Abstract
Background
Deleterious mutations of the BRCA1 and BRCA2 genes confer susceptibility to breast and ovarian cancer. At least 7 models for estimating the probabilities of having a mutation are used widely in clinical and scientific activities; however, the merits and limitations of these models are not fully understood.
Objective
To systematically quantify the accuracy of the following publicly available models to predict mutation carrier status: BRCAPRO, family history assessment tool, Finnish, Myriad, National Cancer Institute, University of Pennsylvania, and Yale University.
Design
Cross-sectional validation study, using model predictions and BRCA1 or BRCA2 mutation status of patients different from those used to develop the models.
Setting
Multicenter study across Cancer Genetics Network participating centers.
Patients
3 population-based samples of participants in research studies and 8 samples from genetic counseling clinics.
Measurements
Discrimination between individuals testing positive for a mutation in BRCA1 or BRCA2 from those testing negative, as measured by the c-statistic, and sensitivity and specificity of model predictions.
Results
The 7 models differ in their predictions. The better-performing models have a c-statistic around 80%. BRCAPRO has the largest c-statistic overall and in all but 2 patient subgroups, although the margin over other models is narrow in many strata. Outside of high-risk populations, all models have high false-negative and false-positive rates across a range of probability thresholds used to refer for mutation testing.
Limitation
Three recently published models were not included.
Conclusions
All models identify women who probably carry a deleterious mutation of BRCA1 or BRCA2 with adequate discrimination to support individualized genetic counseling, although discrimination varies across models and populations.
Deleterious mutations of BRCA1 (MIM 113705) and BRCA2 (MIM 600185) increase the risk for breast and ovarian cancer (1–3). Whereas deleterious variants are relatively rare in the general population, they are common among families with multiple occurrences of breast or ovarian cancer (4–6). When counseling a woman facing decisions about genotyping for BRCA1 and BRCA2, it is important to accurately evaluate the probability that she carries a deleterious mutation (pretest mutation probability) and the probability that a mutation will be found if she is genotyped (which depends on the accuracy of mutation testing). Reliable, evidence-based, individualized counseling strategies can enhance informed decision making, both about whether to pursue BRCA1/BRCA2 testing and what to do with the results (7–9).
The demand for assessment of complex family histories of cancer has led to widespread use of statistical models to estimate mutation probabilities (2, 10–18). Model-based predictions are currently used in counseling about genetic testing, are included in materials distributed to women considering genetic testing (18–21), are used for determining eligibility for screening and prevention studies (22), and are factored into coverage decisions by insurers (23). More than a dozen models exist. They use different statistical methods and source populations, pedigree features, and predicted outcomes. In clinical practice, different models applied to the same person can give a wide range of probabilities that a BRCA1/BRCA2 mutation is present. This degree of variability raises concerns about whether some models are more accurate than others and calls for a careful independent comparative evaluation of the predictive performance of existing models.
We assessed the validity of commonly used models for estimating mutation probabilities of BRCA1 and BRCA2 in individuals identified through the Cancer Genetics Network. We assembled a large set of families with history of breast cancer, ovarian cancer, or both. We used standardized computational methods across contributing institutions to evaluate 7 models. Our main goal was to measure how well these models discriminated between mutation carriers and noncarriers.
METHODS
Study Overview
We conducted a cross-sectional, multicenter analysis. For each family in the study, we identified an individual (the counselee) for whom we collected genetic test results for BRCA1, BRCA2, or both; genotyping methods; pretest estimations of mutation probability using each model; and additional information about family history of cancer. We used genetic test results as the gold standard for judging the sensitivity and specificity of the various models. We evaluated all models on every counselee, except where noted.
Data Collection
Table 1 summarizes the salient data (24–32). Sources include 3 population-based studies and 8 data sets of individuals seen in clinics for women at high risk for a BRCA mutation. In the population-based studies, the participants reflected the demographic characteristics of a defined subpopulation (for example, all breast cancer cases in Orange County in the University of California, Irvine [UCI], study [31]). In contrast, patients from high-risk clinics had been referred because of a family history of cancer or were self-referred because of an interest in genetic testing (inclusion criteria varied across clinics).
Table 1.
Demographic Characteristics of Counselees and Sample Size, by Center *
| Center | Population | Persons Tested, n | Men, % | Mean Age (SD), y | Breast Cancer, n | Ovarian Cancer, n | No Cancer, n | BRCA1 Positive, n | BRCA2 Positive, n | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| BCM | Ashkenazi Jewish volunteers | 282 | 0 | 53.1 (11.8) | 33 | 2 | 247 | 6 | 0 | 25 |
| City of Hope | Counseling clinic | 77 | 0 | 49.8 (13.0) | 47 | 5 | 25 | 13 | 5 | 26 |
| Duke University | Counseling clinic | 275 | 0 | 47.8 (10.7) | 237 | 27 | 22 | 41 | 18 | 27 |
| FHCRC | Population-based study | 583 | 0 | 45.7 (5.1) | 320 | 6 | 263 | 22 | 12 | 28 |
| Georgetown University | Counseling clinic | 242 | 0 | 52.9 (11.4) | 227 | 20 | 3 | 55 | 23 | 21 |
| HCI | Counseling clinic | 61 | 0 | 58.5 (15.2) | 47 | 2 | 12 | 6 | 8 | - |
| JHU | Counseling clinic | 105 | 3 | 50.2 (10.3) | 57 | 7 | 43 | 0 | 6 | 29 |
| Penn | Counseling clinic | 676 | 69 | 50.9 (13.0) | 489 | 82 | 138 | 121 | 53 | 11, 30 |
| UTSW | Counseling clinic | 121 | 7 | 47.9 (12.8) | 67 | 10 | 50 | 23 | 9 | 24 |
| UCI | Population-based study | 803 | 9 | 62.5 (13.6) | 605 | 122 | 87 | 10 | 4 | 31 |
| MDACC | Counseling clinic | 117 | 7 | 51.2 (13.3) | 59 | 8 | 53 | 25 | 18 | 32 |
| Total | - | 3342 | 95 | 52.8 (13.1) | 2188 | 291 | 943 | 332 | 156 | - |
BCM = Baylor College of Medicine; FHCRC = Fred Hutchinson Cancer Research Center; HCI = Huntsman Cancer Institute; JHU = Johns Hopkins University; MDACC = M.D. Anderson Cancer Center; Penn = University of Pennsylvania; UCI = University of California, Irvine (Orange County); UTSW = University of Texas Southwestern.
Each center calculated all of the model probabilities for its own families. We designated the first genotyped person in each family as the counselee and computed predictions by using the genetic counseling software CaGene (University of Texas Southwestern Medical Center, Dallas, Texas) (24). The software version was customized and distributed to participating sites to ensure uniform procedures across all sites. Data entry and computation of model predictions were performed at the sites. This decentralized approach for data entry and probability calculations allowed site investigators to use pedigree information that models required but that centers could not export to a central site because of privacy concerns. In addition to model predictions, a subset of centers also exported the data required for the models to the National Cancer Institute’s (NCI) Cancer Genetics Network Data Coordinating Center. The study population includes 3342 families.
The institutional review boards at each participating institution approved the study protocol. All included counselees gave consent for using their data for research according to local institutional review board requirements. The Cancer Genetics Network steering committee reviewed the study design.
Genetic Testing
Appendix Table 1 (available at www.annals.org) summarizes genotyping methods by center and provides a brief description of each method. Determining whether a person carries a deleterious mutation of BRCA1 or BRCA2 is technically demanding because of the large size of these genes, the wide spectrum of mutations, and the presence of mutations whose clinical significance is unknown (33–35). Commercial testing uses sequencing to search for unknown mutations or to probe for mutations that are commonly found among Ashkenazi Jewish persons. Research settings, particularly in the time in which the study was conducted, have used less expensive and less sensitive techniques (Appendix Table 1, available at www.annals.org). Although sequencing is the most sensitive of the techniques used in our study, recent evidence highlights how it can miss certain mutations, such as large deletions or intronic mutations (3, 36). Therefore, the set of individuals carrying a mutation (the carriers) is not the same as the set of individuals who test positive for a mutation (the positive cases). Thus, Table 1 underestimates the true number of carriers; the size of the error varies according to the method of genotyping.
Models
We studied 7 models: BRCAPRO, the family history assessment tool (FHAT), Finnish, Myriad, NCI, University of Pennsylvania (Penn), and Yale University (Yale). Appendix Table 2 (available at www.annals.org) summarizes the characteristics, input variables, and output of the models. Three broad categories of models have been proposed: empirical (Finnish, Myriad, NCI, and Penn), mendelian (BRCAPRO and Yale), and expert-based (FHAT). The first step in developing an empirical model is to summarize the salient aspects of a family history in some predictor variables. The second step is to apply statistical learning techniques, such as logistic regression, to describe the relationship between these variables and the genotyping results (the dependent variable). Mendelian models represent the known modes of inheritance of deleterious genetic variants by established mathematical relationships between phenotypes (in this case, cancer status of family members) and genotypes (14, 37–41). The mendelian model inputs include cancer incidence curves (penetrance) for both carriers and noncarriers and the prevalence of deleterious variants. Expert-based models calculate scores that summarize degree of risk, using algorithms constructed on the basis of clinical judgment. For example, FHAT (16) uses a 17-question interview to produce a quantitative score (score range, 0 to 45) representing the severity of family history.
Empirical models calculate the probability of a positive test result for a mutation in the counselee (that is, the result of genetic testing), whereas mendelian models directly estimate the probability of carrying a mutation (the true mutation status of the counselee) (37). The 2 types of predictions are therefore not directly comparable, a fact often overlooked in counseling practice. Because genotyping methods are highly specific for the BRCA1 and BRCA2 genes (that is, they have a very low false-positive rate), multiplying the genotype probability by the genotyping sensitivity gives the probability of finding a mutation. Therefore, to compare an empirical model probability of a BRCA mutation with a mendelian model probability, one must know the sensitivity of the genotyping method of the study used to develop the empirical model. Expert-based scores do not have a direct probabilistic interpretation. In our analyses, we rescaled the FHAT score by dividing by its maximum value of 45.
The Penn model (11) estimates the probability of a positive BRCA1 test result in any family member. We adapted it to provide the probability of a positive test in the counselee. We assigned affected counselees the same mutation probability as the family. We assigned unaffected counselees one half the family probability if the closest affected relative of the counselee is a first-degree relative and one quarter of the family probability if the closest relative is a second-degree relative.
We used a version of the BRCAPRO (13, 14) model based on the genetic variables described by Iversen and colleagues (42).
We defined the Yale model by postulating a single gene as reflecting all highly penetrant autosomal dominant breast cancer genes and used genetic variables from a segregation analysis of the Cancer and Steroid Hormone Study (10, 43, 44).
We did not include several models. The LAMBDA model (45) and the Spanish model (46) are empirical models developed on families from Australia and Spain. The Manchester (47) model is an expert-based scoring system. These 3 models became available after the Cancer Genetics Network data collection occurred. We also did not include 2 BRCA1 prediction–only models that are precursors of models considered here (12, 13) and 2 recent mendelian models, the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) and International Breast Cancer Intervention (IBIS) Breast Cancer Risk Evaluation Tool, for which software implementations were not readily available at the time of data collection (48, 49). This omission is a limitation of our study, although a recent study (32) concluded that the BOADICEA, BRCAPRO, and Myriad models performed similarly.
Statistical Analysis
We combined data from all centers to create a matrix in which the rows are individual counselees and the columns include estimated probability of a BRCA1 or BRCA2 mutation using each model; stratification variables, such as age, Ashkenazi ethnicity, and cancer status of the counselee, genetic test results; and genotyping methods. We defined “positive cases” as individuals who test positive for either BRCA1 or BRCA2 and “negative cases” as individuals who had negative results on both tests. We excluded counselees who tested negative for 1 gene and were not tested for the other, which limited our analysis data set to 2240 individuals. Our analytic strategy is to compare predicted testing results for each model with actual testing results for all counselees and within specific counselee strata (for example, age, Ashkenazi ethnicity, or cancer status of the counselee).
Our measurement of discrimination is the c-statistic, which is equal to the area under the receiver-operating characteristic curve. It is also the probability that a randomly chosen test-positive counselee will have a higher probability (or prediction score) of a BRCA mutation than a randomly chosen test-negative counselee (50, 51), which implies a correct rank ordering of the 2 predictions. We computed the c-statistic by using all possible pairs of counselees, one of whom is positive and the other negative. The smallest possible value of the c-statistic is 0.0, wherein all predictions are incorrectly ordered, and the largest is 1.0, wherein all predictions are correctly ordered. A c-statistic of 0.5 means that the model would correctly order half of the pairs and would incorrectly order the other half. This method is equally applicable to empirical, mendelian, and expert-based approaches.
We evaluated all models in each center. Because the NCI model is intended for use in the Ashkenazi population, we evaluated it on Ashkenazi persons only. The applicability of some models is subject to minor restrictions, such that we had to exclude some families for each model (52). We did not evaluate a model on a counselee from a family that we knew had been used to develop the model. We could not follow this rule with the Myriad model because this information was not available. As a result, we may have overestimated the performance of Myriad. However, this overlap is probably limited to individuals genotyped by gene sequencing, who make up less than 25% of all study patients.
To assess the significance of trends observed when comparing all models as a group across strata, we used a Wilcoxon rank-sum test and a 2-sided alternative.
A potential limitation of our analysis plan arises from the imperfect and differing sensitivity of genotyping methods used. Whereas the genotypes of positive counselees are accurate because the false-positive rate of genotyping is negligible (33, 35), those of negative counselees are uncertain because of possible false-negative genotyping results. This discrepancy may result in a bias in favor of empirical models, which estimate the probability of a positive mutation test, and against mendelian models, which estimate the probability of a mutation. In parallel to the analysis presented here, we developed a customized approach that allows one to evaluate the c-statistic and other measurements by comparing predictions with imputed genotypes, therefore adjusting for heterogeneous test sensitivities. Results suggest that the findings reported here penalize mendelian models. Data are not shown, but the details are available from the authors on request. Finally, a limitation of the analysis is that normalized FHAT scores do not provide probabilities, and thus the 10% threshold does not have the same interpretation for this model as it does for the others.
Role of the Funding Sources
The Cancer Genetics Network funded part of the study and is responsible for good research practices and for data storage. The Cancer Genetics Network and the other funding sources, listed under “Grant Support,” had no other role in the design, conduct, and reporting of the study.
RESULTS
Range of Predictions across Models
The models differ widely in their predictions for a given counselee. The largest range of predictions across models was 99 percentage points, reached in families with male breast cancer. On average, the range of predictions was 27 percentage points. Among all possible pairwise comparisons between the predictions of a mendelian and an empirical model for the same counselee, 12% disagreed by 50 percentage points or more.
Model Discrimination
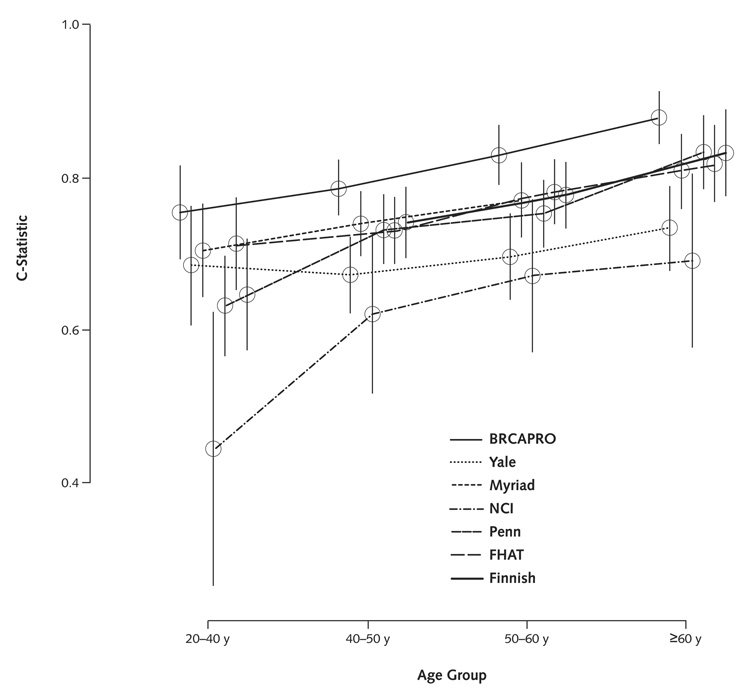
We used the c-statistic to summarize the models’ ability to distinguish individuals testing positive for either gene from those testing negative to both genes. Table 2 presents overall results, as well as results stratified by study type, ethnicity, and counselee cancer status. The Figure presents discrimination for each model, stratified by age. As seen in Table 2, in 82% of the possible comparisons between a positive and a negative counselee, BRCAPRO assigns a higher probability to the positive counselee. By comparison, the c-statistic for the Myriad model is 77%; the 95% CIs for the 2 c-statistics do not overlap.
Table 2.
C-Statistics (95% CIs) of the Models *
| Variable | BRCAPRO | Yale | Myriad | NCI† | Penn‡ | FHAT | Finnish |
|---|---|---|---|---|---|---|---|
| All persons | 0.82 (0.81–0.84) | 0.71 (0.68–0.74) | 0.77 (0.75–0.79) | - | 0.76 (0.74–0.79) | 0.77 (0.75–0.8) | 0.78 (0.75–0.8) |
| High-risk studies§ | 0.76 (0.73–0.79) | 0.64 (0.61–0.67) | 0.71 (0.68–0.74) | - | 0.73 (0.7–0.76) | 0.71 (0.68–0.74) | 0.75 (0.73–0.78) |
| Population-based studies§ | 0.85 (0.81–0.88) | 0.72 (0.65–0.79) | 0.79 (0.72–0.86) | - | 0.75 (0.69–0.81) | 0.79 (0.73–0.85) | 0.75 (0.68–0.82) |
| Not Ashkenazi Jewish∥ | 0.84 (0.82–0.86) | 0.74 (0.71–0.77) | 0.78 (0.75–0.81) | - | 0.76 (0.74–0.79) | 0.8 (0.78–0.83) | 0.8 (0.78–0.82) |
| Ashkenazi Jewish∥ | 0.72 (0.67–0.77) | 0.63 (0.57–0.69) | 0.71 (0.66–0.76) | 0.63 (0.57–0.68) | 0.73 (0.68–0.79) | 0.71 (0.66–0.77) | 0.74 (0.68–0.8) |
| No cancer | 0.74 (0.67–0.8) | 0.62 (0.55–0.69) | 0.73 (0.67–0.8) | - | 0.79 (0.73–0.85) | 0.68 (0.6–0.76) | 0.81 (0.75–0.87) |
| Breast cancer only | 0.84 (0.82–0.85) | 0.76 (0.73–0.78) | 0.76 (0.73–0.79) | - | 0.74 (0.71–0.77) | 0.77 (0.74–0.79) | 0.74 (0.72–0.77) |
| Ovarian cancer only | 0.88 (0.84–0.93) | 0.8 (0.74–0.87) | 0.87 (0.81–0.92) | - | 0.68 (0.57–0.79) | 0.83 (0.76–0.91) | 0.82 (0.73–0.91) |
| Both breast and ovarian cancer | 0.86 (0.78–0.94) | 0.83 (0.75–0.91) | 0.8 (0.7–0.91) | - | 0.8 (0.71–0.89) | 0.78 (0.67–0.89) | 0.8 (0.72–0.89) |
FHAT = family history assessment tool; NCI = National Cancer Institute; Penn = University of Pennsylvania; Yale = Yale University.
The NCI model was applied only to families of Ashkenazi ethnicity.
The Penn model predicts BRCA1 mutations only. In the table, its performance in predicting mutations in either gene is evaluated to facilitate comparisons with other models and to capture a common use of the model.
The “population-based studies” row includes counselees from Fred Hutchinson Cancer Research Center; Baylor College of Medicine; and University of California, Irvine (Orange County). The remainder of counselees are placed in the “high-risk studies” row.
The “Ashkenazi Jewish” row includes counselees of Ashkenazi ancestry from all centers. The remainder of the counselees are placed in the “not Ashkenazi Jewish” row.
Figure 1. C-statistic, by age of the counselee and model.
Points within age groups are slightly spaced horizontally for readability. Vertical bars are 95% CIs. A description of each model is given in Table 3. FHAT = family history assessment tool; NCI = National Cancer Institute; Penn = University of Pennsylvania; Yale = Yale University.
Results shown in Table 2 and the Figure identify areas of strength and weakness of individual models in specific clinical scenarios. Results differ somewhat depending on the stratum, and the 95% CIs often overlap. However, the following general trends emerge. As expected, models perform less well in individuals without breast or ovarian cancer and in younger counselees. Discrimination is generally better in the population-based studies. BRCAPRO has the largest c-statistic in all but 2 strata, although the range of c-statistics across all models is too narrow to choose a clearly superior model.
The methods used to detect mutations in BRCA1 or BRCA2 differed among the study sites, which introduces the possibility that differences among model performances reflect differences in mutation detection. In Appendix Table 3 (available at www.annals.org), we also considered separately 512 cases in which mutation screening was performed by gene sequencing. This analysis addresses the concern about confounding between centers’ characteristics and mutation testing methods, because within the gene sequencing stratum, all centers used the same method for gene sequencing and all 512 cases are from high-risk samples. The model prediction results are consistent with those obtained on all high-risk individuals across the various mutation testing methods used in different centers, suggesting that such confounding is not affecting our conclusions.
Sensitivity, Specificity, and Likelihood Ratios
To calculate sensitivity and specificity, we first specified the probability that defines the threshold between a positive model result and a negative model result. We divided the study population depending on whether each model’s prediction (probability or normalized score) exceeds a threshold of 10%. In the analysis, a mutation probability greater than 10% would be considered a positive model result. To illustrate the consequences of a referral threshold of 10%, we report the sensitivity, specificity, and likelihood ratios for the BRCAPRO model in Table 3 and for all models in Appendix Table 4 (available at www.annals.org). Comparisons across populations indicate a decrease in specificity and an increase in sensitivity as we move from population-based to high-risk studies. The ratio of the proportion of individuals above 10% among positive cases to the same proportion among negative cases is the positive likelihood ratio—the amount by which the odds of a positive test increases when a person’s estimated probability exceeds 10%.
Table 3.
Test Performance Characteristics and Posttest Probabilities for the BRCAPRO Model in 3 Populations*
| Variable | UCI Sample | FHCRC Sample | High-Risk Sample |
|---|---|---|---|
| Overall proportion of individuals testing positive | 0.017 | 0.089 | 0.279 |
| Sensitivity (95% PI)† | 0.429 (0.207–0.656) | 0.706 (0.547–0.834) | 0.824 (0.786–0.857) |
| Specificity (95% PI)† | 0.934 (0.913–0.950) | 0.671 (0.620–0.715) | 0.526 (0.499–0.551) |
| Positive likelihood ratio (95% PI) | 6.503 (2.925–10.24) | 2.148 (1.682–2.71) | 1.739 (1.63–1.87) |
| Negative likelihood ratio (95% PI) | 0.612 (0.372–0.843) | 0.438 (0.242–0.669) | 0.334 (0.269–0.408) |
| Positive predictive value (95% PI), n/n (%)‡ | 6/58 (10.34 [4.435–20.08]) | 24/139 (17.3 [11.69–24.2]) | 352/874 (40.3 [37.1–43.6]) |
| Negative predictive value (95% PI), n/n (%)§ | 737/745 (98.9 [98.0–99.5]) | 235/245 (95.9 [92.9–97.9]) | 579/654 (88.5 [85.9–90.8]) |
95% PI = 95% posterior probability regions (obtained by using Jeffrey noninformative priors); FHCRC = Fred Hutchinson Cancer Research Center; UCI = University of California, Irvine (Orange County).
For a referral threshold probability of 10%.
Proportion of individuals testing positive among those with probability or score >10%.
Proportion of individuals testing negative among those with probability or score ≤10%.
The negative likelihood ratio is the analogous ratio for proportions below or equal to the 10% threshold. According to the Bayes’ theorem, models with higher positive likelihood ratios and lower negative likelihood ratios increase the odds of testing positive more substantially when the probability exceeds 10% and decrease the odds more when the probability is below 10%, respectively. Table 3 and Appendix Table 4 (available at www.annals.org) show how these ratios vary with the model and study population. Exceeding the 10% threshold is stronger evidence for testing positive among unselected breast cancer cases (such as the UCI study [31]) than in a high-risk setting, in which more negative counselees will exceed the threshold as a result of patient selection.
Positive and Negative Predictive Values
The predictive value of a test is the probability of the target condition being present or absent corresponding to a positive or negative test result, respectively. It depends on the prevalence of the target condition in the population and the sensitivity and specificity of the test. In Table 3 and Appendix Table 5 (available at www.annals.org), we compute the proportion of test-positive individuals among those exceeding the 10% threshold (positive predictive value) and the proportion of test-negative individuals among those not exceeding the 10% threshold (negative predictive value). We perform this analysis separately for 3 subsets of the study populations, chosen because the prevalences of BRCA1 and BRCA2 mutations within 1 subset differ from those in the other 2 subsets: the UCI study, the Fred Hutchinson Cancer Research Center study (28), and all high-risk data combined. The UCI data (31) include all breast cancer cases (female and male) diagnosed in Orange County, California, during the year beginning 1 March 1994 and is therefore representative of patients with breast cancer presenting to a general oncology practice. The proportion of positive BRCA mutation tests is 1.74%. Table 4 and Appendix Table 6 (available at www.annals.org) show the consequences of referring patients with breast cancer for genetic testing and counseling if their risk exceeds 10%. For example, using the FHAT model, 4.7% of individuals with a score of 10% or greater will be positive and 99.5% of individuals with a score less than 10% will be negative. Because the predictive value of a test varies with the prevalence of the target condition, when the same referral threshold probability is applied to populations with higher prevalence, the number of test-positive cases missed by using a 10% referral threshold increases. In the high-risk population, where the proportion of positive mutation tests is 27.9%, 32% of those that exceed a 10% threshold on the FHAT score will be test-positive but only 86% of the individuals below the 10% threshold will be test-negative. A similar trend is present with the other models in Appendix Table 5 (available at www.annals.org). The Fred Hutchinson Cancer Research Center population (28) presents an intermediate case: The data are from a case–control study with a mutation prevalence of 8.85%. The cases had either early onset (age <35 years) or a first-degree family history of breast cancer and thus present an intermediate scenario between the low-risk UCI population and the high-risk population. In this population, 11.9% of those with FHAT scores that exceed a 10% threshold will be positive, and 98.2% of the individuals below the 10% threshold will be negative.
Table 4.
Number of Patients per 1000 Referred for Mutation Testing with Different Threshold Probabilities for Referral in the BRCAPRO Model *
| Variable | Proportion Positive | Threshold | Mutation-Negative above Threshold | Mutation-Positive above Threshold | Mutation-Positive below Threshold | Mutation-Negative below Threshold | Total |
|---|---|---|---|---|---|---|---|
| UCI sample | 0.017 | 0.05 | 80.9 | 8.7 | 8.7 | 901.6 | 1000 |
| 0.1 | 64.8 | 7.5 | 10 | 917.8 | 1000 | ||
| 0.2 | 47.3 | 5 | 12.5 | 935.2 | 1000 | ||
| FHCRC sample | 0.089 | 0.05 | 351.6 | 67.7 | 20.8 | 559.9 | 1000 |
| 0.1 | 299.5 | 62.5 | 26 | 612 | 1000 | ||
| 0.2 | 242.2 | 62.5 | 26 | 669.3 | 1000 | ||
| High-risk sample | 0.279 | 0.05 | 418.8 | 241.5 | 38 | 301.7 | 1000 |
| 0.1 | 341.6 | 230.4 | 49.1 | 378.9 | 1000 | ||
| 0.2 | 264.4 | 209.4 | 70 | 456.2 | 1000 |
These calculations consider the hypothetical scenario in which individuals are referred for genetic counseling whenever their predicted probability using BRCAPRO exceeds the threshold indicated in the third column. “Positive” and “negative” refer to the results of genetic testing, as described in the Methods section. Counts are rescaled to add up to 1000 for ease of comparison. See Appendix Table 4 and Appendix Table 5 (available at www.annals.org) for actual sample sizes and Appendix Table 6 (available at www.annals.org) for the similar calculations using the other models. FHCRC = Fred Hutchinson Cancer Research Center; UCI = University of California, Irvine (Orange County).
Effect of Changes in the Threshold
Table 4 illustrates the effect of increasing the threshold to 20% and lowering the threshold to 5% on the classification of patients by the BRCAPRO model. The same information for other models appears in Appendix Table 6 (available at www.annals.org). For example, in the UCI study, a threshold of 5% on the BRCAPRO model predictions leads to referral of half of the test-positive individuals in the population, while still referring less than 10% of the total number of individuals. In general, lowering the threshold will capture a larger proportion of test-positive individuals at the cost of increasing the number of referrals.
DISCUSSION
We provide a comprehensive view of the predictive performance of 7 commonly used, publicly available mutation carrier prediction models for the BRCA1 and BRCA2 genes, across a range of clinically relevant strata. As shown in Table 2, the c-statistic for the better-performing models clusters closely around 80%. Clinicians and counselors can use these results to identify the model that performs best in the strata most relevant to their activities and to weigh the differences in discrimination against practical implementation issues that are specific to their practice. BRCAPRO has the largest c-statistic overall and in all but 2 clinical strata, although the range of c-statistics across all models is too narrow to identify a clearly superior model. If used for referral outside of high-risk groups, all models have high rates of false-negative and false-positive results across a range of thresholds to refer for testing (Appendix Table 6, available at www.annals.org).
A strength of our study is the inclusion of both high-risk and population-based centers. The high-risk samples reflect genetic counseling clinics, and the population-based samples reflect the broader spectrum of patients seen in general oncology practice. The c-statistics reported for the high-risk population are similar to those of previous studies performed in similar settings (53–57). The c-statistic of models is generally greater in the population-based samples than in the high-risk samples (P = 0.036) (Table 2), which suggests that the models can be applied to more broadly representative settings than high-risk clinics. In high-risk populations, a referral threshold of 10% results in relatively high sensitivity with very low specificity. In population-based cohorts, the specificity is higher but the sensitivity is lower, and the 10% threshold misses a large proportion of test-positive cases. Likelihood ratios resulting from the 10% threshold also vary markedly across populations.
Genetic counselors used mutation probability thresholds for referral in the past, and they are still sometimes used for insurance coverage purposes (23). However, guidelines no longer recommend this practice (7). We used a threshold probability to calculate model sensitivity and specificity, which permitted us to illustrate the consequences of using the models to decide on referral for mutation testing. Results in Appendix Table 4 (available at www.annals.org) imply that using a 10% threshold for the pooled high-risk populations will, depending on the model used, exclude 10% to 20% of test-positive individuals from genetic testing. The analogous figure for unselected breast cancer cases is 1% to 8% excluded. Conversely, Table 3 and Table 4 and Appendix Table 5 (available at www.annals.org) show that the models would refer many women who do not carry mutations for testing. The low positive predictive values that we found are consistent with earlier reports (24) comparing BRCAPRO with genetic counselors’ assessments of the same family history. When models are used on unselected breast cancer cases to determine whether to refer a patient to a counseling clinic, using a 10% threshold would still miss women who would test positive, a circumstance more serious than an unnecessary referral. The estimated positive and negative predicted values (Table 3 and Appendix Table 5 [available at www.annals.org]) do not apply to healthy individuals from the general population.
We found that the estimated probability of testing positive differed widely when different models were applied to the same counselee. Genetic counselors may consider using several predictive models, as well as qualitative pedigree analysis (23), because the variation among model predictions may provide an indication of their reliability.
Limitations of our study concern the mutation testing methods used and the lack of representation of minorities. Mutation testing techniques varied across centers generally and across high-risk and population-based studies more specifically. All cases tested using gene sequencing were in high-risk centers, whereas studies of low-risk populations used less sensitive mutation testing methods. However, this potential confounding probably leads to an underestimate of the c-statistics in population-based studies; our conclusion about generalizability is therefore unlikely to be affected. Also, minority populations may be underrepresented in the populations used to develop the models, and existing models do not explicitly take into account the possibility that minority groups, such as African Americans, have a higher probability of carrying genetic variants of uncertain clinical significance, which would be missed by genetic testing. However, independent evidence suggests that BRCAPRO discriminated between carriers and noncarriers in African American (58) and Hispanic (59) families as well as it did in white families.
Genotyping for BRCA1 and BRCA2 is now widespread. Myriad Genetics Laboratories, Salt Lake City, Utah, alone tested more than 100 000 individuals by 2005 (60). Many more women are being counseled about whether to be genotyped. Model-based mutation probabilities have been a critical component of individualized counseling. Our comprehensive evaluation indicates that, overall, the concordance observed between predictions and test results is high. However, relying on model probabilities to decide about referral can cause many false-positive and false-negative referral decisions (Table 4). Decision making about genetic testing and prevention should reflect a broader range of factors, of which carrier probabilities are but one (7, 8). Other factors include the effectiveness and cost of genotyping; the available means and efficacy of measures for early detection and risk reduction; the counselee’s willingness to undergo enhanced surveillance or risk-reducing interventions; and the possible psychological, social, and ethical effects of testing. Physicians should rely on health care professionals who are experienced in cancer genetics to determine the appropriateness of genetic testing. Their evaluation may discover additional reasons for caution because of small family size, few female family members, limited or unconfirmed family history, or family histories that suggest rarer syndromes. In primary care settings for referral to further genetic counseling, setting a referral threshold probability may be a practical approach; however, we do not recommend using a strict 10% threshold, because it may miss a large proportion of clinically appropriate cases.
Acknowledgments
Data were collected within the framework of the NCI’s Cancer Genetics Network, combining data previously collected at each Cancer Genetics Network center and at City of Hope National Medical Center. Most data predated Cancer Genetics Network activities, but the Cancer Genetics Network provided the venue for the pooled analysis. The authors thank Connie Griffin for her Cancer Genetics Network leadership at the Johns Hopkins University, Kelly Qu for support with database management at Johns Hopkins University, Jihong Zong for collecting and transmitting data at M.D. Anderson Cancer Center, and Neil Malloy for assistance with data coordination at Massachusetts General Hospital.
Grant Support: In part by the NCI Cancer Genetics Network. Work of the Cancer Genetics Network Statistical Coordinating Center was supported by National Cancer Institute grant CA78284. Work of Drs. Parmigiani and Chen and Ms. Friebel was also supported in part by National Cancer Institute grants P50CA88843, P50CA62924-05, and 5P30 CA06973-39, R01CA105090-01A1; National Institutes of Health grant HL 99-024; and the Hecht Fund. Work of investigators at the Fred Hutchinson Cancer Research Center was supported in part by National Institutes of Health grants R01 CA 36397, R01 CA 63705, and K05 CA-90754. The work of Dr. Weitzel and Ms. Sand was supported in part by California Cancer Research Program of the University of California (grant no. 99-86874) and in part by a General Clinical Research Center grant from National Institutes of Health (M01 RR00043) awarded to the City of Hope National Medical Center. Data from Georgetown University were provided by the Familial Cancer Registry Shared Resource of Lombardi Comprehensive Cancer Center, which is supported in part by the National Institutes of Health (grant P30-CA-51008).
Appendix Tables
Appendix Table 1.
Number of Counselees, by Genotyping Method for Each Gene and Center *
| Genotyping Method |
Duke Univ. |
Penn | George- town Univ. |
JHU | FHCRC | BCM | MDACC | UTSW | HCI | UCI | COH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | |||||||||||
| SSCP | 1 | 61 | 170 | 0 | 583 | 0 | 0 | 1 | 0 | 9 | 0 |
| Seq | 72 | 127 | 28 | 68 | 0 | 0 | 105 | 105 | 61 | 0 | 33 |
| ASO | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 688 | 0 |
| Targeted mut screening | 37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Seq for 185delAG and 5382insC | 41 | 91 | 4 | 33 | 0 | 0 | 11 | 15 | 0 | 0 | 25 |
| Seq for 185delAG only | 0 | 0 | 0 | 0 | 0 | 282 | 0 | 0 | 0 | 106 | 0 |
| CSGE | 3 | 310 | 41 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| SSCP + ASO | 0 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Targeted mut screening + Seq | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Targeted mut screening + CSGE | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other† | 4 | 42 | 14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 19 |
| None‡ | 1 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BRCA2 | |||||||||||
| SSCP | 0 | 0 | 171 | 0 | 384 | 0 | 0 | 0 | 0 | 0 | 0 |
| Seq | 64 | 242 | 28 | 66 | 0 | 0 | 104 | 101 | 61 | 0 | 32 |
| CSGE | 5 | 199 | 40 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASO | 55 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Seq for 6174delT | 47 | 0 | 6 | 33 | 0 | 0 | 11 | 14 | 0 | 688 | 25 |
| SSCP + ASO | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Seq + ASO | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASO + Seq | 36 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other† | 0 | 76 | 13 | 1 | 0 | 0 | 0 | 0 | 0 | 115 | 19 |
| None‡ | 11 | 159 | 0 | 3 | 199 | 282 | 2 | 6 | 0 | 0 | 1 |
ASO = allele-specific oligonucleotide hybridization assay (33, 61); BCM = Baylor College of Medicine; COH = City of Hope; CSGE = confirmation-sensitive gel electrophoresis (33); FHCRC = Fred Hutchinson Cancer Research Center; HCI = Huntsman Cancer Institute; JHU = Johns Hopkins University; MDACC = M.D. Anderson Cancer Center; Penn = University of Pennsylvania; Seq = full sequencing of the coding regions of the gene, as implemented by Myriad Genetics Laboratories at the time of testing (18, 61, 62); Seq for 185delAG = sequencing for Ashkenazi founder mutation 185delAG in BRCA1 (1, 63); Seq for 185delAG and 5382insC = sequencing for Ashkenazi founder mutations 185delAG and 5382insC in BRCA1 (1, 63); Seq for 6174delT = sequencing for Ashkenazi founder mutation 6174delT in BRCA1 (1, 63); SSCP = single-strand conformation polymorphism (62); targeted mut screening = sequencing for a panel of 8 known deleterious mutation in BRCA1 and 4 deleterious mutations in BRCA2 (64); UCI = University of California, Irvine (Orange County); UTSW = University of Texas Southwestern.
”Other” includes all genotyping methods that were used in a sample that was too small to be worth reporting in detail, as well as some cases whose genotyping method was missing.
“None” indicates that individuals were tested for 1 gene and not the other, contributing to the main analysis only if they test positive. This occurs either by design or because genes are tested sequentially and the second gene is not tested after a mutation is found in the first.
Appendix Table 2.
Input Variables and Features of Each Model *
| Variable | BRCAPRO | FHAT | Finnish | Myriad | NCI† | Penn | Yale |
|---|---|---|---|---|---|---|---|
| Model characteristics | |||||||
| Mendelian | • | • | |||||
| Empirical | • | • | • | • | |||
| Expert-based | • | ||||||
| Requires a computer | • | • | • | • | • | ||
| Requires pedigree | • | • | |||||
| Available in the CaGene (67) package‡ | • | • | • | • | • | • | • |
| Trained on high-risk data | • | • | • | • | • | ||
| Size of training sample (for empirical models), n | 148 | 238§ | 5318 | 169 | |||
| Updated periodically | • | • | |||||
| References: development | 13, 14, 65 | 16 | 17 | 18, 61, 66 | 15 | 11 | |
| References: validation | 24, 46, 58, 32, 53?57 | 16, 54 | 46, 55 | 46, 32, 53–55, 57 | 46, 32, 54, 55 | 55 | |
| Model input∥ | |||||||
| Mendelian transmission | • | • | |||||
| Exact family structure | • | • | |||||
| Age of unaffected counselee | • | • | • | ||||
| Age of unaffected relatives | • | • | |||||
| Ashkenazi Jewish ethnicity of family | • | • | • | • | • | ||
| Breast cancer status, counselee | • | • | • | • | • | • | |
| Breast cancer status, relatives | • | • | • | • | • | • | |
| Breast cancer age of onset, counselee | • | • | • | • | • | • | • |
| Breast cancer age of onset, relatives | • | • | • | • | • | • | |
| Ovarian cancer status, counselee | • | • | • | • | • | • | |
| Ovarian cancer status, relatives | • | • | • | • | • | • | |
| Ovarian cancer age of onset, counselee | • | • | • | ||||
| Ovarian cancer age of onset, relatives | • | • | |||||
| Male breast cancer status, counselee | • | • | • | • | |||
| Male breast cancer status, relatives | • | • | • | • | |||
| Both breast and ovarian cancer in counselee | • | • | • | • | |||
| Both breast and ovarian cancer in a single relative | • | • | • | ||||
| Bilateral breast cancer status, counselee | • | • | • | ||||
| Bilateral breast cancer status, relatives | • | • | • | ||||
| Bilateral breast cancer, both ages of onset, counselee | • | • | |||||
| Bilateral breast cancer, both ages of onset, relatives | • | • | |||||
| Prostate cancer in relatives | • | ||||||
| Colon cancer in relatives | • | ||||||
| Model output | |||||||
| Predictions for BRCA1 and BRCA2 separately | • | ||||||
| Predictions for BRCA1 and BRCA2 together | • | • | • | • | |||
| Predictions for BRCA1 only | • | ||||||
| Probability of carrying a mutation | • | • | |||||
| Probability of finding a mutation if tested | • | • | • | • | |||
| Nonprobabilistic score | • |
FHAT = family history assessment tool; NCI = National Cancer Institute; Penn = University of Pennsylvania; Yale = Yale University.
Intended for use with Ashkenazi Jewish women only.
CaGene manufactured by University of Texas Southwestern Medical Center, Dallas, Texas (accessed at www4.utsouthwestern.edu/breasthealth/cagene on 1 August 2007).
Classes were originally selected by using 238 breast cancer cases. Subsequently, chances of finding a mutation in each risk class have been updated by using empirical frequencies from additional genotyping at Myriad. We used the January 2002 version of BRCAPRO (accessed at http://astor.som.jhmi.edu/BayesMendel/brcapro.html on 1 August 2007).
Models encode and utilize these in different ways
Appendix Table 3.
C-Statistics (95% CIs) of the Models, by Cancer Status of the Counselee, Study Type, Ashkenazi Ethnicity, and Overall, for Persons Tested by Gene Sequencing *
| Model | All Persons |
Not Ashkenazi Jewish |
Ashkenazi Jewish |
No Cancer |
Breast Cancer Only |
Ovarian Cancer Only |
Both Breast and Ovarian Cancer |
|---|---|---|---|---|---|---|---|
| BRCAPRO | 0.74 (0.69–0.79) | 0.75 (0.7–0.81) | 0.66 (0.49–0.83) | 0.61 (0.49–0.72) | 0.76 (0.71–0.81) | 0.91 (0.8–1.0) | 0.81 (0.63–0.99) |
| Yale | 0.64 (0.58–0.69) | 0.65 (0.59–0.71) | 0.61 (0.46–0.77) | 0.57 (0.46–0.67) | 0.67 (0.61–0.74) | 0.82 (0.68–0.95) | 0.71 (0.46–0.96) |
| Myriad | 0.67 (0.62–0.72) | 0.68 (0.62–0.73) | 0.66 (0.49–0.82) | 0.61 (0.49–0.73) | 0.65 (0.58–0.72) | 0.87 (0.74–0.99) | 0.67 (0.43–0.91) |
| NCI† | – | – | 0.6 (0.45–0.75) | – | – | – | – |
| Penn‐ | 0.69 (0.63–0.75) | 0.69 (0.63–0.74) | 0.76 (0.61–0.92) | 0.7 (0.57–0.83) | 0.64 (0.57–0.71) | 0.7 (0.45–0.95) | 0.62 (0.38–0.86) |
| FHAT | 0.68 (0.63–0.73) | 0.69 (0.64–0.75) | 0.66 (0.49–0.82) | 0.58 (0.45–0.71) | 0.66 (0.59–0.73) | 0.84 (0.7–0.98) | 0.72 (0.49–0.94) |
| Finnish | 0.71 (0.66–0.77) | 0.69 (0.64–0.75) | 0.78 (0.6–0.95) | 0.8 (0.69–0.91) | 0.63 (0.56–0.69) | 0.87 (0.69–1) | 0.69 (0.47–0.91) |
FHAT = family history assessment tool; NCI = National Cancer Institute; Penn = University of Pennsylvania; Yale = Yale University.
The NCI model was applied only to families of Ashkenazi ethnicity.
The Penn model predicts BRCA1 mutations only. In the table, its performance in predicting mutations at either gene is evaluated to facilitate comparisons with other models and to capture a common use of the model.
Appendix Table 4.
Sensitivity, Specificity, and Likelihood Ratios of All Predictive Models *
| Model | UCI Sample | FHCRC Sample | High-Risk Sample |
|---|---|---|---|
| Sensitivity (95% PI)† | |||
| BRCAPRO | 0.429 (0.207–0.656) | 0.706 (0.547–0.834) | 0.824 (0.786–0.857) |
| Yale | 0.071 (0.010–0.260) | 0.765 (0.627–0.866) | 0.639 (0.592–0.677) |
| Myriad | 0.286 (0.126–0.540) | 0.853 (0.734–0.936) | 0.775 (0.732–0.813) |
| NCI | 0.400 (0.102–0.775) | 1.000 (0.354–1.000) | 0.625 (0.522–0.697) |
| FHAT | 0.786 (0.536–0.931) | 0.941 (0.837–0.987) | 0.885 (0.852–0.913) |
| Finnish | 0.273 (0.082–0.563) | 0.853 (0.712–0.931) | 0.728 (0.686–0.777) |
| Specificity (95% PI)† | |||
| BRCAPRO | 0.934 (0.913–0.950) | 0.671 (0.6201–0.715) | 0.526 (0.499–0.551) |
| Yale | 0.928 (0.910–0.943) | 0.597 (0.5443–0.639) | 0.574 (0.543–0.605) |
| Myriad | 0.864 (0.840–0.888) | 0.683 (0.6390–0.730) | 0.479 (0.449–0.512) |
| NCI | 0.766 (0.627–0.857) | 0.125 (0.0193–0.486) | 0.655 (0.600–0.711) |
| FHAT | 0.720 (0.694–0.749) | 0.320 (0.2669–0.375) | 0.271 (0.249–0.299) |
| Finnish | 0.879 (0.850–0.904) | 0.447 (0.3954–0.505) | 0.653 (0.628–0.682) |
| Positive LR (95% PI)† | |||
| BRCAPRO | 6.503 (2.925–10.24) | 2.148 (1.682–2.71) | 1.739 (1.63–1.87) |
| Yale | 0.989 (0.125–3.67) | 1.898 (1.500–2.23) | 1.501 (1.37–1.65) |
| Myriad | 2.107 (0.899–4.13) | 2.689 (2.127–3.29) | 1.487 (1.39–1.60) |
| NCI | 1.709 (0.445–3.84) | 1.143 (0.429–1.68) | 1.813 (1.42–2.20) |
| FHAT | 2.805 (1.981–3.45) | 1.384 (1.210–1.55) | 1.214 (1.16–1.27) |
| Finnish | 2.259 (0.679–4.46) | 1.543 (1.282–1.80) | 2.097 (1.89–2.34) |
| Negative LR (95% PI)† | |||
| BRCAPRO | 0.612 (0.3722–0.843) | 0.438 (0.24249–0.669) | 0.334 (0.269–0.408) |
| Yale | 1.001 (0.8018–1.075) | 0.394 (0.22660–0.629) | 0.628 (0.548–0.708) |
| Myriad | 0.826 (0.5284–1.018) | 0.215 (0.09054–0.395) | 0.47 (0.388–0.556) |
| NCI | 0.783 (0.3129–1.229) | 0 (0–9.819) | 0.572 (0.454–0.750) |
| FHAT | 0.298 (0.0957–0.632) | 0.184 (0.03927–0.547) | 0.424 (0.315–0.555) |
| Finnish | 0.827 (0.5017–1.045) | 0.329 (0.14274–0.637) | 0.416 (0.340–0.485) |
For each model, we divided the study population into 2 groups depending on whether the model’s prediction for a positive for either gene is >10%; crosstabulated this information with the genetic testing results; and computed the sensitivity, specificity, and LRs. The 95% PIs are 95% posterior probability regions (obtained by using Jeffrey noninformative priors). The PIs are necessary to account for skewness and small sample sizes in some cells. FHAT = family history assessment tool; FHCRC = Fred Hutchinson Cancer Research Center; LR = likelihood ratio; NCI = National Cancer Institute; UCI = University of California, Irvine (Orange County); Yale = Yale University.
For a referral threshold probability of 10%.
Appendix Table 5.
Predictive Performance of Models*
| Model | UCI Sample | FHCRC Sample | High-Risk Sample |
|---|---|---|---|
| Individuals testing positive among those with probability >10% [95% PI], n/n (%) † | |||
| BRCAPRO | 6/58 (10.34 [4.435–20.08]) | 24/139 (17.3 [11.69–24.2]) | 352/874 (40.3 [37.1–43.6]) |
| Yale | 1/58 (1.72 [0.187–7.78]) | 26/167 (15.6 [10.68–21.6]) | 273/742 (36.8 [33.4–40.3]) |
| Myriad | 4/111 (3.60 [1.228–8.34]) | 29/140 (20.7 [14.64–28.0]) | 331/905 (36.6 [33.5–39.8]) |
| NCI | 2/13 (15.38 [3.341–40.90]) | 2/9 (22.2 [4.93–54.4]) | 75/176 (42.6 [35.5–50.0]) |
| FHAT | 11/232 (4.74 [2.545–8.06]) | 32/270 (11.9 [8.41–16.1]) | 378/1181 (32.0 [29.4–34.7]) |
| Finnish | 3/77 (3.90 [1.109–10.03]) | 29/218 (13.3 [9.29–18.3]) | 284/642 (44.2 [40.4–48.1]) |
| Individuals testing negative among those with probability ≤10% [95% PI], n/n (%) ‡ | |||
| BRCAPRO | 737/745 (98.9 [98.0–99.5]) | 235/245 (95.9 [92.9–97.9]) | 579/654 (88.5 [85.9–90.8]) |
| Yale | 732/745 (98.3 [97.1–99.0]) | 209/217 (96.3 [93.2–98.2]) | 632/786 (80.4 [77.5–83.1]) |
| Myriad | 682/692 (98.6 [97.5–99.3]) | 239/244 (98.0 [95.6–99.2]) | 527/623 (84.6 [81.6–87.3]) |
| NCI | 36/39 (92.3 [80.9–97.8]) | 1/1 (100.0 [14.7–100.0]) | 192/237 (81.0 [75.7–85.6]) |
| FHAT | 568/571 (99.5 [98.6–99.9]) | 112/114 (98.2 [94.5–99.6]) | 298/347 (85.9 [81.9–89.2]) |
| Finnish | 539/547 (98.5 [97.3–99.3]) | 153/158 (96.8 [93.2–98.8]) | 673/779 (86.4 [83.9–88.7]) |
For each model, we divided the study population into 2 groups depending on whether the model’s prediction for a positive result for either gene is >10%, crosstabulated this information with the genetic testing results, and computed the positive and negative predictive values. The 95% PIs are 95% posterior probability regions (obtained by using Jeffrey noninformative priors). The PIs are necessary to account for skewness and small sample sizes in some cells. FHAT = family history assessment tool; FHCRC = Fred Hutchinson Cancer Research Center; UCI = University of California, Irvine (Orange County); Yale = Yale University.
Positive predictive value.
Negative predictive value.
Appendix Table 6.
Number of Patients per 1000 Referred for Mutation Testing, with Different Threshold Probabilities for Referral*
| Variable | Proportion Positive |
Threshold | Mutation-Negative above Threshold |
Mutation-Positive above Threshold |
Mutation-Positive below Threshold |
Mutation-Negative below Threshold |
Total |
|---|---|---|---|---|---|---|---|
| Yale | |||||||
| UCI sample | 0.017 | 0.05 | 102.1 | 2.5 | 14.9 | 880.4 | 1000 |
| 0.1 | 71 | 1.2 | 16.2 | 911.6 | 1000 | ||
| 0.2 | 43.6 | 1.2 | 16.2 | 939 | 1000 | ||
| FHCRC sample | 0.089 | 0.05 | 455.7 | 75.5 | 13 | 455.7 | 1000 |
| 0.1 | 367.2 | 67.7 | 20.8 | 544.3 | 1000 | ||
| 0.2 | 291.7 | 59.9 | 28.6 | 619.8 | 1000 | ||
| High-risk sample | 0.279 | 0.05 | 376.3 | 197 | 82.5 | 344.2 | 1000 |
| 0.1 | 306.9 | 178.7 | 100.8 | 413.6 | 1000 | ||
| 0.2 | 234.9 | 158.4 | 121.1 | 485.6 | 1000 | ||
| Myriad | |||||||
| UCI sample | 0.017 | 0.05 | 459.5 | 11.2 | 6.2 | 523 | 1000 |
| 0.1 | 133.3 | 5 | 12.5 | 849.3 | 1000 | ||
| 0.2 | 64.8 | 3.7 | 13.7 | 917.8 | 1000 | ||
| FHCRC sample | 0.089 | 0.05 | 750 | 88.5 | 0 | 161.5 | 1000 |
| 0.1 | 289.1 | 75.5 | 13 | 622.4 | 1000 | ||
| 0.2 | 80.7 | 36.5 | 52.1 | 830.7 | 1000 | ||
| High-risk sample | 0.279 | 0.05 | 549.1 | 255.2 | 24.2 | 171.5 | 1000 |
| 0.1 | 375.7 | 216.6 | 62.8 | 344.9 | 1000 | ||
| 0.2 | 138.7 | 138.7 | 140.7 | 581.8 | 1000 | ||
| NCI | |||||||
| UCI sample | 0.096 | 0.05 | 211.5 | 38.5 | 57.7 | 692.3 | 1000 |
| 0.1 | 211.5 | 38.5 | 57.7 | 692.3 | 1000 | ||
| 0.2 | 76.9 | 0 | 96.2 | 826.9 | 1000 | ||
| FHCRC sample | 0.2 | 0.05 | 700 | 200 | 0 | 100 | 1000 |
| 0.1 | 700 | 200 | 0 | 100 | 1000 | ||
| 0.2 | 500 | 100 | 100 | 300 | 1000 | ||
| High-risk sample | 0.291 | 0.05 | 247 | 181.6 | 109 | 462.5 | 1000 |
| 0.1 | 244.6 | 181.6 | 109 | 464.9 | 1000 | ||
| 0.2 | 104.1 | 82.3 | 208.2 | 605.3 | 1000 | ||
| FHAT | |||||||
| UCI sample | 0.017 | 0.05 | 638.9 | 17.4 | 0 | 343.7 | 1000 |
| 0.1 | 275.2 | 13.7 | 3.7 | 707.3 | 1000 | ||
| 0.2 | 51.1 | 3.7 | 13.7 | 931.5 | 1000 | ||
| FHCRC sample | 0.089 | 0.05 | 849 | 88.5 | 0 | 62.5 | 1000 |
| 0.1 | 619.8 | 83.3 | 5.2 | 291.7 | 1000 | ||
| 0.2 | 177.1 | 54.7 | 33.9 | 734.4 | 1000 | ||
| High-risk sample | 0.279 | 0.05 | 678 | 269 | 10.5 | 42.5 | 1000 |
| 0.1 | 525.5 | 247.4 | 32.1 | 195 | 1000 | ||
| 0.2 | 174.7 | 158.4 | 121.1 | 545.8 | 1000 | ||
| Finnish | |||||||
| UCI sample | 0.018 | 0.05 | 216.3 | 9.6 | 8 | 766 | 1000 |
| 0.1 | 118.6 | 4.8 | 12.8 | 863.8 | 1000 | ||
| 0.2 | 60.9 | 4.8 | 12.8 | 921.5 | 1000 | ||
| FHCRC sample | 0.09 | 0.05 | 656.9 | 87.8 | 2.7 | 252.7 | 1000 |
| 0.1 | 502.7 | 77.1 | 13.3 | 406.9 | 1000 | ||
| 0.2 | 170.2 | 39.9 | 50.0 | 739.4 | 1000 | ||
| High-risk sample | 0.274 | 0.05 | 373.7 | 233.6 | 40.8 | 351.9 | 1000 |
| 0.1 | 251.9 | 199.9 | 74.6 | 473.6 | 1000 | ||
| 0.2 | 145 | 145.7 | 128.8 | 580.6 | 1000 |
Continued from Table 4. FHAT = family history assessment tool; FHCRC = Fred Hutchinson Cancer Research Center; NCI = National Cancer Institute; UCI = University of California, Irvine (Orange County); Yale = Yale University.
Footnotes
Current Author Addresses: Dr. Parmigiani: The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, 550 North Broadway, Suite 1103, Baltimore, MD 21205-2011.
Dr. Chen: Department of Environmental Health Sciences, Johns Hopkins School of Public Health, 615 North Wolfe Street, Baltimore, MD 21205.
Dr. Iversen: Department of Statistical Sciences, Duke University, Box 90251, Durham, NC 27708.
Ms. Friebel: Center for Clinical Epidemiology and Biostatistics, University of Pennsylvania School of Medicine, 909 Blockley Hall, 423 Guardian Drive, Philadelphia, PA 19104-6021.
Dr. Finkelstein: Biostatistics Center, Massachusetts General Hospital, 50 Staniford Street, Suite 560, Boston, MA 02114.
Drs. Anton-Culver and Ziogas: University of California, 224 IH, Mail Code 7550, Irvine, CA 92697-7550.
Dr. Weber: GlaxoSmithKline, 2301 Renaissance Boulevard, Building 510, Mailcode RN0510, King of Prussia, PA 19406-2772.
Dr. Eisen: Department of Medicine, University of Toronto, Suite RFE 3-805, 190 Elizabeth Street, Toronto, Ontario M5G 2C4, Canada.
Drs. Malone, Daling, and Hsu: Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109-1024.
Dr. Ostrander: Cancer Genetics Branch, National Human Genome Research Institute, National Institutes of Health, 50 South Drive, MSC 8000, Building 50, Room 5351, Bethesda, MD 20892-8000.
Dr. Peterson: The Methodist Hospital, 6550 Fannin Street, SM-1299, Houston, TX 77030.
Dr. Schildkraut: Duke University Medical Center, Box 2949, Durham, NC 27510.
Dr. Isaacs, Ms. Corio, Ms. Leondaridis, and Ms. Peshkin: Georgetown University, 2233 Wisconsin Avenue NW, Suite 317, Washington, DC 20007.
Drs. Tomlinson and Euhus: University of Texas Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, TX 75390-9155.
Drs. Amos, Strong, and Berry: M.D. Anderson Cancer Center, 1515 Holcombe Boulevard, Box 189, Houston, TX 77030.
Dr. Weitzel and Ms. Sand: City of Hope National Medical Center, 1500 East Duarte Road, Duarte, CA 91001.
Ms. Dutson and Dr. Kerber: Huntsman Cancer Institute, 2000 Circle of Hope, Salt Lake City, UT 84112-5550.
Author Contributions: Conception and design: G. Parmigiani, D.M. Finkelstein, H. Anton-Culver, B.L. Weber, G. Tomlinson, D.M. Euhus.
Analysis and interpretation of the data: G. Parmigiani, S. Chen, E.S. Iversen Jr., A. Ziogas, B.L. Weber, L.E. Peterson, R. Kerber, B.N. Peshkin, D.M. Euhus.
Drafting of the article: G. Parmigiani, H. Anton-Culver, L.E. Peterson.
Critical revision of the article for important intellectual content: S. Chen, E.S. Iversen Jr., K.E. Malone, J.R. Daling, L. Hsu, E.A. Ostrander, C. Isaacs, C.I. Amos, D.A. Berry, J.N. Weitzel, B.N. Peshkin, D.M. Euhus.
Final approval of the article: G. Parmigiani, S. Chen, E.S. Iversen Jr., D.M. Finkelstein, B.L. Weber, A. Eisen, K.E. Malone, J.R. Daling, E.A. Ostrander, J.M. Schildkraut, C. Isaacs, C. Corio, G. Tomlinson, J.N. Weitzel, B.N. Peshkin.
Provision of study materials or patients: D.M. Finkelstein, B.L. Weber, A. Eisen, K.E. Malone, J.R. Daling, J.M. Schildkraut, C. Isaacs, C. Corio, G. Tomlinson, C.I. Amos, L.C. Strong, J.N. Weitzel, D. Dutson, B.N. Peshkin, D.M. Euhus.
Statistical expertise: G. Parmigiani, S. Chen, E.S. Iversen Jr., A. Ziogas, L. Hsu, L. Leondaridis, C.I. Amos, S. Sand, R. Kerber.
Obtaining of funding: G. Parmigiani, H. Anton-Culver, B.L. Weber, J.R. Daling, E.A. Ostrander, J.M. Schildkraut, L.C. Strong.
Administrative, technical, or logistic support: G. Parmigiani, T.M. Friebel, D.M. Finkelstein, A. Ziogas, B.L. Weber, J.R. Daling, D.A. Berry, J.N. Weitzel, D. Dutson, D.M. Euhus.
Collection and assembly of data: T.M. Friebel, D.M. Finkelstein, H. Anton-Culver, B.L. Weber, K.E. Malone, J.R. Daling, E.A. Ostrander, J.M. Schildkraut, C. Isaacs, C. Corio, L. Leondaridis, G. Tomlinson, L.C. Strong, D.M. Euhus.
Potential Financial Conflicts of Interest: Employment: B.L. Weber (GlaxoSmithKline). Stock ownership or options (other than mutual funds): B.L. Weber (GlaxoSmithKline).
Current author addresses and author contributions are available at www.annals.org.
References
- 1.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. PMID: 9145676. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. PMID: 12677558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. PMID: 14576434. [DOI] [PubMed] [Google Scholar]
- 4.Newman B, Millikan RC, King MC. Genetic epidemiology of breast and ovarian cancers. Epidemiol Rev. 1997;19:69–79. doi: 10.1093/oxfordjournals.epirev.a017948. PMID: 9360904. [DOI] [PubMed] [Google Scholar]
- 5.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. PMID: 9497246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber BL. Genetic testing for breast cancer. Sci Med. 1996;3:12–21. [Google Scholar]
- 7.American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J Clin Oncol. 2003;21:2397–2406. doi: 10.1200/JCO.2003.03.189. PMID: 12692171. [DOI] [PubMed] [Google Scholar]
- 8.Trepanier A, Ahrens M, McKinnon W, Peters J, Stopfer J, Grumet SC, et al. Genetic cancer risk assessment and counseling: recommendations of the national society of genetic counselors. J Genet Couns. 2004;13:83–114. doi: 10.1023/B:JOGC.0000018821.48330.77. PMID: 15604628. [DOI] [PubMed] [Google Scholar]
- 9.Nelson HD, Huffman LH, Fu R, Harris EL. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: systematic evidence review for the U.S. Preventive Services Task Force. Ann Intern Med. 2005;143:362–379. doi: 10.7326/0003-4819-143-5-200509060-00012. PMID: 16144895. [DOI] [PubMed] [Google Scholar]
- 10.Claus EB, Risch N, Thompson WD. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet. 1991;48:232–242. PMID: 1990835. [PMC free article] [PubMed] [Google Scholar]
- 11.Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997;336:1409–1415. doi: 10.1056/NEJM199705153362002. PMID: 9145677. [DOI] [PubMed] [Google Scholar]
- 12.Shattuck-Eidens D, Oliphant A, McClure M, McBride C, Gupte J, Rubano T, et al. BRCA1 sequence analysis in women at high risk for susceptibility mutations. Risk factor analysis and implications for genetic testing. JAMA. 1997;278:1242–1250. PMID: 9333265. [PubMed] [Google Scholar]
- 13.Berry DA, Parmigiani G, Sanchez J, Schildkraut J, Winer E. Probability of carrying a mutation of breast-ovarian cancer gene BRCA1 based on family history. J Natl Cancer Inst. 1997;89:227–238. doi: 10.1093/jnci/89.3.227. PMID: 9017003. [DOI] [PubMed] [Google Scholar]
- 14.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–158. doi: 10.1086/301670. PMID: 9443863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartge P, Struewing JP, Wacholder S, Brody LC, Tucker MA. The prevalence of common BRCA1 and BRCA2 mutations among Ashkenazi Jews. Am J Hum Genet. 1999;64:963–970. doi: 10.1086/302320. PMID: 10090881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilpin CA, Carson N, Hunter AG. A preliminary validation of a family history assessment form to select women at risk for breast or ovarian cancer for referral to a genetics center. Clin Genet. 2000;58:299–308. doi: 10.1034/j.1399-0004.2000.580408.x. PMID: 11076055. [DOI] [PubMed] [Google Scholar]
- 17.Vahteristo P, Eerola H, Tamminen A, Blomqvist C, Nevanlinna H. A probability model for predicting BRCA1 and BRCA2 mutations in breast and breast-ovarian cancer families. Br J Cancer. 2001;84:704–708. doi: 10.1054/bjoc.2000.1626. PMID: 11237395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. PMID: 11896095. [DOI] [PubMed] [Google Scholar]
- 19.Bluman LG, Rimer BK, Berry DA, Borstelmann N, Iglehart JD, Regan K, et al. Attitudes, knowledge, and risk perceptions of women with breast and/or ovarian cancer considering testing for BRCA1 and BRCA2. J Clin Oncol. 1999;17:1040–1046. doi: 10.1200/JCO.1999.17.3.1040. PMID: 10071299. [DOI] [PubMed] [Google Scholar]
- 20.Skinner CS, Schildkraut JM, Berry D, Calingaert B, Marcom PK, Sugarman J, et al. Pre-counseling education materials for BRCA testing: does tailoring make a difference? Genet Test. 2002;6:93–105. doi: 10.1089/10906570260199348. PMID: 12215248. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz MD, Peshkin BN, Hughes C, Main D, Isaacs C, Lerman C. Impact of BRCA1/BRCA2 mutation testing on psychologic distress in a clinic-based sample. J Clin Oncol. 2002;20:514–520. doi: 10.1200/JCO.2002.20.2.514. PMID: 11786581. [DOI] [PubMed] [Google Scholar]
- 22.Hartman AR, Daniel BL, Kurian AW, Mills MA, Nowels KW, Dirbas FM, et al. Breast magnetic resonance image screening and ductal lavage in women at high genetic risk for breast carcinoma. Cancer. 2004;100:479–489. doi: 10.1002/cncr.11926. PMID: 14745863. [DOI] [PubMed] [Google Scholar]
- 23.Domchek SM, Eisen A, Calzone K, Stopfer J, Blackwood A, Weber BL. Application of breast cancer risk prediction models in clinical practice. J Clin Oncol. 2003;21:593–601. doi: 10.1200/JCO.2003.07.007. PMID: 12586794. [DOI] [PubMed] [Google Scholar]
- 24.Euhus DM, Smith KC, Robinson L, Stucky A, Olopade OI, Cummings S, et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. J Natl Cancer Inst. 2002;94:844–851. doi: 10.1093/jnci/94.11.844. PMID: 12048272. [DOI] [PubMed] [Google Scholar]
- 25.Richards CS, Ward PA, Roa BB, Friedman LC, Boyd AA, Kuenzli G, et al. Screening for 185delAG in the Ashkenazim. Am J Hum Genet. 1997;60:1085–1098. PMID: 9150156. [PMC free article] [PubMed] [Google Scholar]
- 26.Weitzel JN, McCaffrey SM, Nedelcu R, MacDonald DJ, Blazer KR, Cullinane CA. Effect of genetic cancer risk assessment on surgical decisions at breast cancer diagnosis. Arch Surg. 2003;138:1323–1328. doi: 10.1001/archsurg.138.12.1323. discussion 1329. [PMID: 14662532] [DOI] [PubMed] [Google Scholar]
- 27.Miron A, Schildkraut JM, Rimer BK, Winer EP, Sugg Skinner C, Futreal PA, et al. Testing for hereditary breast and ovarian cancer in the southeastern United States. Ann Surg. 2000;231:624–634. doi: 10.1097/00000658-200005000-00002. PMID: 10767783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone KE, Daling JR, Thompson JD, O’Brien CA, Francisco LV, Ostrander EA. BRCA1 mutations and breast cancer in the general population: analyses in women before age 35 years and in women before age 45 years with first-degree family history. JAMA. 1998;279:922–929. doi: 10.1001/jama.279.12.922. PMID: 9544766. [DOI] [PubMed] [Google Scholar]
- 29.Friebel TM, Beutler RA, Lee SM, Bernhardt BA, Helzlsouer KJ, Griffin CA. Active recruitment increased enrollment in a hereditary cancer registry. J Clin Epidemiol. 2004;57:1172–1176. doi: 10.1016/j.jclinepi.2004.04.007. PMID: 15567634. [DOI] [PubMed] [Google Scholar]
- 30.Martin AM, Blackwood MA, Antin-Ozerkis D, Shih HA, Calzone K, Colligon TA, et al. Germline mutations in BRCA1 and BRCA2 in breast-ovarian families from a breast cancer risk evaluation clinic. J Clin Oncol. 2001;19:2247–2253. doi: 10.1200/JCO.2001.19.8.2247. PMID: 11304778. [DOI] [PubMed] [Google Scholar]
- 31.Anton-Culver H, Cohen PF, Gildea ME, Ziogas A. Characteristics of BRCA1 mutations in a population-based case series of breast and ovarian cancer. Eur J Cancer. 2000;36:1200–1208. doi: 10.1016/s0959-8049(00)00110-6. PMID: 10882857. [DOI] [PubMed] [Google Scholar]
- 32.Barcenas CH, Hosain GM, Arun B, Zong J, Zhou X, Chen J, et al. Assessing BRCA carrier probabilities in extended families. J Clin Oncol. 2006;24:354–360. doi: 10.1200/JCO.2005.02.2368. PMID: 16421416. [DOI] [PubMed] [Google Scholar]
- 33.Kristensen VN, Kelefiotis D, Kristensen T, Børresen-Dale AL. High-throughput methods for detection of genetic variation. Biotechniques. 2001;30:318–322. 324, 326. doi: 10.2144/01302tt01. passim. [PMID: 11233601] [DOI] [PubMed] [Google Scholar]
- 34.Andrulis IL, Anton-Culver H, Beck J, Bove B, Boyd J, Buys S, et al. Comparison of DNA- and RNA-based methods for detection of truncating BRCA1 mutations. Hum Mutat. 2002;20:65–73. doi: 10.1002/humu.10097. PMID: 12112659. [DOI] [PubMed] [Google Scholar]
- 35.Myriad Genetics Laboratories. Salt Lake City, UT: Myriad Genetics Laboratories; [updated 4 May 2006]. BRACAnalysis technical specifications. Accessed at www.myriadtests.com/provider/doc/BRACAnalysis-Technical-Specifications.pdf on 26 July 2007. [Google Scholar]
- 36.Yan H, Kinzler KW, Vogelstein B. Tech.sight. Genetic testing–present and future. Science. 2000;289:1890–1892. doi: 10.1126/science.289.5486.1890. PMID: 11012364. [DOI] [PubMed] [Google Scholar]
- 37.Murphy EA, Mutalik GS. The application of Bayesian methods in genetic counseling. Hum Hered. 1969;19:126–151. [Google Scholar]
- 38.Elston RC, Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21:523–542. doi: 10.1159/000152448. PMID: 5149961. [DOI] [PubMed] [Google Scholar]
- 39.Szolovits P, Pauker SP. Pedigree analysis for genetic counseling. In: Lun KC, Degoulet P, Piemme TE, Rienhoff O, editors. MEDINFO 92: Proceedings of the Seventh Conference on Medical Informatics. New York: Elsevier; 1992. pp. 679–683. [Google Scholar]
- 40.Offit K, Brown K. Quantitating familial cancer risk: a resource for clinical oncologists. J Clin Oncol. 1994;12:1724–1736. doi: 10.1200/JCO.1994.12.8.1724. PMID: 8040684. [DOI] [PubMed] [Google Scholar]
- 41.Antoniou AC, Gayther SA, Stratton JF, Ponder BA, Easton DF. Risk models for familial ovarian and breast cancer. Genet Epidemiol. 2000;18:173–190. doi: 10.1002/(SICI)1098-2272(200002)18:2<173::AID-GEPI6>3.0.CO;2-R. PMID: 10642429. [DOI] [PubMed] [Google Scholar]
- 42.Iversen ES, Jr, Parmigiani G, Berry DA, Schildkraut JM. Genetic susceptibility and survival: application to breast cancer. J Am Stat Assoc. 2000;95:28–42. [Google Scholar]
- 43.Claus EB, Risch NJ, Thompson WD. Age at onset as an indicator of familial risk of breast cancer. Am J Epidemiol. 1990;131:961–972. doi: 10.1093/oxfordjournals.aje.a115616. PMID: 2188501. [DOI] [PubMed] [Google Scholar]
- 44.Claus EB, Risch N, Thompson WD. Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction. Cancer. 1994;73:643–651. doi: 10.1002/1097-0142(19940201)73:3<643::aid-cncr2820730323>3.0.co;2-5. PMID: 8299086. [DOI] [PubMed] [Google Scholar]
- 45.Apicella C, Andrews L, Hodgson SV, Fisher SA, Lewis CM, Solomon E, et al. Log odds of carrying an ancestral mutation in BRCA1 or BRCA2 for a defined personal and family history in an Ashkenazi Jewish woman (LAMBDA) Breast Cancer Res. 2003;5:R206–R216. doi: 10.1186/bcr644. PMID: 14580256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de la Hoya M, Díez O, Pérez-Segura P, Godino J, Fernández JM, Sanz J, et al. Pre-test prediction models of BRCA1 or BRCA2 mutation in breast/ovarian families attending familial cancer clinics. J Med Genet. 2003;40:503–510. doi: 10.1136/jmg.40.7.503. PMID: 12843322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans DG, Eccles DM, Rahman N, Young K, Bulman M, Amir E, et al. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41:474–480. doi: 10.1136/jmg.2003.017996. PMID: 15173236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antoniou AC, Pharoah PP, Smith P, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–1590. doi: 10.1038/sj.bjc.6602175. PMID: 15381934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23:1111–1130. doi: 10.1002/sim.1668. PMID: 15057881. [DOI] [PubMed] [Google Scholar]
- 50.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. PMID: 7069920. [PubMed] [Google Scholar]
- 51.Harrell FE., Jr . New York: Springer; 2001. Regression Modeling Strategies with Application to Linear Models, Logistic Regression, and Survival Analysis. [Google Scholar]
- 52.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. PMID: 12509396. [DOI] [PubMed] [Google Scholar]
- 53.Berry DA, Iversen ES, Jr, Gudbjartsson DF, Hiller EH, Garber JE, Peshkin BN, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20:2701–2712. doi: 10.1200/JCO.2002.05.121. PMID: 12039933. [DOI] [PubMed] [Google Scholar]
- 54.James PA, Doherty R, Harris M, Mukesh BN, Milner A, Young MA, et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006;24:707–715. doi: 10.1200/JCO.2005.01.9737. PMID: 16446345. [DOI] [PubMed] [Google Scholar]
- 55.Marroni F, Aretini P, D’Andrea E, Caligo MA, Cortesi L, Viel A, et al. Evaluation of widely used models for predicting BRCA1 and BRCA2 mutations [Letter] J Med Genet. 2004;41:278–285. doi: 10.1136/jmg.2003.013623. PMID: 15060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antoniou AC, Durocher F, Smith P, Simard J, Easton DF. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2006;8:R3. doi: 10.1186/bcr1365. PMID: 16417652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Capalbo C, Ricevuto E, Vestri A, Sidoni T, Buffone A, Cortesi E, et al. Improving the accuracy of BRCA1/2 mutation prediction: validation of the novel country-customized IC software. Eur J Hum Genet. 2006;14:49–54. doi: 10.1038/sj.ejhg.5201511. PMID: 16288312. [DOI] [PubMed] [Google Scholar]
- 58.Nanda R, Schumm LP, Cummings S, Fackenthal JD, Sveen L, Ademuyiwa F, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–1933. doi: 10.1001/jama.294.15.1925. PMID: 16234499. [DOI] [PubMed] [Google Scholar]
- 59.Vogel KJ, Atchley DP, Erlichman J, Broglio KR, Ready KJ, Valero V, et al. BRCA1 and BRCA2 genetic testing in Hispanic patients: mutation prevalence and evaluation of the BRCAPRO risk assessment model. J Clin Oncol. doi: 10.1200/JCO.2006.10.4703. [Forthcoming] [DOI] [PubMed] [Google Scholar]
- 60.Myriad Genetics Laboratories. Salt Lake City, UT: Myriad Genetics Laboratories; 2006. Myriad Genetics 2006 annual report. Accessed at www.myriad.com/downloads/MYGN_AR2006.pdf on 26 July 2007. [Google Scholar]
- 61.Frank TS, Manley SA, Olopade OI, Cummings S, Garber JE, Bernhardt B, et al. Sequence analysis of BRCA1 and BRCA2: correlation of mutations with family history and ovarian cancer risk. J Clin Oncol. 1998;16:2417–2425. doi: 10.1200/JCO.1998.16.7.2417. PMID: 9667259. [DOI] [PubMed] [Google Scholar]
- 62.Gross E, Arnold N, Goette J, Schwarz-Boeger U, Kiechle M. A comparison of BRCA1 mutation analysis by direct sequencing, SSCP and DHPLC. Hum Genet. 1999;105:72–78. doi: 10.1007/s004399900092. PMID: 10480358. [DOI] [PubMed] [Google Scholar]
- 63.Warner E, Foulkes W, Goodwin P, Meschino W, Blondal J, Paterson C, et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999;91:1241–1247. doi: 10.1093/jnci/91.14.1241. PMID: 10413426. [DOI] [PubMed] [Google Scholar]
- 64.Peshkin BN, Lerman C, Isaacs C, Brown KM, de Leon A, Abbaszadegan MR. A detection panel of prevalent mutations in BRCA1/2 genes is sensitive and cost-effective in an initial screen of high risk patients [Abstract] Proc Am Assoc Cancer Res. 1998;39:3232. [Google Scholar]
- 65.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1063. Article21. [PMID: 16646800] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Myriad Genetics Laboratories. Salt Lake City, UT: Myriad Genetics Laboratories; 2006. BRCA risk calculator and mutation prevalence tables. Accessed at www.myriadtests.com/provider/brca-mutation-prevalence.htm on 26 July 2007. [Google Scholar]
- 67.Euhus DM. Understanding mathematical models for breast cancer risk assessment and counseling. Breast J. 2001;7:224–232. doi: 10.1046/j.1524-4741.2001.20012.x. PMID: 11678799. [DOI] [PubMed] [Google Scholar]