Abstract
Cellular morphogenesis is a complex process and molecular studies in the last few decades have amassed a large amount of information that is difficult to grasp in any completeness. Fungal systems, in particular the budding and fission yeasts, have been important players in unravelling the basic structural and regulatory elements involved in a wide array of cellular processes. In this article, we address the design principles underlying the various processes of yeast and fungal morphogenesis. We attempt to explain the apparent molecular complexity from the perspective of the evolutionary theory of “facilitated variation”. Following a summary of some of the most studied morphogenetic phenomena, we discuss, using recent examples, the underlying core processes and their associated “weak” regulatory linkages that bring about variation in morphogenetic phenotypes.
Keywords: yeast, fungi, morphogenesis, cell polarity, facilitated variation
Introduction
Fungi exist in nature in a rich variety of shapes and forms suitable to their distinctive life styles. How these relatively simple organisms develop diverse morphologies at the single cell level is of significant interest to both cell and evolutionary biologists. Powerful molecular genetics techniques in budding and fission yeasts have brought forth an era where most of the genes and the pathways they constitute are known, but the emerging molecular networks exhibit a daunting complexity, and the logic and design principles are not apparent. As all living systems are built through an evolutionary process, the ability to evolve must be a fundamental design principle. Thus, a useful perspective for understanding complex cellular systems is the relationship between the molecular network architecture and the ability to generate rapid phenotypic variation.
The recent “Theory of Facilitated Variation” [1] proposes that variation in form and function is brought about through variation in the regulatory linkages that impinge upon highly conserved core processes. The premise of this theory is that the core processes of biological systems are built from conserved core elements in a highly adaptable way, which enables “weak regulatory linkages” to release their output in response to diverse signal input. The term “weak” refers to those regulatory connections that are not directly involved in output generation and can thus be changed with little constraints in the course of evolution under different selective conditions. In this review, we intend to use this theory as a guiding principle to analyse and interpret some of the recent data from the study of yeast and fungal morphogenesis. We focus on the budding yeast Saccharomyces cerevisae, as this model organism has provided the majority of the basic insights into the establishment of cell polarity – a core process required for all morphogenetic processes. However, due to the limited range of morphogenetic features in budding yeast, we will broaden the scope to include other types of fungi where new molecular details of cellular morphogenesis have been unravelled in recent years. Our purpose is not to provide a comprehensive review on all the detailed molecular interactions, which have been expertly covered in a recent article [2]. Moreover, due to the large volume of the literature touched upon, we apologize for often citing review articles where primary citations can be found instead of the primary articles themselves.
Fungal morphogenesis – an overview
The kingdom of fungi is visible to cell biologists mostly through groundbreaking discoveries in the model organisms S. cerevisiae and Schizosaccharomyces pombe (fission yeast). It is rarely appreciated that fungi constitute a large group of organisms that diverged from animals around one billion years ago and encompass a vast range of morphological forms [3]. In addition to the aforementioned budding and fission yeasts there are a number of other fungal models currently studied to address such topics as plant and animal pathogenicity, and antibiotics biosynthesis [4]. Furthermore, a growing number of studies address basic cell biological questions in diverse fungal species. These studies exploit the powerful genetic and molecular manipulations possible in fungi and often uncover fascinating variations on common themes as well as novel regulatory mechanisms.
A defining property of fungi is that they are surrounded by a stable cell wall made up of proteins, glucans and chitin [5]. The presence of a cell wall puts significant constraint on the way fungal cells grow but also provide a rigid stabilizing scaffold to the cell shape that forms through polarized growth [6,7]. The two major modes of growth in fungi are filamentous growth, such as formation of hyphae, and yeast-like growth [8]. Hyphae are typical for most fungi and are characterized by continuous tip growth leading to the formation of an elongated tube consisting of a chain of single nucleate cells divided by septa. In some fungi, the cells along the hyphae communicate through septal pores that result from incomplete cytokinesis [9,10]. The yeast forms are characterized by discrete cells that divide either by budding or by fission where daughter cells dissociate from the mother cell after division. Many human or plant pathogenic fungi, such as Candida albicans or the corn smut fungus Ustilago maydis, are capable of a “dimorphic switch” between yeast and hyphal growth forms, but the filamentous form is most often associated with pathogenicity [11]. A distinct growth mode in filamentous fungi is the simultaneous maintenance of more than one axis of polarized growth during sub-apical branching or tip splitting [12]. In addition there are several more specialized morphological states in fungi such as pseudohyphae, various types of spores and mating projections, which develop in response to well defined environmental cues [13]. Even more specialized morphologies are adopted during interaction of pathogenic fungi with their host plants or animals. For example, infection of corn plants by U. maydis occurs through a single dikaryotic, highly elongated cell. This cell results after two compatible haploid yeast-like sporidia form mating protrusions and fuse with each other [14]. Thus, fungal species are a rich platform for studying the development of forms and shapes in response to physiological cues.
The budding yeast S. cerevisiae is naturally found in soil or the bark of trees [15] but has probably adopted new hosts such as grapes or rice as a result of domestication and its use in the production of wine and sake [16]. Nearly all studies of budding yeast are limited to laboratory strains that have lost morphogenetic features important in the natural environment. In particular, naturally occurring S. cerevisae colonies exhibit a structured pattern that seems related to an extracellular matrix material and is rapidly lost under laboratory conditions [17]. Despite their smooth colony morphology in the lab, budding yeast cells are also capable of forming biofilms [18] and multicellular stalk-like structures that might be linked to spore dissemination [19,20]. Budding yeast can stably exist as either haploid or diploid cells, which, under favourable growth conditions, divide by budding. In this process a daughter cell (bud) forms through polarized growth from a particular site on the mother cell surface and remains connected to the mother cell through the bud neck that is stabilized by a ring of chitin in the cell wall [21]. After nuclear division, cytokinesis occurs through constriction of an actomyosin ring at the bud neck coupled with formation of the primary septum catalyzed by chitin synthase II [22,23]. Cell separation is accomplished through a cohort of cell wall remodelling enzymes [24,25]. The position of an incipient bud site on the cortex is specified through a process termed “bud site selection” and depends on the previous sites of cell division known as bud scars (on mother cells) or birth scars (on daughter cells) [26]. Budding is also temporally controlled by the cell cycle: buds are initiated at the G1-S transition upon establishment of growth polarity [27]. The bud continues to enlarge through polarized growth during S and G2 phases, and cytokinesis occurs subsequent to mitotic exit [23].
Haploid yeast cells can be either of a or the α mating type. If cells of opposing mating types are in sufficient proximity to each other, a distinct morphogenetic program is engaged through pheromone signalling [28]. Both cells form shmoos, which are mating projections that grow in a direction guided by the pheromone gradient elicited by the opposing mating cell. Cell fusion occurs at the shmoo tip, followed by nuclear fusion (karyogamy). The direction of shmoo formation is guided through the pheromone gradient elicited by the opposing mating cell. Diploid S. cerevisiae cells of the a/α type cannot form shmoos but undergo another growth form, termed pseudohyphal growth, when the cells are starved for nitrogen [29]. Pseudohyphae are chains of slightly elongated cells that have separated cytoplasms but are connected through their cell walls. This growth mode enables the cells to cover long distances and is thought to aid in the search for an improved nutritional environment.
The fission yeast S. pombe cells have a regular, rod-shaped morphology. In contrast to budding yeast, they do not form buds but grow by polarized tip elongation, first at the old end and then also at the new end that resulted from the previous cell division [30]. Cells divide in the middle through membrane constriction driven by an actomyosin-based contractile ring, followed by septum formation and cell separation [31]. There has been considerable work on the regulation of polarized growth and cell division in S. pombe. One major difference relative to S. cerevisiae is the prominent role for microtubules and microtubule-associated motors in the determination of cell shape.
The conserved core process in the generation of cell polarity
Whereas many studies are focused on specific morphogenetic processes in yeast or fungi, it may be difficult to understand how regulatory linkages affect specific morphogenetic outcomes without insights into the core processes upon which regulatory linkages exert their effects. A hallmark of the core processes is the highly conserved structural or regulatory elements that are shared not just among fungal species but also among other eukaryotic kingdoms such as animals and plants. However, core processes are also adaptable, enabling their own regulation by a variety of signals. Whether in budding or hyphal growth, the origin of fungal morphogenesis, as in most morphogenetic processes, is the establishment of cell polarity. This core process is constructed through several conserved core elements, as first discussed below, and enables cell surface growth to be localized in defined patterns.
Rho family GTPases
The central regulator of cell polarity in S. cerevisiae –and nearly all other eukaryotic organisms studied so far– is a small GTPase of the Rho family called Cdc42 [32]. Rho GTPases hydrolyze GTP and cycle between two states, either bound to GDP (“off” state), or to GTP (“on” state). The latter enables interaction with downstream effector molecules. This GTPase cycle is slow on its own, but can be drastically accelerated by two types of enzymes: the guanine nucleotide exchange factors (GEF), which assists the release of GDP and the GTPase activating proteins (GAP), which stimulate the rate of GTP hydrolysis. Rho GTPases are anchored into the membrane through C-terminal prenylation but can also exist in a soluble cytosolic form due to binding to GDP dissociation inhibitors (GDI), which shield the hydrophobic prenyl group and at the same time block access of GEFs and GAPs. This way, Rho GTPases are also capable of cycling between membrane and cytosolic compartments. While Cdc42 is essential for cell polarization in S. cerevisiae, in other fungi such as U. maydis [33] Cdc42 has a less prominent role, partly due to other Rho type GTPases taking over Cdc42's functions, in particular in the organization of the actin cytoskeleton.
Cytoskeletal polymers
While the GTPase module is central to the control of cell polarity, structures containing polar cytoskeletal elements, actin filaments and microtubules, are almost always the work horse for this process. In budding yeast, only the actin cytoskeleton is involved in cell polarity, while microtubules are mainly involved in chromosome segregation and nuclear migration. S. cerevisiae have three types of F-actin structures [34]: 1) actin patches, which contain a dendritic network of actin filaments nucleated by the Arp2/3 complex and are the major sites of endocytosis; 2) actin cables, made up of parallel bundles of short actin filaments nucleated by formin family proteins, which are directly activated by Rho family GTPases, and responsible for intracellular transport of organelles and RNAs through type V myosin motors; and 3) the actomyosin ring, which contains formin-nucleated actin filament and type II myosin and is involved in cytokinesis. These three types of structures are comprised largely of distinct sets of actin-associated proteins but are regulated in a coordinated fashion. The Rho GTPases are known to exert control over the assembly of all three types of actin structures; however, the molecular mechanisms underlying this control remain poorly understood and are still an area of active research.
In contrast to the situation in budding yeast, microtubules play an important role in polarized growth and cell shape determination in the fission yeast S. pombe and in many filamentous or dimorphic fungi, such as U. maydis. These fungal species possess the same actin structures as budding yeast and these structures are equally important for polarized growth. However, in the case of fission yeast, microtubules exert control over the location of actin assembly, thus dictating the pattern of growth and cell shape [35], whereas in U. maydis microtubules have taken over part of actin's role in vesicular transport [36]. In fission yeast, anti-parallel interphase microtubule arrays originating from the interphase microtubule organizing centres at the cell centre, deliver factors, such as the Kelch-repeat-containing protein Tea1 and its interacting proteins, which restrict actin cable nucleating proteins to the growing cell cortex [30]. In order to ensure growth only at cell ends, the organization and dynamics of microtubules must be precisely controlled. Recent work suggests that the antiparallel microtubule arrays are established through coordinated actions of a kinesin family microtubule motor Klp2 and a conserved microtubules bundling protein Ase1 [30,37]. Microtubules grow continuously until the plus ends reach the polar cortex (but not the lateral cortex) where depolymerization is triggered. This pattern of microtubule dynamics requires several highly conserved microtubule plus end-tracking proteins (Tip1/CLIP170, Mal3/EB1), a kinesin-like motor protein (Tea2), as well as a nuclear rim protein Amo1 [30].
In U. maydis, while polarized growth still happens in the absence of microtubules or microtubule dependent motors, the remaining actin cytoskeleton can only support growth, at a reduced rate, up to 40-60 μm compared to 100-150 μm long wildtype hyphae [36]. The role of microtubules in efficient polarized growth may be three fold. First, like in all other studied filamentous fungi, nuclei in U. maydis are exclusively transported via microtubules and they fail to enter the hyphae in the absence of microtubules [38]. Therefore, if any signal is generated from the nucleus to sustain tip growth, nuclear positioning in the hyphae would be required for fast signal transmission. Second, it was shown recently that several RNA binding proteins are transported along microtubules in U. maydis and some of these seem to be involved in polarized growth [39,40]. Finally, endosomes are transported bidirectionally along microtubules, which is required for efficient recycling and the generation of fast polar growth [41,42]. Interestingly, depletion of a type V myosin (essential in budding yeast) also only reduces hyphal growth rates, and only the combined disruption of actin- and microtubule dependent transport completely blocks growth [43]. This is an example of multiple pathways contributing to a similar goal, where individual pathways might be non-essential but their collaboration results in a higher efficiency, robustness or fine-tuning of the outcome.
In addition to actin and microtubules, the septins are a group of conserved GTP-binding proteins that assemble into filaments and play important roles in morphogenesis, cytokinesis and membrane trafficking [44,45]. In budding yeast the septins form a ring around the bud neck and define the typical morphology of a budding yeast cell [46]. The septins also serve as a diffusion barrier between the bud and mother cortex, which in turn is required for the asymmetric distribution of several proteins involved in mitotic exit and cell polarization [47,48]. Interestingly, a recent paper reported that the septins are localized at the bases of “bud-like” dendritic spines in hippocampal neurons and are important for dendritic spine development [49]. This finding suggests that septins' role in morphogenesis may be conserved between fungi and animal cells. In S. pombe, septins are not essential for cell division but are required for efficient cell separation. Interestingly, haploid U. maydis cells, which are elongated but divide by budding, switch to a fission-like division process upon deletion of the septin Sep3 [50]. The septins also seem to have important roles in hyphal growth [51] and the localization of the septin ring can be used as morphological distinction between hyphae and pseudohyphae [52].
Exocytosis
Cell surface growth ultimately requires addition of new surface materials. Exocytosis is the process of delivering newly synthesized or recycled components to the plasma membrane or the extracellular milieu, and as such, encompasses many different steps, from trafficking of exocytic vesicles through the ER and Golgi to cytoskeleton-based transport to the sites of growth, and finally, fusion with the plasma membrane. This discussion emphasizes the final vesicle docking and fusion step once delivery via the actin cytoskeleton has been accomplished. Six subunits of the so-called exocyst complex are preassembled on secretory vesicles that contain the Rab GTPase Sec4 [53]. Other exocyst components, Sec3 and Exo70, are localized at the target site possibly via direct interaction with Rho GTPases [54,55], although the extent to which this localization contributes to polarized secretion is still being debated [56]. Completion of exocyst formation could thus tether secretory vesicles at the plasma membrane to enable fusion. There appears to be some separation of duty between Sec3 and Exo70, where the latter is required for secretion of a specific class of secretory vesicles that are important early steps in bud formation [57], whereas Sec3 has an important role in inheritance of the cortical ER in the bud [55]. In the vesicle fusion reaction Sec4 facilitates the formation of a SNARE-complex between the v-SNAREs Snc1 and Snc2 and the t-SNAREs Sso1, Sso2 and Sec9 [58]. Recently, Sro7 and Sro77, members of the conserved Lethal Giant Larvae (Lgl) family, were shown to play an important role in SNARE regulation. Sro7 and Sro77 interact with the exocyst and Sec4-GTP, as well as the t-SNARE Sec9 [58-60]. A new structure study suggests that Sro7 allosterically regulates the formation of the SNARE complex through its interaction with Sec9 [61].
Many filamentous fungi contain an intriguing structure in their growing hyphal tips called the “Spitzenkörper” (spk) [62]. This structure has been proposed to function as a vesicle supply centre that supports sustained and rapid polar growth at the apex of the highly elongated hyphal tip cell [63]. It has been estimated that in fast growing hyphae of N. crassa up to 38000 vesicles per minute fuse with the apical area of the plasma membrane [64]. As this study did not take recycling processes into account this may even be an underestimation [65]. Markers localized to the spk in C. albicans include the endocytic dye FM4-64, the myosin light chain Mlc1 and the formin Bni1 [66]. The integrity of the spk is dependent on the actin cytoskeleton and on several proteins that interact with Bni1 [66]. One possible interpretation of the spk is simply as the terminal site of secretory vesicle transport. Vesicles might accumulate there because they get delivered faster than they can fuse with their target sites in the hyphal tip. This would be consistent with observations that the spk is most visible in hyphae that grow rapidly (and where more vesicles get delivered to the tip) and is often absent from slow or non-growing hyphae [62].
The core process in the establishment of cell polarity
From an evolutionary perspective, an intrinsic core process of symmetry breaking that allows polarity establishment, and is not completely dependent on external or developmental signals, can be highly advantageous for diverse morphogenetic purposes. From the standpoint of fungi, the most crucial and perhaps ancient function is vegetative proliferation through either bud formation or hyphal elongation. Additional morphogenetic patterns, such as bud site selection, chemotactic shmoo formation or pseudohyphal formation, are likely to be related to sexual reproduction or nutrient scavenging and might have evolved under different selective conditions through the emergence of “weak regulatory linkages” that could bias the existing core process.
Work in S. cerevisiae during the past several years has indeed revealed at least two intrinsic mechanisms for cell polarization without recourse to external signals [67-69] (Figure 1). One of these mechanisms is constituted by cyclic interactions between three core elements: the Cdc42 GTPase module, the actin-based transport system and exocytosis. Experimental evidence and mathematical modelling implicate positive feedback between these elements. Thus, Cdc42 recruits the actin cytoskeleton and is itself delivered to the membrane via actin dependent transport and exocytosis, and this is sufficient to drive spontaneous cell polarity establishment [68]. Recent work demonstrates that the resulting polarized state is dynamic and its maintenance requires active recycling through processes such as endocytosis [70]. The second mechanism for polarization does not require actin but appears to simply involve the Cdc42 GTPase module and a protein called Bem1 [67,69]. Bem1 homologs have been found in many fungal species but not yet in animal organisms. However, Bem1 appears to share several domains with proteins in animals that are directly regulated by Rho family GTPases, such as p47Phox [71] and the conserved polarity protein Par6 [72]. Precisely how Bem1 functions in cell polarization remains an open question, but existing data suggest that Bem1 interacts with Cdc42GTP, the Cdc42 GEF Cdc24 and the Cdc42 effectors Cla4 and Ste20 [73,74] and may mediate a feedback loop linking Cdc42 activation and GEF activation. During bud formation, the two polarization mechanisms described above are partially redundant, but their coupling brings both temporal precision and stability to polarity establishment [69,75]. Other morphogenetic processes may exhibit different levels of emphasis on each of these polarization mechanisms. It is interesting that a recent model capturing the characteristics of S. pombe morphogenesis relies on an assumption that microtubules exert their effect on polarized growth through delivery of a hypothetical factor that activates an actin assembly feedback loop, not just actin nucleation [76].
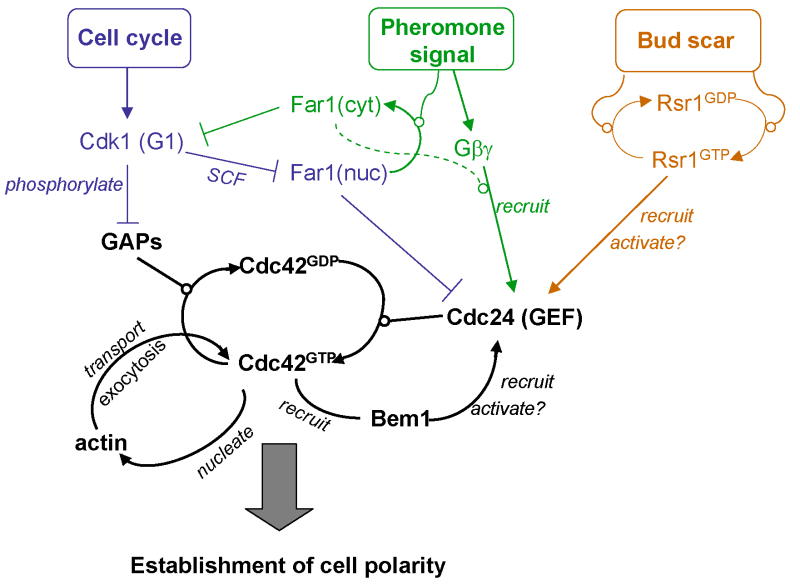
Figure 1.
Regulatory linkages exert spatial and temporary control over the core process for cell polarization. Black letters and thick line represent the core process; colored letters and thin lines denote the upstream regulatory events responding to cell cycle (blue), pheromone (green) and bud scar (brown) signals.
Regulatory linkages affecting the Cdc42 module
The ability to generate phenotypic variation through modification of core processes requires that regulatory linkages can be easily attached to the various elements that constitute the core processes. The Rho GTPase modules are well suited to be the malleable core elements that accommodate a wide array of morphogenetic signals. Rho GTPases are not autonomous switches but are tightly regulated by GEFs, GAPs and GDIs. This separation of GTPase regulation into different components could be regarded as an extreme form of allostery. It allows unconstrained evolution of the different elements in the module and increases the range of possible regulatory interactions. Consequently, a multitude of signals are transmitted to Rho GTPases via GEFs and GAPs (relatively little is known about the function of GDIs) while few act directly on the GTPases themselves.
The Cdc42 module is the best understood example of GTPase-mediated regulation of distinct morphogenetic signals (Figure 1). As the only GEF for Cdc42 in S. cerevisiae, Cdc24 is not only essential for cell polarization but also a target of temporal and spatial regulation by a variety of signals. One level of control that differentiates budding versus mating response occurs through differential regulation of nuclear sequestration of Cdc24 [77]. During M and early G1 phase Cdc24 is retained in the nucleus via interaction with Far1, an inhibitor of the yeast Cdk1 Cdc28. At the G1-S transition Cdk1 triggers the degradation of nuclear Far1 through SCF-mediated ubiquitination, and consequently, the release of Cdc24 from the nucleus and its subsequent localization to the presumptive bud site. The same mechanism of sequestration, with a different twist, is used in pheromone response, where pheromone signalling leads to Far1 export from the nucleus. Cytoplasmic Far1 is stable and inhibits Cdk1 to cause cell cycle arrest.
Recruitment of Cdc24 to the site of pheromone receptor activation through complex formation with Far1 and subunits of the heterotrimeric G protein serves the important function of orienting the direction of shmoo formation toward the pheromone gradient [78-80]. During budding, Cdc24 is also a target of regulation dictating the orientation of bud formation relative to the previous bud scar (Figure 1). Cdc24 is activated and recruited to the plasma membrane partly through interaction with the GTP-bound Ras type GTPase Rsr1 [81-83]. Involvement of GTPase cascades is a recurring theme in spatial organization. Another example in yeast is the Rab GTPase cascade that regulates exocytosis [84,85]. As with the Rsr1GTP-Cdc24 interaction the connection between the different GTPase modules occurs mainly between the GTPase of one module and a regulator (GAP or GEF) of the other module [84]. Interestingly, Rsr1GTP also directly binds Cdc42 [86], which might further increase the efficiency of local Cdc42 activation through Cdc24. Importantly, the function of Rsr1 in activation of the Cdc42 module requires its GTPase cycle, which is in turn regulated through the GEF Bud5 and GAP Bud2 [87]. Rapid cycling may enable multiple binding and release events, allowing the GTPase to act catalytically to promote the downstream reactions. Bud5 and Bud2 are localized in turn through interactions with proteins associated with the bud scars. Thus the Rsr1/Bud5/ Bud2 GTPase module biases Cdc42-driven polarization toward the cortical landmark (bud scar). Consistent with the concept of “weak regulatory linkages”, Rsr1 is not essential for Cdc24 activation or cortical recruitment and may be partially redundant with Bem1 in these activities [73,88].
Whereas budding yeast only has a single GEF for Cdc42, three GAPs have been identified so far as Rga1, Rga2 and Bem3. Another GAP, Bem2, shows GAP activity towards Rho1 instead of Cdc42 in vitro [89]. However, genetic analysis of cell polarity indicates that Bem2 might still act on Cdc42 in living cells without relying on its GAP activity [90,91]. Genetic studies showed that the Cdc42 GAPs have overlapping but non-identical functions. Recent work from several labs suggests that the GAPs are important targets for cell cycle kinases to control the timing of polarization (Figure 1). Two proteomics searches for proteins associated with specific Cdc28-cyclin complexes identified Rga1 and Bem3 as targets of Cdc28 in complex with G1 cyclin Gln2 [92,93]. Three recent reports involving S. cerevisae and one involving C. albicans provided evidence that the GAPs are phosphorylated by Cdc28 and Pho85 Cdks in late G1 [90,94-96]. Interestingly, overexpression of a GAP-deficient allele of Bem3 acts in a dominant-negative fashion and induces spontaneous polarization of G1 cells in a manner similar to constitutively active Cdc42 [90]. This suggests that Cdc42's intrinsic nucleotide exchange is sufficient to achieve a threshold level of Cdc42GTP once the GAPs are inactivated. A non-phosphorylatable mutant of Rga2 acts in a dominant-active fashion and inhibits cell polarization upon overexpression [95]. The same holds true for the respective Bem3 mutants even for low levels of expression [90]. These results provide compelling evidence that Cdk1 activation at G1-S induces polarization at least in part through inhibition of Cdc42 GAP.
Regulatory linkages downstream of Cdc42
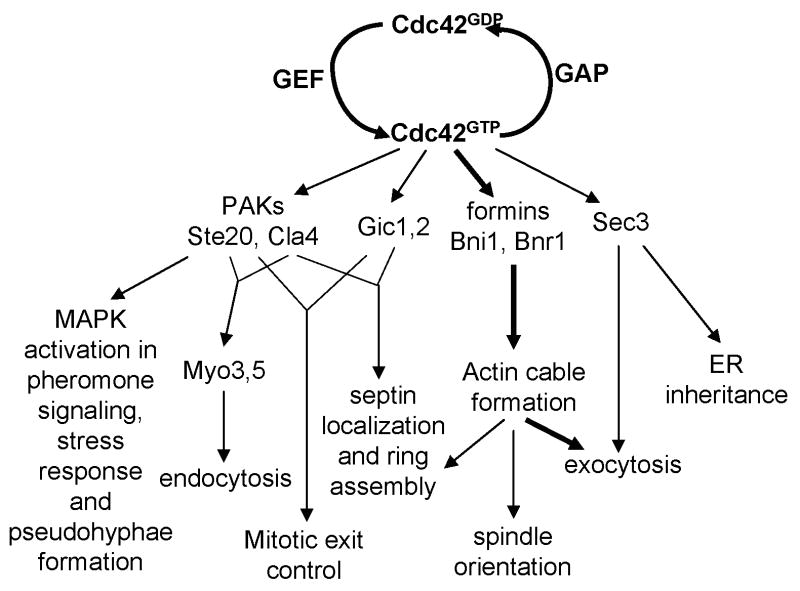
Variation in morphogenetic responses may also be accomplished through diverse and often redundant Cdc42 effectors (Figure 2). A conserved family of Cdc42 effectors are the p21 activated kinases (PAK), including Cla4, Ste20 and Skm1 [97]. These kinases, again, are partially redundant, but many results point to both shared and divergent roles of these kinases in budding yeast morphogenesis. Ste20 and Cla4 have been shown to phosphorylate and activate type I myosins Myo3 and Myo5 [98], which in turn have a role in the internalization step of endocytosis via actin patches [99,100]. Cla4 is also the major kinase that phosphorylates the GEF Cdc24, although the exact role for this phosphorylation is still controversial [74,101]. Cla4 plays a critical role in the assembly of the septin ring at the bud neck [102]. Whereas Cla4's function is more important during vegetative growth, Ste20 is essential during pheromone signalling, stress response and pseudohyphal growth [97,103]. The major role for Ste20 during these processes is to transmit signal input at the plasma membrane to the MAP kinase cascades that leads to transcriptional activation of the downstream morphogenetic programs. Whereas Ste20 has its own set of interacting partners promoting these processes, Cla4 appears to have a negative effect on the pheromone response [104], further indicating functional divergence of these homologous kinases.
Figure 2.
Regulatory linkages downstream of Cdc42 GTPase cycle elicit a multitude of events in signaling and structure assembly required for various morphogenetic processes. The events that lead to actin cable formation and exocytosis, denoted with thick lines, are part of the core process in the establishment of cell polarity.
Cdc42 interacts with a pair of yeast-specific effectors, Gic1 and Gic2, which bind active Cdc42 through the conserved CRIB domain [105,106]. These homologous proteins are largely redundant and are important for polarized actin assembly and the recruitment and assembly of the septin ring at elevated temperatures [107]. Both proteins interact specifically with the Cdc12 septin. Interestingly, Borg3, a Cdc42 effector in mammalian cells that bears no similarity to the Gic proteins, also regulates septin assembly [108,109]. Although the exact mechanism is unclear in either process, this might be another example of flexible regulatory linkages between conserved core elements to implement specific morphogenetic responses. In addition to the role in bud formation, the Gic proteins, as well as the PAKs, also play a role in mitotic exit control, which senses spindle orientation relative to the axis of polarity [110-112].
While S. cerevisiae and S. pombe only express the Rho and Cdc42 classes of Rho GTPases most filamentous fungi also express the third type, Rac1. Interestingly, in one of these fungi, U. maydis, Cdc42 is not essential but limited to a function in septum formation and cell separation [33]. Instead, the Rac1 homologue is required for proper bud formation and cells without Rac1 divide by fission instead [33]. Rac1 overexpression, on the other hand, results in hypha formation suggesting that Rac1 specifically induces downstream events required for filamentous growth [33]. Fitting the concept of flexible linkages it seems that Rac1 in U. maydis partially acts through some of the same effectors used by Cdc42 in S. cerevisiae, namely Cla4 [113].
Scaffolds as potential flexible linkages
The use of protein scaffolds to link regulators within defined signalling pathways or at particular locations in the cell is a frequently inferred mechanism to explain the role of protein complex formation [114,115]. However, the term scaffold has not been used in a consistent manner and can imply distinct mechanistic scenarios. In processes that emphasize spatial localization, scaffold often implies more static structures that serve to restrict mobility of interacting proteins. For example, the S. cerevisiae bud scars are scaffolds that immobilize associated proteins, such as Rax2, to form a cortical landmark. Rax2 remains immobilized for several consecutive cell divisions and can be used to trace the history of a cell [116]. Similarly, eisosomes are immobile structures in the yeast plasma membrane that exhibit no lateral motion over time spans of more than 30 minutes and could therefore act as scaffolds for other proteins [117]. Static scaffolds may be large multiprotein complexes or macromolecular assemblies that are not subject to the effects of Brownian diffusion.
In contrast, most proposed scaffold proteins implicated in signal transduction or during cell polarization are dynamic. Through multiple protein interaction domains, these proteins may facilitate transient protein complex formation but are unlikely to act as static reference site for the localization of the bound proteins. One example is Bem1, which has binding sites for a number of proteins involved in cell polarization, including Cdc42GTP, Cdc24, Cla4 and Boi1,2 proteins [67,118]. Whereas Bem1 may engage in hetero-oligomeric interactions, it has been shown to rapidly cycle on and off the plasma membrane and is therefore unlikely to provide any significant anchorage for its binding partners [69]. Similarly the MAP kinase scaffold protein Ste5 has been shown to dynamically interact with the shmoo tip as well as the MAP kinase cascade proteins [119]. In both of these examples, significant interactions also exist among the binding partners of the scaffold proteins. Thus, it may be reasonable to speculate that the function of such dynamic scaffolds is to promote cooperative protein complex assembly, enabling weak interacting partners to form strong and possibly self-enhancing functional units under a variety of conditions. Through such effects, dynamic scaffolds can be particularly versatile linkages to generate pathway diversity.
Bem1 is potentially such an example: whereas during vegetative growth it links Cla4 to active Cdc42 and the GEF Cdc24 to promote bud formation, during pheromone response Bem1 binds Ste20 through the same domain and this interaction is required for optimal signalling through the MAP kinase cascade [120]. Dynamic scaffolding could also account for assembly of polarity complexes on the cortex without the involvement of transmembrane domains. Cdc42 and other Rho proteins are reversibly linked to the membrane via their prenylated C-termini. A number of polar cortical components contain weak membrane binding motifs, including Cdc24, Cla4, Bem1, and Bem1-interacting proteins Boi1 and Boi2, GAP proteins for Cdc42 and Rho and the exocyst component Exo84. Dynamic scaffolding to allow assembly of protein complex on the membrane through multiple weak interactions may provide a flexible means for polarity regulation and adaptation to changing conditions. A similar mechanism has been proposed for Ste5 in MAPK signalling [121]. Studies of dynamic scaffolds in other fungal morphogenetic systems could lend further insights into their role as flexible linkages in the evolution of morphological diversity.
Acknowledgments
The authors thank Michael Sixt and Susanne Wedlich for critical reading of the manuscript. This work was supported by the Max-Planck Society (RWS) and NIH RO1-057063 (RL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gerhart J, Kirschner M. The theory of facilitated variation. Proc Natl Acad Sci U S A. 2007;104 1:8582–9. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–22. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 4.Hamer L, Tanzer M. Fungal role models: A bouquet of foes and friends. Fifth European conference on fungal genetics; Arachon, France. March 25-29, 2000; [DOI] [PubMed] [Google Scholar]; Fungal Genet Biol. 2000;30:163–5. doi: 10.1006/fgbi.2000.1220. [DOI] [PubMed] [Google Scholar]
- 5.Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 6.Harold FM. To shape a cell: and inquiry into the causes of morphogenesis of microorganisms. Microbiol Rev. 1990;54:381–431. doi: 10.1128/mr.54.4.381-431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slaughter B, Li R. Toward a molecular interpretation of the surface stress theory for yeast morphogenesis. Curr Opin Cell Biol. 2006;18:47–53. doi: 10.1016/j.ceb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–24. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 9.van Driel KG, Boekhout T, Wosten HA, Verkleij AJ, Muller WH. Laser microdissection of fungal septa as visualised by scanning electron microscopy. Fungal Genet Biol. 2007;44:466–73. doi: 10.1016/j.fgb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Shepherd VA, Orlovich DA, Ashford AE. Cell-to-cell transport via motile tubules in growing hyphae of a fungus. J Cell Sci. 1993;105(Pt 4):1173–8. doi: 10.1242/jcs.105.4.1173. [DOI] [PubMed] [Google Scholar]
- 11.Klosterman SJ, Perlin MH, Garcia-Pedrajas M, Covert SF, Gold SE. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv Genet. 2007;57:1–47. doi: 10.1016/S0065-2660(06)57001-4. [DOI] [PubMed] [Google Scholar]
- 12.Momany M. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr Opin Microbiol. 2002;5:580–5. doi: 10.1016/s1369-5274(02)00368-5. [DOI] [PubMed] [Google Scholar]
- 13.Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007 doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldbrugge M, Kamper J, Steinberg G, Kahmann R. Regulation of mating and pathogenic development in Ustilago maydis. Curr Opin Microbiol. 2004;7:666–72. doi: 10.1016/j.mib.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 16.Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuthan M, Devaux F, Janderova B, Slaninova I, Jacq C, Palkova Z. Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol Microbiol. 2003;47:745–54. doi: 10.1046/j.1365-2958.2003.03332.x. [DOI] [PubMed] [Google Scholar]
- 18.Reynolds TB, Fink GR. Bakers' yeast, a model for fungal biofilm formation. Science. 2001;291:878–81. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 19.Engelberg D, Mimran A, Martinetto H, Otto J, Simchen G, Karin M, et al. Multicellular stalk-like structures in Saccharomyces cerevisiae. J Bacteriol. 1998;180:3992–6. doi: 10.1128/jb.180.15.3992-3996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherz R, Shinder V, Engelberg D. Anatomical analysis of Saccharomyces cerevisiae stalk-like structures reveals spatial organization and cell specialization. J Bacteriol. 2001;183:5402–13. doi: 10.1128/JB.183.18.5402-5413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J Biol Chem. 2001;276:19679–82. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- 22.Silverman SJ, Sburlati A, Slater ML, Cabib E. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988;85:4735–9. doi: 10.1073/pnas.85.13.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolliday N, Bouquin N, Li R. Assembly and regulation of the cytokinetic apparatus in budding yeast. Curr Opin Microbiol. 2001;4:690–5. doi: 10.1016/s1369-5274(01)00270-3. [DOI] [PubMed] [Google Scholar]
- 24.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–50. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 25.Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–86. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- 27.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–20. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorner J. Pheromonal Regulation of Development in Saccharomyces cerevisiae. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1981. pp. 143–80. [Google Scholar]
- 29.Gancedo JM. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:107–23. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 30.La Carbona S, Le Goff C, Le Goff X. Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex. Biol Cell. 2006;98:619–31. doi: 10.1042/BC20060048. [DOI] [PubMed] [Google Scholar]
- 31.Feierbach B, Chang F. Cytokinesis and the contractile ring in fission yeast. Curr Opin Microbiol. 2001;4:713–9. doi: 10.1016/s1369-5274(01)00273-9. [DOI] [PubMed] [Google Scholar]
- 32.Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bolker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2006;59:567–78. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- 34.Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–45. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang F, Feierbach B, Martin S. Regulation of actin assembly by microtubules in fission yeast cell polarity. Novartis Found Symp. 2005;269:59–66. discussion -72, 223-30. [PubMed] [Google Scholar]
- 36.Steinberg G. Tracks for traffic: microtubules in the plant pathogen Ustilago maydis. New Phytol. 2007;174:721–33. doi: 10.1111/j.1469-8137.2007.02072.x. [DOI] [PubMed] [Google Scholar]
- 37.Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, et al. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–68. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 38.Fuchs U, Manns I, Steinberg G. Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol Biol Cell. 2005;16:2746–58. doi: 10.1091/mbc.E05-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarnack K, Feldbrugge M. mRNA trafficking in fungi. Mol Genet Genomics. 2007;278:347–59. doi: 10.1007/s00438-007-0271-8. [DOI] [PubMed] [Google Scholar]
- 40.Becht P, Konig J, Feldbrugge M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci. 2006;119:4964–73. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- 41.Wedlich-Soldner R, Bolker M, Kahmann R, Steinberg G. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 2000;19:1974–86. doi: 10.1093/emboj/19.9.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wedlich-Soldner R, Straube A, Friedrich MW, Steinberg G. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 2002;21:2946–57. doi: 10.1093/emboj/cdf296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuchardt I, Assmann D, Thines E, Schuberth C, Steinberg G. Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol Biol Cell. 2005;16:5191–201. doi: 10.1091/mbc.E05-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindsey R, Momany M. Septin localization across kingdoms: three themes with variations. Curr Opin Microbiol. 2006;9:559–65. doi: 10.1016/j.mib.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Kinoshita M. Diversity of septin scaffolds. Curr Opin Cell Biol. 2006;18:54–60. doi: 10.1016/j.ceb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Longtine MS, Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–9. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 47.Faty M, Fink M, Barral Y. Septins: a ring to part mother and daughter. Curr Genet. 2002;41:123–31. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- 48.Castillon GA, Adames NR, Rosello CH, Seidel HS, Longtine MS, Cooper JA, et al. Septins have a dual role in controlling mitotic exit in budding yeast. Curr Biol. 2003;13:654–8. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 49.Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, Sheng M. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–8. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyce KJ, Chang H, D'Souza CA, Kronstad JW. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize. Eukaryot Cell. 2005;4:2044–56. doi: 10.1128/EC.4.12.2044-2056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13:2732–46. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berman J, Sudbery PE. Candida Albicans: a molecular revolution built on lessons from budding yeast. Nature reviews. 2002;3:918–30. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- 53.Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Zajac A, Zhang J, Wang P, Li M, Murray J, et al. The critical role of Exo84p in the organization and polarized localization of the exocyst complex. J Biol Chem. 2005;280:20356–64. doi: 10.1074/jbc.M500511200. [DOI] [PubMed] [Google Scholar]
- 55.Wiederkehr A, Du Y, Pypaert M, Ferro-Novick S, Novick P. Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Mol Biol Cell. 2003;14:4770–82. doi: 10.1091/mbc.E03-04-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roumanie O, Wu H, Molk JN, Rossi G, Bloom K, Brennwald P. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J Cell Biol. 2005;170:583–94. doi: 10.1083/jcb.200504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He B, Xi F, Zhang J, TerBush D, Zhang X, Guo W. Exo70p mediates the secretion of specific exocytic vesicles at early stages of the cell cycle for polarized cell growth. J Cell Biol. 2007;176:771–7. doi: 10.1083/jcb.200606134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novick P, Medkova M, Dong G, Hutagalung A, Reinisch K, Grosshans B. Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem Soc Trans. 2006;34:683–6. doi: 10.1042/BST0340683. [DOI] [PubMed] [Google Scholar]
- 59.Grosshans BL, Andreeva A, Gangar A, Niessen S, Yates JR, 3rd, Brennwald P, et al. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Wang P, Gangar A, Zhang J, Brennwald P, TerBush D, et al. Lethal giant larvae proteins interact with the exocyst complex and are involved in polarized exocytosis. J Cell Biol. 2005;170:273–83. doi: 10.1083/jcb.200502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hattendorf DA, Andreeva A, Gangar A, Brennwald PJ, Weis WI. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature. 2007;446:567–71. doi: 10.1038/nature05635. [DOI] [PubMed] [Google Scholar]
- 62.Virag A, Harris SD. The Spitzenkorper: a molecular perspective. Mycol Res. 2006;110:4–13. doi: 10.1016/j.mycres.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Bartnicki-Garcia S, Bartnicki DD, Gierz G, Lopez-Franco R, Bracker CE. Evidence that Spitzenkorper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp Mycol. 1995;19:153–9. doi: 10.1006/emyc.1995.1017. [DOI] [PubMed] [Google Scholar]
- 64.Collinge AJ, Trinci AP. Hyphal tips of wild-type and spreading colonial mutants of Neurospora crassa. Arch Microbiol. 1974;99:353–68. doi: 10.1007/BF00696249. [DOI] [PubMed] [Google Scholar]
- 65.Wessels JGH. International Review of Cytology. Academic Press; 1986. Cell wall synthesis in apical hyphal growth; pp. 37–79. [Google Scholar]
- 66.Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, et al. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118:2935–47. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- 67.Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–70. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- 68.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–5. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 69.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–22. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson JM, Ohira T, Badwey JA. Regulation of the NADPH-oxidase complex of phagocytic leukocytes. Recent insights from structural biology, molecular genetics, and microscopy. Histochem Cell Biol. 2004;122:293–304. doi: 10.1007/s00418-004-0672-2. [DOI] [PubMed] [Google Scholar]
- 72.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 73.Butty AC, Perrinjaquet N, Petit A, Jaquenoud M, Segall JE, Hofmann K, et al. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–76. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–86. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- 75.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–8. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Csikasz-Nagy A, Gyorffy B, Alt W, Tyson JJ, Novak B. Spatial controls for growth zone formation during the fission yeast cell cycle. Yeast. 2007 doi: 10.1002/yea.1571. [DOI] [PubMed] [Google Scholar]
- 77.O'Shea EK, Herskowitz I. The ins and outs of cell-polarity decisions. Nat Cell Biol. 2000;2:E39–41. doi: 10.1038/35004065. [DOI] [PubMed] [Google Scholar]
- 78.Nern A, Arkowitz RA. A GTP-exchange factor required for cell orientation. Nature. 1998;391:195–8. doi: 10.1038/34458. [DOI] [PubMed] [Google Scholar]
- 79.Nern A, Arkowitz RA. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J Cell Biol. 1999;144:1187–202. doi: 10.1083/jcb.144.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiget P, Shimada Y, Butty AC, Bi E, Peter M. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J. 2004;23:1063–74. doi: 10.1038/sj.emboj.7600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park HO, Bi E, Pringle JR, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci U S A. 1997;94:4463–8. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang PJ, Sanson A, Lee B, Park HO. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science. 2001;292:1376–8. doi: 10.1126/science.1060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–62. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ortiz D, Medkova M, Walch-Solimena C, Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol. 2002;157:1005–15. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang W, Ferro-Novick S. A Ypt32p exchange factor is a putative effector of Ypt1p. Mol Biol Cell. 2002;13:3336–43. doi: 10.1091/mbc.01-12-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kozminski KG, Beven L, Angerman E, Tong AH, Boone C, Park HO. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol Biol Cell. 2003;14:4958–70. doi: 10.1091/mbc.E03-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marston AL, Chen T, Yang MC, Belhumeur P, Chant J. A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr Biol. 2001;11:803–7. doi: 10.1016/s0960-9822(01)00230-5. [DOI] [PubMed] [Google Scholar]
- 88.Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–62. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng Y, Hart MJ, Shinjo K, Evans T, Bender A, Cerione RA. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J Biol Chem. 1993;268:24629–34. [PubMed] [Google Scholar]
- 90.Knaus M, Pelli-Gulli MP, van Drogen F, Springer S, Jaquenoud M, Peter M. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 2007;26:4501–13. doi: 10.1038/sj.emboj.7601873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marquitz AR, Harrison JC, Bose I, Zyla TR, McMillan JN, Lew DJ. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J. 2002;21:4012–25. doi: 10.1093/emboj/cdf416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 93.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 94.Zheng XD, Lee RT, Wang YM, Lin QS, Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26:3760–9. doi: 10.1038/sj.emboj.7601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sopko R, Huang D, Smith JC, Figeys D, Andrews BJ. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 2007;26:4487–500. doi: 10.1038/sj.emboj.7601847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McCusker D, Denison C, Anderson S, Egelhofer TA, Yates JR, 3rd, Gygi SP, et al. Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol. 2007;9:506–15. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- 97.Hofmann C, Shepelev M, Chernoff J. The genetics of Pak. J Cell Sci. 2004;117:4343–54. doi: 10.1242/jcs.01392. [DOI] [PubMed] [Google Scholar]
- 98.Wu C, Lytvyn V, Thomas DY, Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J Biol Chem. 1997;272:30623–6. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- 99.Jonsdottir GA, Li R. Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles. Curr Biol. 2004;14:1604–9. doi: 10.1016/j.cub.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 100.Toret CP, Drubin DG. The budding yeast endocytic pathway. J Cell Sci. 2006;119:4585–7. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- 101.Gulli MP, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol Cell. 2000;6:1155–67. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 102.Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–45. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leberer E, Thomas DY, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 104.Heinrich M, Kohler T, Mosch HU. Role of Cdc42-Cla4 interaction in the pheromone response of Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:317–27. doi: 10.1128/EC.00102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen GC, Kim YJ, Chan CS. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–71. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown JL, Jaquenoud M, Gulli MP, Chant J, Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–82. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, et al. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–25. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Joberty G, Perlungher RR, Sheffield PJ, Kinoshita M, Noda M, Haystead T, et al. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat Cell Biol. 2001;3:861–6. doi: 10.1038/ncb1001-861. [DOI] [PubMed] [Google Scholar]
- 109.Joberty G, Perlungher RR, Macara IG. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol Cell Biol. 1999;19:6585–97. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hofken T, Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;21:4851–62. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiroli E, Fraschini R, Beretta A, Tonelli M, Lucchini G, Piatti S. Budding yeast PAK kinases regulate mitotic exit by two different mechanisms. J Cell Biol. 2003;160:857–74. doi: 10.1083/jcb.200209097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hofken T, Schiebel E. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J Cell Biol. 2004;164:219–31. doi: 10.1083/jcb.200309080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leveleki L, Mahlert M, Sandrock B, Bolker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- 114.Elion EA. The Ste5p scaffold. J Cell Sci. 2001;114:3967–78. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- 115.Pullikuth AK, Catling AD. Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell Signal. 2007;19:1621–32. doi: 10.1016/j.cellsig.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen T, Hiroko T, Chaudhuri A, Inose F, Lord M, Tanaka S, et al. Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science. 2000;290:1975–8. doi: 10.1126/science.290.5498.1975. [DOI] [PubMed] [Google Scholar]
- 117.Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 118.Bender L, Lo HS, Lee H, Kokojan V, Peterson V, Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in Yeast. J Cell Biol. 1996;133:879–94. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Drogen F, Peter M. MAP kinase dynamics in yeast. Biol Cell. 2001;93:63–70. doi: 10.1016/s0248-4900(01)01123-6. [DOI] [PubMed] [Google Scholar]
- 120.Winters MJ, Pryciak PM. Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20. Mol Cell Biol. 2005;25:2177–90. doi: 10.1128/MCB.25.6.2177-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lamson RE, Takahashi S, Winters MJ, Pryciak PM. Dual role for membrane localization in yeast MAP kinase cascade activation and its contribution to signaling fidelity. Curr Biol. 2006;16:618–23. doi: 10.1016/j.cub.2006.02.060. [DOI] [PubMed] [Google Scholar]