increase in apoptosis |
| Ribozyme-hTERT | Doxorubicin | Increase in sensitivity |
| RNAi-hTERT | Topoisomerase inhibitors/bleomycin/radiation | Increase in sensitivity |
| hTR-NAT | [131I]MIBG | Induced uptake |
| AZT | Paclitaxel | Increased activity and effect |
NAT = noradrenaline transporter gene; MIBG = [131I]meta-iodobenzylguanidine; AZT = azidothymidine.
POTENTIAL DRAWBACKS OF TELOMERASE INHIBITORS
Despite the obvious benefits that could be garnered from the development of this class of novel therapeutic agents, there are a few risks that warrant mentioning. While telomerase is not significantly expressed in normal cells, some, such as hematopoietic stem cells, intestinal crypt cells, and cells lining the endometrium do express telomerase and could theoretically be adversely affected by treatment with telomerase inhibitors. It is thought, however, that adverse effects would be minimal, as these normal cells express telomerase infrequently and at lower levels as compared to cancer cells. The telomeres are also generally of greater length in the rare telomerase-positive normal cells which makes these cells less susceptible to telomerase inhibition. These cells also proliferate at a slower rate than most cancer cells and should therefore incur less telomeric attrition. Another concern is that treatment with telomerase inhibitors might incur a clinically significant lag time before telomeric erosion initiates cell senescence or apoptosis. This appears to be less of a concern with the recent appearance of numerous reports of rapid apoptotic responses upon telomerase inhibition [37, 62, 63] resulting from possible telomere uncapping or the induction of a novel gene expression pathway [63]. Also, standard chemotherapeutic agents such as paclitaxel, a known telomere-shortening agent [93], could be used to offset any lag time to therapeutic effect.
As previously mentioned, telomerase inhibitors may exert a delayed effect on cells dependent upon telomeric attrition or they can act rapidly without bulk telomere shortening [35-37, 62, 63, 69]. Interestingly, this phenomenon cannot be explained solely by initial telomere length or p53 status (see [62]). Perhaps these alternate outcomes are in some way related to the putative extratelomeric functions of telomerase. These functions may include cell protection [96, 97], anti-apoptotic function [98], and support of the tumorigenic phenotype in a manner beyond that of simple telomere maintenance [99] (see [100] for review). It may be the case that targeting either hTR or hTERT in various ways affects the telomerase enzyme at different levels, resulting in slightly different outcomes. Not all telomerase inhibitors cause telomeric attrition (and consequently a delayed effect on the cell) by abrogating telomerase activity. Perhaps some of the rapid responses to telomerase inhibition may be the result of telomere uncapping as a consequence of inhibition, since telomerase is known to play a role in the capping of telomeres as well as their elongation [101]. It is desirable for these different outcomes to be further studied and the mechanisms of action of the inhibitors to be better understood before advancing into clinical trials, as the extratelomeric effects of telomerase are not yet well understood. Better understanding of the mechanisms of action will allow better targeting of drug therapies, design of more effective combination therapies, and minimize undesirable side effects of treatment.
A significant consideration when contemplating using telomerase inhibitors is the potential for selecting for the alternative lengthening of telomeres (ALT) pathway [102]. Cells that maintain their telomere lengths while proliferating indefinitely must use either telomerase or the ALT pathway. Eliminating telomerase activity may promote the development of the ALT pathway in cancer cells. Such a phenomenon was observed after telomerase inhibition in a colon cancer cell line having a mismatch repair defect [103]. Efforts should therefore be made to further examine the relationship between telomerase inhibition and induction of ALT and to develop mechanisms to block the ALT pathway.
The studies reviewed here, in addition to others outside the scope of this article, demonstrate the considerable therapeutic potential of molecular-based methods of telomerase inhibition. Either alone or in conjunction with existing cancer therapies, telomerase inhibition promises to be a targeted, specific means of treatment with minimal side effects.
ACKNOWLEDGEMENTS
This work was supported in part by grants from the National Cancer Institute, the University of Alabama at Birmingham Ovarian SPORE, and the American Institute for Cancer Research.
ABBREVIATIONS
- hTR
Human telomerase RNA
- hTERT
Human telomerase reverse transcriptase
- PS
Phosphorothioatecd
- PNA
Peptide nucleic acid
- 2-5A
2'-5'-oligoadenylate
- LNA
Locked nucleic acid
- NP
N3'-P5' phosphoramidates
- NPS
N3'-P5' thio-phosphoramidates
- TRAP
Telomeric repeat amplification protocol
- RNAi
RNA interference
- siRNA
Small interfering RNA
- PD
Population doubling
- nt
Nucleotide
- shRNA
Short hairpin RNA
- DN-hTERT
Dominant negative human telomerase re verse transcriptase
- MT-hTR
Mutant template human telomerase RNA
- TRAD
Tumor- or telomerase-specific replication competent adenovirus
- ALT
Alternative lengthening of telomeres
REFERENCES
- 1.Blackburn EH. Nature. 1991;350:569. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW. Curr. Opin. Cell Biol. 1991;3:444. doi: 10.1016/0955-0674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- 3.Harley CB. Mutat. Res. 1991;256:271. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Nature. 1990;345:458. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Hayflick L. Exp. Cell Res. 1965;37:614. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 6.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Science. 1998;279:349. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 7.Olovnikov AM. Dokl. Akad. Nauk. SSSR. 1971;201:1496. [PubMed] [Google Scholar]
- 8.Watson JD. Nat. New Biol. 1972;239:197. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 9.Shay JW, Pereira-Smith OM, Wright WE. Exp. Cell Res. 1991;196:33. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 10.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, Weinberg RA, Stewart SA, Hahn WC. Cell. 2003;114:241. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 11.Shay JW, Bacchetti S. Eur. J. Cancer. 1997;33:787. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 12.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Mol. Cell Biol. 1990;10:518. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelhardt M, Drullinsky P, Guillem J, Moore MA. Clin. Cancer Res. 1997;3:1931. [PubMed] [Google Scholar]
- 14.Engelhardt M, Ozkaynak MF, Drullinsky P, Sandoval C, Tugal O, Jayabose S, Moore MA. Leukemia. 1998;12:13. doi: 10.1038/sj.leu.2400889. [DOI] [PubMed] [Google Scholar]
- 15.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Nature. 1990;346:866. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 16.Melana SM, Holland JF, Pogo BG. Clin. Cancer Res. 1998;4:693. [PubMed] [Google Scholar]
- 17.Pendino F, Flexor M, Delhommeau F, Buet D, Lanotte M, Segal-Bendirdjian E. Proc. Natl. Acad. Sci. USA. 2001;98:6662. doi: 10.1073/pnas.111464998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldous WK, Marean AJ, DeHart MJ, Matej LA, Moore KH. Cancer. 1999;85:1523. [PubMed] [Google Scholar]
- 19.Naasani I, Seimiya H, Tsuruo T. Biochem. Biophys. Res. Commun. 1998;249:391. doi: 10.1006/bbrc.1998.9075. [DOI] [PubMed] [Google Scholar]
- 20.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Nucleic Acids Res. 2004;32:371. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu W, Begley JG, Killen MW, Mattson MP. J. Biol. Chem. 1999;274:7264. doi: 10.1074/jbc.274.11.7264. [DOI] [PubMed] [Google Scholar]
- 22.Smogorzewska A, de Lange T. Annu. Rev. Biochem. 2004;73:177. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 23.Stansel RM, de Lange T, Griffith JD. EMBO J. 2001;20:5532. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lange T. Oncogene. 2002;21:532. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 25.Braasch DA, Corey DR. Biochemistry. 2002;41:4503. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 26.Geary RS, Yu RZ, Levin AA. Curr. Opin. Investig. Drugs. 2001;2:562. [PubMed] [Google Scholar]
- 27.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 28.Orr RM. Curr. Opin. Mol. Ther. 2001;3:288. [PubMed] [Google Scholar]
- 29.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. Science. 1995;269:1236. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 30.Zhang FX, Zhang XY, Fan DM, Deng ZY, Yan Y, Wu HP, Fan JJ. World J. Gastroenterol. 2000;6:430. doi: 10.3748/wjg.v6.i3.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo S, Tanaka Y, Kondo Y, Hitomi M, Barnett GH, Ishizaka Y, Liu J, Haqqi T, Nishiyama A, Villeponteau B, Cowell JK, Barna BP. FASEB J. 1998;12:801. doi: 10.1096/fasebj.12.10.801. [DOI] [PubMed] [Google Scholar]
- 32.Bisoffi M, Chakerian AE, Fore ML, Bryant JE, Hernandez JP, Moyzis RK, Griffith JK. Eur. J. Cancer. 1998;34:1242. doi: 10.1016/s0959-8049(98)00049-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Chen Z, Chen Y, Tong T. Oncogene. 2003;22:2405. doi: 10.1038/sj.onc.1206317. [DOI] [PubMed] [Google Scholar]
- 34.Kraemer K, Fuessel S, Schmidt U, Kotzsch M, Schwenzer B, Wirth MP, Meye A. Clin. Cancer Res. 2003;9:3794. [PubMed] [Google Scholar]
- 35.Cao Y, Li H, Deb S, Liu JP. Oncogene. 2002;21:3130. doi: 10.1038/sj.onc.1205419. [DOI] [PubMed] [Google Scholar]
- 36.Cao Y, Li H, Mu F-T, Ebisui O, Funder JW, Liu J-P. FASEB J. 2001;01 doi: 10.1096/cj.01-0447fje. [DOI] [PubMed] [Google Scholar]
- 37.Saretzki G, Ludwig A, von Zglinicki T, Runnebaum IB. Cancer Gene Ther. 2001;8:827. doi: 10.1038/sj.cgt.7700383. [DOI] [PubMed] [Google Scholar]
- 38.Fu XH, Zhang JS, Zhang N, Zhang YD. World J. Gastroenterol. 2005;11:785. doi: 10.3748/wjg.v11.i6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo S, Kondo Y, Li G, Silverman RH, Cowell JK. Oncogene. 1998;16:3323. doi: 10.1038/sj.onc.1201885. [DOI] [PubMed] [Google Scholar]
- 40.Kondo Y, Koga S, Komata T, Kondo S. Oncogene. 2000;19:2205. doi: 10.1038/sj.onc.1203538. [DOI] [PubMed] [Google Scholar]
- 41.Pitts AE, Corey DR. Proc. Natl. Acad. Sci. USA. 1998;95:11549. doi: 10.1073/pnas.95.20.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthes E, Lehmann C. Nucleic Acids Res. 1999;27:1152. doi: 10.1093/nar/27.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye J, Wu YL, Zhang S, Chen Z, Guo LX, Zhou RY, Xie H. World J. Gastroenterol. 2005;11:2230. doi: 10.3748/wjg.v11.i15.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shammas MA, Liu X, Gavory G, Raney KD, Balasubramanian S, Shmookler Reis RJ. Exp. Cell Res. 2004;295:204. doi: 10.1016/j.yexcr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Shammas MA, Simmons CG, Corey DR, Shmookler Reis RJ. Oncogene. 1999;18:6191. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- 46.Folini M, Berg K, Millo E, Villa R, Prasmickaite L, Daidone MG, Benatti U, Zaffaroni N. Cancer Res. 2003;63:3490. [PubMed] [Google Scholar]
- 47.Elayadi AN, Braasch DA, Corey DR. Biochemistry. 2002;41:9973. doi: 10.1021/bi025907j. [DOI] [PubMed] [Google Scholar]
- 48.Gryaznov S, Pongracz K, Matray T, Schultz R, Pruzan R, Aimi J, Chin A, Harley C, Shea-Herbert B, Shay J, Oshima Y, Asai A, Yamashita Y. Nucleosides Nucleotides Nucleic Acids. 2001;20:401. doi: 10.1081/NCN-100002314. [DOI] [PubMed] [Google Scholar]
- 49.Herbert BS, Pongracz K, Shay JW, Gryaznov SM. Oncogene. 2002;21:638. doi: 10.1038/sj.onc.1205064. [DOI] [PubMed] [Google Scholar]
- 50.Pruzan R, Pongracz K, Gietzen K, Wallweber G, Gryaznov S. Nucleic Acids Res. 2002;30:559. doi: 10.1093/nar/30.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gryaznov S, Asai A, Oshima Y, Yamamoto Y, Pongracz K, Pruzan R, Wunder E, Piatyszek M, Li S, Chin A, Harley C, Akinaga S, Yamashita Y. Nucleosides Nucleotides Nucleic Acids. 2003;22:577. doi: 10.1081/NCN-120021958. [DOI] [PubMed] [Google Scholar]
- 52.Akiyama M, Hideshima T, Shammas MA, Hayashi T, Hamasaki M, Tai YT, Richardson P, Gryaznov S, Munshi NC, Anderson KC. Cancer Res. 2003;63:6187. [PubMed] [Google Scholar]
- 53.Wang ES, Wu K, Chin AC, Chen-Kiang S, Pongracz K, Gryaznov S, Moore MA. Blood. 2004;103:258. doi: 10.1182/blood-2003-02-0546. [DOI] [PubMed] [Google Scholar]
- 54.Asai A, Oshima Y, Yamamoto Y, Uochi TA, Kusaka H, Akinaga S, Yamashita Y, Pongracz K, Pruzan R, Wunder E, Piatyszek M, Li S, Chin AC, Harley CB, Gryaznov S. Cancer Res. 2003;63:3931. [PubMed] [Google Scholar]
- 55.Herbert BS, Gellert GC, Hochreiter A, Pongracz K, Wright WE, Zielinska D, Chin AC, Harley CB, Shay JW, Gryaznov SM. Oncogene. 2005;24:5262. doi: 10.1038/sj.onc.1208760. [DOI] [PubMed] [Google Scholar]
- 56.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 57.Kosciolek BA, Kalantidis K, Tabler M, Rowley PT. Mol. Cancer Ther. 2003;2:209. [PubMed] [Google Scholar]
- 58.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Nat. Med. 1999;5:1164. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 59.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD. RNA. 2003;9:493. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura M, Masutomi K, Kyo S, Hashimoto M, Maida Y, Kanaya T, Tanaka M, Hahn WC, Inoue M. Hum. Gene Ther. 2005;16:859. doi: 10.1089/hum.2005.16.859. [DOI] [PubMed] [Google Scholar]
- 61.Shammas MA, Koley H, Batchu RB, Bertheau RC, Protopopov A, Munshi NC, Goyal RK. Mol. Cancer. 2005;4:24. doi: 10.1186/1476-4598-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Cancer Res. 2004;64:4833. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Crothers J, Haqq CM, Blackburn EH. J. Biol. Chem. 2005;280:23709. doi: 10.1074/jbc.M502782200. [DOI] [PubMed] [Google Scholar]
- 64.Weil N. Running Interference. 2004 http://www.benitec.com/PRDownloads/Running%20Interference% 20RNAi%20021805.pdf.
- 65.Kanazawa Y, Ohkawa K, Ueda K, Mita E, Takehara T, Sasaki Y, Kasahara A, Hayashi N. Biochem. Biophys. Res. Commun. 1996;225:570. doi: 10.1006/bbrc.1996.1213. [DOI] [PubMed] [Google Scholar]
- 66.Folini M, Colella G, Villa R, Lualdi S, Daidone MG, Zaffaroni N. J. Invest. Dermatol. 2000;114:259. doi: 10.1046/j.1523-1747.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- 67.Yeo M, Rha SY, Jeung HC, Hu SX, Yang SH, Kim YS, An SW, Chung HC. Int. J. Cancer. 2005;114:484. doi: 10.1002/ijc.20720. [DOI] [PubMed] [Google Scholar]
- 68.Yokoyama Y, Takahashi Y, Shinohara A, Wan X, Takahashi S, Niwa K, Tamaya T. Biochem. Biophys. Res. Commun. 2000;273:316. doi: 10.1006/bbrc.2000.2939. [DOI] [PubMed] [Google Scholar]
- 69.Ludwig A, Saretzki G, Holm PS, Tiemann F, Lorenz M, Emrich T, Harley CB, von Zglinicki T. Cancer Res. 2001;61:3053. [PubMed] [Google Scholar]
- 70.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Genes Dev. 1999;13:2388. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Misawa M, Tauchi T, Sashida G, Nakajima A, Abe K, Ohyashiki JH, Ohyashiki K. Int. J. Oncol. 2002;21:1087. [PubMed] [Google Scholar]
- 72.Nakajima A, Tauchi T, Sashida G, Sumi M, Abe K, Yamamoto K, Ohyashiki JH, Ohyashiki K. Leukemia. 2003;17:560. doi: 10.1038/sj.leu.2402825. [DOI] [PubMed] [Google Scholar]
- 73.Preto A, Singhrao SK, Haughton MF, Kipling D, Wynford-Thomas D, Jones CJ. Oncogene. 2004;23:4136. doi: 10.1038/sj.onc.1207564. [DOI] [PubMed] [Google Scholar]
- 74.Levine AJ. Cell. 1997;88:323. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 75.Yu GL, Bradley JD, Attardi LD, Blackburn EH. Nature. 1990;344:126. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 76.McEachern MJ, Blackburn EH. Nature. 1995;376:403. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 77.Fang G, Cech TR. Telomere proteins. In: Greider E.H.B.a.C.W., editor. Telomeres. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1995. pp. 69–105. [Google Scholar]
- 78.Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Mol. Cell. Biol, 1997;17:6394. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. Proc. Natl. Acad. Sci. USA. 2001;98:7982. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawashima T, Kagawa S, Kobayashi N, Shirakiya Y, Umeoka T, Teraishi F, Taki M, Kyo S, Tanaka N, Fujiwara T. Clin. Cancer Res. 2004;10:285. doi: 10.1158/1078-0432.ccr-1075-3. [DOI] [PubMed] [Google Scholar]
- 81.O'Shea CC. Oncogene. 2005;24:7640. doi: 10.1038/sj.onc.1209047. [DOI] [PubMed] [Google Scholar]
- 82.Abdul-Ghani R, Ohana P, Matouk I, Ayesh S, Ayesh B, Laster M, Bibi O, Giladi H, Molnar-Kimber K, Sughayer MA, de Groot N, Hochberg A. Mol. Ther. 2000;2:539. doi: 10.1006/mthe.2000.0196. [DOI] [PubMed] [Google Scholar]
- 83.Gu J, Kagawa S, Takakura M, Kyo S, Inoue M, Roth JA, Fang B. Cancer Res. 2000;60:5359. [PubMed] [Google Scholar]
- 84.Komata T, Koga S, Hirohata S, Takakura M, Germano IM, Inoue M, Kyo S, Kondo S, Kondo Y. Int. J. Oncol. 2001;19:1015. doi: 10.3892/ijo.19.5.1015. [DOI] [PubMed] [Google Scholar]
- 85.Komata T, Kondo Y, Kanzawa T, Hirohata S, Koga S, Sumiyoshi H, Srinivasula SM, Barna BP, Germano IM, Takakura M, Inoue M, Alnemri ES, Shay JW, Kyo S, Kondo S. Cancer Res. 2001;61:5796. [PubMed] [Google Scholar]
- 86.Koga S, Hirohata S, Kondo Y, Komata T, Takakura M, Inoue M, Kyo S, Kondo S. Hum. Gene Ther. 2000;11:1397. doi: 10.1089/10430340050057477. [DOI] [PubMed] [Google Scholar]
- 87.Majumdar AS, Hughes DE, Lichtsteiner SP, Wang Z, Lebkowski JS, Vasserot AP. Gene Ther. 2001;8:568. doi: 10.1038/sj.gt.3301421. [DOI] [PubMed] [Google Scholar]
- 88.Song JS, Kim HP, Yoon WS, Lee KW, Kim MH, Kim KT, Kim HS, Kim YT. Biosci. Biotechnol. Biochem. 2003;67:2344. doi: 10.1271/bbb.67.2344. [DOI] [PubMed] [Google Scholar]
- 89.Plumb JA, Bilsland A, Kakani R, Zhao J, Glasspool RM, Knox RJ, Evans TR, Keith WN. Oncogene. 2001;20:7797. doi: 10.1038/sj.onc.1204954. [DOI] [PubMed] [Google Scholar]
- 90.Bilsland AE, Anderson CJ, Fletcher-Monaghan AJ, McGregor F, Evans TR, Ganly I, Knox RJ, Plumb JA, Keith WN. Oncogene. 2003;22:370. doi: 10.1038/sj.onc.1206168. [DOI] [PubMed] [Google Scholar]
- 91.Chen Z, Koeneman KS, Corey DR. Cancer Res. 2003;63:5917. [PubMed] [Google Scholar]
- 92.Kondo Y, Kondo S, Tanaka Y, Haqqi T, Barna BP, Cowell JK. Oncogene. 1998;16:2243. doi: 10.1038/sj.onc.1201754. [DOI] [PubMed] [Google Scholar]
- 93.Multani AS, Li C, Ozen M, Imam AS, Wallace S, Pathak S. Oncol. Rep. 1999;6:39. doi: 10.3892/or.6.1.39. [DOI] [PubMed] [Google Scholar]
- 94.Mo Y, Gan Y, Song S, Johnston J, Xiao X, Wientjes MG, Au JL. Cancer Res. 2003;63:579. [PubMed] [Google Scholar]
- 95.Boyd M, Mairs RJ, Mairs SC, Wilson L, Livingstone A, Cunningham SH, Brown MM, Quigg M, Keith WN. Oncogene. 2001;20:7804. doi: 10.1038/sj.onc.1204955. [DOI] [PubMed] [Google Scholar]
- 96.Lu C, Fu W, Mattson MP. Brain Res. Dev. Brain Res. 2001;131:167. doi: 10.1016/s0165-3806(01)00237-1. [DOI] [PubMed] [Google Scholar]
- 97.Zhu H, Fu W, Mattson MP. J. Neurochem. 2000;75:117. doi: 10.1046/j.1471-4159.2000.0750117.x. [DOI] [PubMed] [Google Scholar]
- 98.Luiten RM, Pene J, Yssel H, Spits H. Blood. 2003;101:4512. doi: 10.1182/blood-2002-07-2018. [DOI] [PubMed] [Google Scholar]
- 99.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, Popescu NC, Weinberg RA. Pro.c Natl. Acad. Sci. USA. 2002;99:12606. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chung HK, Cheong C, Song J, Lee HW. Curr. Mol. Med. 2005;5:233. doi: 10.2174/1566524053586635. [DOI] [PubMed] [Google Scholar]
- 101.Chan SW, Blackburn EH. Oncogene. 2002;21:553. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 102.Henson JD, Neumann AA, Yeager TR, Reddel RR. Oncogene. 2002;21:598. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 103.Bechter OE, Zou Y, Walker W, Wright WE, Shay JW. Cancer Res. 2004;64:3444. doi: 10.1158/0008-5472.CAN-04-0323. [DOI] [PubMed] [Google Scholar]