Abstract
Objectives
Pancreatitis is an inflammatory condition of the pancreas. However, it often shares many molecular features with pancreatic cancer. Biomarkers present in pancreatic cancer frequently occur in the setting of pancreatitis. The efforts to develop diagnostic biomarkers for pancreatic cancer have thus been complicated by the false-positive involvement of pancreatitis.
Methods
In an attempt to develop protein biomarkers for pancreatic cancer, we previously use quantitative proteomics to identify and quantify the proteins from pancreatic cancer juice. Pancreatic juice is a rich source of proteins that are shed by the pancreatic ductal cells. In this study, we used a similar approach to identify and quantify proteins from pancreatitis juice.
Results
In total, 72 proteins were identified and quantified in the comparison of pancreatic juice from pancreatitis patients versus pooled normal control juice. Nineteen of the juice proteins were overexpressed, and 8 were underexpressed in pancreatitis juice by at least 2-fold compared with normal pancreatic juice. Of these 27 differentially expressed proteins in pancreatitis, 9 proteins were also differentially expressed in the pancreatic juice from pancreatic cancer patient.
Conclusions
Identification of these differentially expressed proteins from pancreatitis juice provides useful information for future study of specific pancreatitis-associated proteins and to eliminate potential false-positive biomarkers for pancreatic cancer.
Keywords: pancreatic juice, pancreatitis, ICAT, biomarker, pancreatic cancer, proteomics
Pancreatic cancer is a highly lethal disease.1,2 The death rate nearly matches the incidence because the diagnosis usually occurs late, after metastases have occurred, and the only chance for a cure (ie, surgical excision) has been eliminated. The problem of early diagnosis is complicated by the obscure location of the pancreas, the absence of reliable symptoms, and the insensitivity and expense of current tests. Better methods of detecting early stages of cancer or precancerous lesions are needed.
In the efforts to develop biomarkers for the early detection of pancreatic cancer, one of the problems is the false-positive involvement of pancreatitis patients. Pancreatitis is an inflammatory condition of the pancreas that shares many molecular features with pancreatic cancer. Thus, biomarkers present in the setting of pancreatic cancer frequently occur in pancreatitis, providing an unacceptably low level of specificity for screening. It is therefore important to understand the proteins that underlie pancreatitis, as they could be a source of false-positive biomarkers for pancreatic cancer. Moreover, chronic pancreatitis is risk factor for eventual neoplastic progression; thus, understanding the proteins involved in both diseases may yield some insights into the mechanisms that link these events.
Recently, there has been substantial interest in applying proteomic methods for the discovery of new targets for therapeutics and new biomarkers for diagnosis and early detection.3 In particular, quantitative proteomics has enabled researchers to use a combination of biochemistry, biology, and bioinformatics to detect proteins that are differentially expressed in cancer. In pancreatic cancer, recent studies using proteomics approach have focused on pancreatic cancer tissues.4–6 However, from a biomarker’s standpoint, pancreatic juice is an excellent starting specimen for the identification of protein biomarkers. Pancreatic juice is a rich source for cancer-specific proteins because the highly proliferative cancer cells are shed into the juice, as they undergo cellular turnover and degradation.7
Pancreatic juice was extensively studied in late 1970s and 1980s, primarily by early 2-dimensional electrophoresis analyses, which led to the discovery and description of several pancreatic enzymes.8–12 Recently, Gronborg and colleagues13 used a mass spectrometry-based proteomic approach for the analysis of pancreatic juice which used 1-dimensional electrophoresis and liquid chromatography (LC) tandem mass spectrometry (MS/MS). We previously used an isotope-coded affinity tag (ICAT)–based quantitative proteomic approach to identify and characterize potential biomarkers from pancreatic cancer juice.14 A total of 30 proteins were identified that exhibited greater than 2-fold abundance change in pancreatic cancer juice compared with normal pancreatic juice. Given the false-positive role of pancreatitis in pancreatic cancer, it is important to discover possible pancreatitis specific proteins that can be used to differentiate pancreatic cancer and pancreatitis. In addition, discovery of the proteins in pancreatitis could help identify proteins that might contribute to false-positive findings of pancreatic cancer.
Isotope-coded affinity tag (ICAT) technology provides a comprehensive approach for quantitative proteomic analysis.15,16 This methodology demonstrates a significant improvement over gel-based methods in identifying low-abundant proteins, and it minimizes problems associated with solubility and extremes of pH.17 In this study, we used ICAT technology to perform comprehensive quantitative protein profiling of the pancreatitis juice. We performed the analyses by comparing pooled normal pancreatic juice with pancreatic juice from a chronic pancreatitis patient. Identification and quantification of the proteins from pancreatic juice were accomplished by differentially labeling the target proteins (pancreatitis) with heavy ICAT reagents and the normal comparator proteins with light ICAT reagents. The isotopically labeled proteins were then combined, purified, and followed by LC MS/MS analysis. Protein identification and quantification were then accomplished by using a suite of bioinformatics software. The proteomic analysis of pancreatitis juice was then compared with the analysis of pancreatic cancer juice.
MATERIALS AND METHODS
Specimens
Pancreatic juice samples were collected during endoscopic retrograde cholangiopancreatography and immediately stored at −80°C. Pancreatic tissue specimens were collected at surgery and stored in freezing media (10% dimethyl sulfoxide) at −80°C. The specimens were collected in accordance with approved human subject’s guidelines at the University of Washington. Pancreatic juice from a chronic pancreatitis patient and 10 normal controls were included in this study. Pancreatic juices from 10 normal controls were pooled together by equal quantity of total protein. Because endoscopic retrograde cholangiopancreatography is an invasive procedure, it is uncommon to obtain pancreatic juice from patients with a completely normal pancreas. In this study, the normal juices were from the patients who have benign pancreatic diseases such as benign cystic neoplasms. Samples from potentially neoplastic precursor lesions, such as intrapapillary mucinous neoplasia, were not included as normal control. The pancreatitis sample was from a patient with chronic pancreatitis and history of alcoholism, without evidence of malignancy.
Isotope-Coded Affinity Tag Labeling and Mass Spectrometry
The ICAT labeling and mass spectrometry were performed as previously described.14 Briefly, protease inhibitor, phenyl methyl sulfonyl fluoride, was added to pancreatic juice at a final concentration of 0.5 mmol/L to prevent protein degradation. For each sample, 0.5-mg protein was labeled with the acid cleavable ICAT reagents, either in an isotopically light 12C (pooled normal pancreatic juice) or in heavy 13C (pancreatitis juice) isoforms (Applied Biosystems, Foster City, Calif) as previously described. The labeled 2 samples were then combined and digested into peptides by trypsin (Promega, Madison, Wis). Isotope-Coded affinity tag–labeled peptides were subsequently fractionated by cation-exchange chromatography and purified by avidin-affinity chromatography. The resulting 50 fractions were then combined into 21 fractions and analyzed by microcapillary high-performance LC–electrospray ionization–MS/MS using an ion-trap mass spectrometer (LCQ-DecaXP, ThermoFinnigan, San Jose, Calif) as previously described.4,14
Data Analysis
Tandem mass spectrometry spectra were searched against the National Cancer Institute human sequence database using SEQUEST.18 The database search results were validated using the PeptideProphet program.19 Peptide-Prophet uses various SEQUEST scores and a number of other parameters to calculate a probability score for each identified peptide. The identified peptides were then assigned a protein identification using the ProteinProphet software.20 Protein-Prophet allows filtering of large-scale data sets with predictable sensitivity and false-positive identification error rates. In this study, we used ProteinProphet probability score more than or equal to 0.9 as a cutoff value for protein identification. This will ensure that the false-positive rate (error rate) for protein identification in this study less than 0.9%. Quantification of the ratio of each protein (isotopically heavy vs light) was calculated using the ASAPRatio program.21 Information on the software can be found on line at http://www.systemsbiology.org/Default.aspx?pagename=FullList. The identified proteins were classified based on GO (Gene Ontology) consortium.22
RESULTS AND DISCUSSION
Proteins Identified in Pancreatitis Juice
In this study, we used the ICAT approach to identify and quantify proteins in a pair of pancreatic juices: pooled normal pancreatic juice versus pancreatic juice from a patient with pancreatitis. The approach was to label the comparator (pooled normal juice) and target (pancreatitis juice) proteins with the isotopically light and heavy ICAT reagents, respectively. The labeled proteins from the comparator and target proteomes were then combined, digested, fractionated, and purified by multidimensional chromatography and analyzed by MS/MS. The protein identification and quantification were accomplished by using a suite of bioinformatic software tools.18–21 All together, 72 proteins were identified and quantified in the comparison of pancreatitis juice to pooled normal pancreatic juice, with an error rate of less than 0.9%.
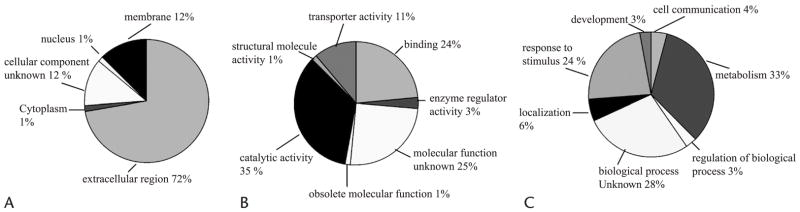
The 72 proteins identified in the pancreatic juices were examined by cellular component, molecular function, and biological process classified accordingly based on GO22 nomenclature. The distribution of these proteins in cellular component, molecular function, and biological process are presented in Figure 1. Most of the identified proteins was from the extracellular region (72%) or bound to the plasma membrane (12%). This is consistent with the fact that proteins from pancreatic juice are primarily secreted and with a previous study in pancreatic cancer juice and normal pancreatic juice.14 In addition, about 3% of proteins were from the ductal cell cytoplasm and nucleus. Because pancreatic juice is mainly cell-free, the existent of these cellular proteins supports the assumption that cellular proteins may be shed into pancreatic juice by cell turnover or secretion.14
FIGURE 1.
Distribution of the identified proteins from pancreatic juice. The 72 proteins identified and quantified in pancreatic juice were classified into categories based on A, cellular component, B, molecular function, and C, biological process. The assignments were based on GO consortium.
Many of the proteins identified in pancreatic juice were enzymes (catalytic activity, 35%). Other molecular functions identified included: binding function 24%, enzyme regulation 3%, obsolete molecular function 1%, structural molecules 1%, and transporter activity 11%. The molecular functions for 25% of the proteins are still unknown at the present time.
In classifying the proteins by biological process, one third of the proteins identified were involved with metabolism; this finding is consistent with the fact that the major role of pancreatic juice is to provide enzymes for digestion. Other biological functions attributed to the discovered proteins include response to stimulus (24%) (not surprising in the setting of chronic inflammation), developmental (3%), cell communication (4%), regulators of biological processes (3%), and localization (6%).
We have previously identified a total of 105 proteins from pancreatic cancer juice.14 Comparison of the 2 disease states (pancreatitis and cancer) reveals that 31 of the 72 pancreatitis proteins identified in this pancreatitis study have not been detected in previous studies and are unique to this study of pancreatitis juice. In total, 136 proteins were identified in pancreatic juices in the 2 studies (current and previous) (Table 1).
TABLE 1.
List of Proteins Identified in the Pancreatic Juices
| Database ID | Protein Name | Identified in CA/nl | Identified in CP/nl | Identified in nl/nl |
|---|---|---|---|---|
| 3D:1clyH | 1clyH IGG FAB (human IgG1, κ) | ✓ | ||
| SW:W70T_HUMAN | 70 kDa WD-repeat tumor rejection antigen | ✓ | ||
| SW:A1AG_HUMAN | α1-Acid glycoprotein 1 precursor | ✓ | ✓ | |
| SW:A1AT_HUMAN | α1-Antitrypsin precursor | ✓ | ||
| SW:A1BG_HUMAN | α1b-Glycoprotein precursor | ✓ | ✓ | |
| SW:A2MG_HUMAN | α2-Macroglobulin precursor | ✓ | ✓ | |
| SW:AMYC_HUMAN | α-Amylase 2b precursor | ✓ | ✓ | |
| SW:AMYP_HUMAN | α-Amylase, pancreatic precursor | ✓ | ✓ | |
| IPI00025476 | Amylase, α2A | ✓ | ✓ | |
| SW:APOD_HUMAN | Apolipoprotein D precursor | ✓ | ||
| GP:AF332505_1 | Apoptosis inhibitor–like protein mRNA | ✓ | ||
| SW:CORI_HUMAN | Atrial natriuteric peptide–converting enzyme | ✓ | ✓ | |
| SW:AXU1_HUMAN | Axin 1 up-regulated 1 protein (TGF-β–induced apoptosis protein 3) | ✓ | ||
| IPI00179390 | β-actin | ✓ | ||
| SW:B2MG_HUMAN | β2-microglobulin | ✓ | ✓ | |
| SW:APOH_HUMAN | β2–Glycoprotein 1 precursor (apolipoprotein H) | ✓ | ✓ | |
| GP:BC042510_1 | Bile salt–stimulated lipase | ✓ | ||
| SW:BAL_HUMAN | Bile salt–activated lipase precursor | ✓ | ✓ | ✓ |
| SW:BMP8_HUMAN | Bone morphogenetic protein 8 precursor | ✓ | ||
| SW:C4BP_HUMAN | C4b-binding protein α chain precursor | ✓ | ||
| GP:AF127036_1 | Calcium-activated chloride channel protein 1 | ✓ | ||
| SW:CRTC_HUMAN | Calreticulin precursor | ✓ | ||
| SW:CBP1_HUMAN | Carboxypeptidase a1 precursor | ✓ | ✓ | ✓ |
| SW:CPB2_HUMAN | Carboxypeptidase a2 precursor | ✓ | ✓ | ✓ |
| SW:CBPB_HUMAN | Carboxypeptidase b precursor | ✓ | ✓ | ✓ |
| SW:CATO_HUMAN | Cathepsin o precursor | ✓ | ||
| SW:CD5L_HUMAN | CD5 antigen–like precursor | ✓ | ✓ | |
| SW:CERU_HUMAN | Ceruloplasmin precursor | ✓ | ||
| GP:AF287894_1 | CFTR-associated ligand (CAL) | ✓ | ||
| GP:AL512883_3 | Chymotrypsin C (caldecrin) | ✓ | ✓ | ✓ |
| SW:CTRL_HUMAN | Chymotrypsin-like protease ctrl-1 precursor | ✓ | ✓ | ✓ |
| SW:CTRB_HUMAN | Chymotrypsinogen b precursor | ✓ | ✓ | ✓ |
| GP:BC005951_1 | Clone MGC:14588 | ✓ | ||
| IPI00219642 | Clusterin precursor | ✓ | ||
| SW:COL_HUMAN | Colipase precursor | ✓ | ✓ | ✓ |
| SW:CO3_HUMAN | Complement C3 precursor | ✓ | ✓ | |
| SW:CO4_HUMAN | Complement C4 precursor | ✓ | ||
| SW:CFAH_HUMAN | Complement factor H precursor | ✓ | ||
| SW:FHR3_HUMAN | Complement factor H-related protein 3 precursor | ✓ | ||
| IPI00032293 | Cystatin C precursor | ✓ | ||
| SW:EL2A_HUMAN | Elastase 2a precursor | ✓ | ✓ | |
| SW:EL2B_HUMAN | Elastase 2b precursor | ✓ | ✓ | |
| IPI00240986 | Elastase 3, pancreatic (protease E) | ✓ | ||
| SW:EL3A_HUMAN | Elastase iiia precursor | ✓ | ✓ | |
| SW:EL3B_HUMAN | Elastase iiib precursor | ✓ | ✓ | ✓ |
| SW:FIBA_HUMAN | Fibrinogen α/αe chain precursor | ✓ | ||
| SW:FIBB_HUMAN | Fibrinogen β chain precursor | ✓ | ✓ | |
| SW:FIBG_HUMAN | Fibrinogen γ chain precursor | ✓ | ✓ | |
| SW:FGL1_HUMAN | Fibrinogen-like protein 1 precursor | ✓ | ✓ | |
| GP:AK074044_1 | FLJ00102 protein | ✓ | ||
| SW:FOL3_HUMAN | Folate receptor γ precursor | ✓ | ||
| IPI00023673 | Galectin 3 binding protein | ✓ | ||
| SW:SGCG_HUMAN | γ-Sarcoglycan (35 kDa dystrophin-associated glycoprotein) | ✓ | ||
| SW:KLK1_HUMAN | Glandular kallikrein 1 precursor | ✓ | ✓ | |
| SW:VGLH_HSV6U | Glycoprotein H precursor | ✓ | ||
| SW:HPT2_HUMAN | Haptoglobin 2 precursor | ✓ | ✓ | |
| SW:HPTR_HUMAN | Haptoglobin-related protein precursor | ✓ | ||
| SW:HBB_HUMAN | Hemoglobin β chain | ✓ | ✓ | |
| SW:HEMO_HUMAN | Hemopexin precursor | ✓ | ✓ | |
| GP:M19233_1 | Human α-amylase-1 gene | ✓ | ✓ | |
| GP:U88581_1 | Human transferrin mRNA, C2 allele | ✓ | ||
| GP:AL136795_1 | Hypothetical protein DKFZp434G131 | ✓ | ||
| GP:AB083068_1 | Hypothetical protein DKFZp434N1415 | ✓ | ||
| GP:BC027590_1 | Hypothetical protein FLJ10261 | ✓ | ||
| IPI00168679 | Hypothetical protein Tr:Q8NEJ1 | ✓ | ||
| IPI00152428 | Hypothetical protein Tr:Q8TCS3 | ✓ | ||
| SW:ALC1_HUMAN | Ig-α1 chain c region | ✓ | ✓ | ✓ |
| PIR1:A2HU | Ig-α2 chain C region | ✓ | ✓ | |
| SW:GC2_HUMAN | Ig-γ2 chain c region | ✓ | ||
| SW:KAC_HUMAN | Ig-κ chain c region | ✓ | ✓ | |
| IPI00004574 | Ig-κ chain C region | ✓ | ||
| SW:MUC_HUMAN | Ig-μ chain C region | ✓ | ✓ | |
| GP:D84239_1 | IgG Fc binding protein | ✓ | ||
| SW:KAC_HUMAN | IGK mRNA for immunoglobulin κ light chain VLJ region | ✓ | ✓ | |
| GP:AJ294732_1 | Immunoglobulin heavy chain constant region γ3 (IGHG3) | ✓ | ||
| GP:AJ294733_1 | Immunoglobulin heavy chain constant region γ4 | ✓ | ||
| GP:AB021510_1 | Immunoglobulin heavy chain variable region | ✓ | ||
| SW:GC1_HUMAN | Immunoglobulin heavy constant γ1 | ✓ | ||
| SW:IBP2_HUMAN | Insulin-like growth factor binding protein | ✓ | ✓ | |
| SW:ITH4_HUMAN | Inter–α-trypsin inhibitor heavy chain h4 precursor | ✓ | ||
| GP:AB040939_1 | KIAA1506 protein | ✓ | ||
| SW:LITA_HUMAN | Lithostathine 1 α precursor pancreatic stone protein) | ✓ | ✓ | ✓ |
| SW:LITB_HUMAN | Lithostathine 1β precursor | ✓ | ||
| GP:AY044164_1 | Lymphocyte α kinase | ✓ | ||
| SW:LYC_HUMAN | Lysozyme c precursor | ✓ | ✓ | |
| GP:AK009737_1 | Moderately similar to gallus gallus syndesmos | ✓ | ||
| GP:AC003682_1 | Most similar to zinc finger protein ZNF132 | ✓ | ||
| IPI00103397 | Mucin 5 | ✓ | ||
| PIR2:S53362 | Mucin 5AC | ✓ | ✓ | ✓ |
| GP:AF244548_1 | Na+ and H+ coupled amino acid transport system N mRNA | ✓ | ||
| SW:CAML_HUMAN | NCAM L1 (CD171 antigen) | ✓ | ||
| IPI00220513 | P21359 Neurofibromin | ✓ | ||
| SW:PAX7_HUMAN | Paired box protein pax-7 (hup1). | ✓ | ||
| PIR2:A29934 | Pancreatic elastase (EC 3.4.21.36) IIIA precursor | ✓ | ||
| SW:LIPP_HUMAN | Pancreatic lipase | ✓ | ✓ | ✓ |
| SW:LIP1_HUMAN | Pancreatic lipase–related protein 1 precursor | ✓ | ✓ | |
| SW:LIP2_HUMAN | Pancreatic lipase–related protein 2 precursor | ✓ | ✓ | ✓ |
| SW:GP2_HUMAN | Pancreatic secretory granule membrane major glycoprotein gp2 precursor | ✓ | ✓ | ✓ |
| SW:IPK1_HUMAN | Pancreatic secretory trypsin inhibitor precursor (tumor-associated trypsin inhibitor) | ✓ | ✓ | |
| GP:AB035542_1 | Pancreatic zymogen granule membrane–associated protein GP2β | ✓ | ✓ | |
| SW:PAP1_HUMAN | Pancreatitis-associated protein 1 precursor | ✓ | ✓ | |
| IPI00022543 | Phosphatidylinositol glycan | ✓ | ||
| SW:PA21_HUMAN | Phospholipase a2 precursor | ✓ | ✓ | ✓ |
| SW:PLMN_HUMAN | Plasminogen precursor | ✓ | ||
| SW:PIGR_HUMAN | Polymeric-immunoglobulin receptor precursor | ✓ | ✓ | |
| SW:SAP_HUMAN | Proactivator polypeptide precursor (contains saposin a) | ✓ | ✓ | |
| IPI00022213 | Progastricsin (pepsinogen C) | ✓ | ||
| IPI00011694 | Protease, serine, 1 preproprotein | ✓ | ||
| IPI00011695 | Protease, serine, 2 preproprotein | ✓ | ||
| SW:CLPP_HUMAN | Putative ATP-dependent clp protease proteolytic subunit | ✓ | ||
| PIR2:S36262 | Rearranged Ig-κ region V-domain | ✓ | ||
| IPI00009197 | Regenerating islet–derived 1β precursor | ✓ | ||
| IPI00019176 | Retinoic acid receptor responder | ✓ | ||
| SW:RNP_HUMAN | Ribonuclease pancreatic precursor | ✓ | ✓ | ✓ |
| GP:AK090123_1 | RNA binding protein homolog | ✓ | ||
| IPI00032179 | Serine (or cysteine) proteinase inhibitor, clade C (antithrombin) | ✓ | ||
| SW:TRFE_HUMAN | Serotransferrin precursor (transferrin) | ✓ | ✓ | ✓ |
| SW:ALBU_HUMAN | Serum albumin | ✓ | ✓ | ✓ |
| IPI00156237 | Similar to chymotrypsinogen B precursor | ✓ | ||
| IPI00027722 | Similar to elastase 1, pancreatic | ✓ | ||
| IPI00247169 | Similar to syncollin | ✓ | ||
| SW:SODC_HUMAN | Superoxide dismutase | ✓ | ||
| GP:U66061_9 | T-cell receptor β chain (human germline) | ✓ | ||
| GP:AY190093_1 | T-cell receptor β chain (clone PSA.S.20) | ✓ | ||
| IPI00010675 | Trefoil factor 2 (spasmolytic protein 1) | ✓ | ||
| IPI00181107 | Triacylglycerol lipase | ✓ | ||
| SW:TRY1_HUMAN | Trypsin I precursor | ✓ | ✓ | ✓ |
| SW:TRY2_HUMAN | Trypsin II precursor | ✓ | ✓ | |
| IPI00015614 | Trypsin III precursor | ✓ | ||
| SW:TRY4_HUMAN | Trypsin IV precursor | ✓ | ||
| IPI00169276 | Trypsinogen C | ✓ | ||
| GP:AF305835_1 | Uterus-ovary specific putative transmembrane protein UO | ✓ | ✓ | ✓ |
| SW:VTDB_HUMAN | Vitamin D–binding protein precursor | ✓ | ||
| GP:U06117_1 | Xanthine dehydrogenase | ✓ | ||
| SW:ZA2G_HUMAN | Zinc α2-glycoprotein precursor | ✓ | ✓ |
GP indicates GenPept; PIR, Protein information resource; IPI, International Protein Index; SW, SWISS-PROT; CA, cancer; CP, chronic pancreatitis; nl, normal.
Proteins With at Least 2-Fold Change in Abundance in Pancreatitis Juice
The use of ICAT technology allows us to perform protein identification and quantification studies in the same experiment. The quantification of each protein is presented as a protein ratio between 2 samples tested. In the comparison of pancreatitis juice/pooled normal juice, 27 proteins showed differential expression in pancreatitis juice by at least 2-fold: 19 were overexpressed, and 8 were underexpressed in pancreatitis juice (Table 2). Below, we will discuss some of the differentially regulated proteins.
TABLE 2.
Proteins With at Least 2-fold Change in Abundance in Pancreatic Juice From Pancreatitis
| Database ID | Protein Name | Ratio (pancreatitis/nl) | SD | Unique Peptide Identified |
|---|---|---|---|---|
| More abundant by at least 2-fold | ||||
| 3D:1clyH | 1clyH IGG FAB (human IGG1 | 7.0 | 5.1 | 4 |
| SW:A1BG_HUMAN | α1b-Glycoprotein precursor | 3.1 | 0.9 | 3 |
| SW:A2MG_HUMAN | α2-Macroglobulin precursor | 2.8 | 0.3 | 12 |
| SW:APOH_HUMAN | β2-Glycoprotein I precursor (apolipoprotein H) | 6.5 | 6.1 | 7 |
| SW:CTRB_HUMAN | Chymotrypsinogen b precursor | 2.1 | 0.3 | 19 |
| SW:FIBB_HUMAN | Fibrinogen β chain precursor | 3.0 | 0.9 | 13 |
| SW:HPT2_HUMAN | Haptoglobin 2 precursor | 2.4 | 0.3 | 16 |
| SW:HBB_HUMAN | Hemoglobin β chain | 6.8 | 6.6 | 12 |
| GP:AF542069_1 | Human serum albumin | 3.9 | 1.7 | 267 |
| SW:ALC1_HUMAN | Ig-α1 chain c region | 4.7 | 1.1 | 7 |
| SW:MUC_HUMAN | Ig-μ chain c region | 2.5 | 2.1 | 1 |
| GP:AJ294732_1 | Immunoglobulin heavy chain constant region γ3 | 11.1 | 8.9 | 10 |
| GP:AB021510_1 | Immunoglobulin heavy chain variable region (IgM) | 9.6 | 1.0 | 1 |
| SW:CAML_HUMAN | NCAM L1 precursor | 34.5 | 4.8 | 1 |
| SW:PLMN_HUMAN | Plasminogen/plasmin | 2.7 | 1.3 | 1 |
| SW:RNP_HUMAN | Ribonuclease pancreatic precursor | 2.3 | 6.40 | 2 |
| SW:TRFE_HUMAN | Serotransferrin precursor (transferrin) | 2.7 | 0.4 | 29 |
| SW:TRY1_HUMAN | Trypsin I precursor | 3.8 | 1.7 | 16 |
| SW:TRY2_HUMAN | Trypsin II precursor | 2.2 | 0.5 | 26 |
| Less abundant by at least 2-fold | ||||
| SW:BMP8_HUMAN | Bone morphogenetic protein 8 precursor | 0.2 | 0.17 | 1 |
| SW:CLCR_HUMAN | Caldecrin precursor | 0.4 | 0.19 | 3 |
| SW:EL2A_HUMAN | Elastase 2a precursor | 0.5 | 0.06 | 6 |
| SW:EL2B_HUMAN | Elastase 2b precursor | 0.4 | 0.18 | 4 |
| SW:SGCG_HUMAN | γ-Sarcoglycan | 0.1 | 0.00 | 1 |
| GP:D84239_1 | IgG Fc binding protein | 0.3 | 0.03 | 5 |
| PIR2:A29934 | Pancreatic elastase IIIA | 0.3 | 0.03 | 27 |
| GP:AY190093_1 | T-cell receptor β chain (clone PSA.S.20) | 0.1 | 0.02 | 1 |
GP indicates GenPept; PIR, Protein information resource; SW, SWISS-PROT.
Fibrinogen β Chain
Fibrinogen was found to be 3.0-fold up-regulated in pancreatitis juice compared with normal pancreatic juice in this study. The MS/MS spectrum of a unique peptide (PVVSGKECEEIIR) derived from fibrinogen β chain is presented in Figure 2A. Twelve other peptides from this protein were also identified in the experiment (MS/MS spectra not shown). The identification of these 13 peptides gave an explicit identification of fibrinogen β chain in the samples. The relative abundance of each peptide pair was calculated based on the signal intensities for each peptide using ASAPRatio software. The relative abundance data revealed that this peptide was 4-fold more abundant in the pancreatitis sample (heavy) than in the normal sample (light). The ratio of fibrinogen β chain between pancreatitis and pooled normal was eventually calculated to be 3.0 (Fig. 2B) by incorporation of the intensities of all 13 peptides identified.
FIGURE 2.
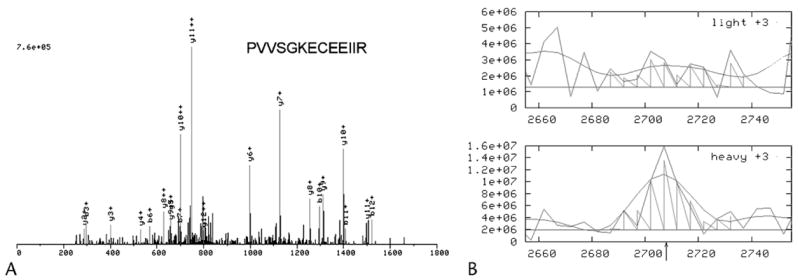
Identification and quantification of fibrinogen beta chain in pancreatic juice by ICAT labeling and MS/MS analysis. A, MS/MS spectrum of a unique peptide (PVVSGKECEEIIR) derived from fibrinogen beta chain. The peptide was identified by sequence database searching using SEQUEST. Twelve other peptides from the same protein were also identified (MS/MS spectra not shown) which lead to conclusive identification of fibrinogen beta chain. B, The relative abundance of each peptide pair was cacculated based on the signal intensities for each peptide using ASAPRatio software. The relative abundance data revealed that this peptide was more abundant in pancreatitis sample (heavy) than in normal sample (light): ratio = 4.0. The ratio of fibrinogen beta chain between pancreatitis and pooled normal was calculated to be 3.0 by incorporation of the intensities of all 13 peptides identified.
Blood coagulation/fibrinolytic systems are commonly activated in pancreatic cancer. Immunohistochemical staining of pancreatic cancer sections indicate that fibrinogen exists throughout the tumor stroma, and tumor cells are surrounded by fibrin.23 Our previous studies also detected overexpression of fibrinogen β chain in pancreatic cancer tissue4 and elevation of fibrinogen β chain in pancreatic cancer juice.24 It has been postulated that local coagulation activation may regulate growth of pancreatic cancer.23 Alternatively, the up-regulation of fibrinogen in pancreatitis juice discovered in this study may be consistent with the fact that fibrogen is a major acute response protein involved in the inflammation of pancreas. Our study verifies that pancreatic cancer and pancreatitis have some common protein alterations. As such, the consideration of proteins in these 2 diseases should be considered concurringly to avoid false-positive findings in the development of diagnostic biomarkers for cancer.
Plasmin/Plasminogen
Identification of a peptide NYCRNPDGDVGGPW-CYTTNPR derived from plasminogen or plasmin suggested the presence of plasminogen or/and plasmin in the juice samples; however, the exact form of the protein from which the peptide was derived cannot be determined. It is clear that plasminogen/plasmin is elevated in pancreatitis juice by 2.7-fold. In the plasminogen/plasmin system, the inactive proenzyme plasminogen can be transformed into the proteolytically active plasmin mainly by plasminogen activators. Plasmin is involved in fibrinolysis and inflammation. The pathophysiological importance of the plasminogen/plasmin system in classical inflammation diseases has been well established by several studies.25 Increase level of plasmin activator (uPA) and plasmin–α2-antiplasmin complexes were found in synovial fluid as well as in plasma in patients with rheumatic diseases.25–27 Involvement of plasmin in inflammatory process has also been demonstrated in plasminogen-deficient mice. In light of these findings, novel approaches were proposed for the treatment of chronic inflammatory diseases. For one example, induction of adenoviral vectors encoding an engineered cell surface-targeted plasmin inhibitor seems to reduce synovial fibroblast-dependent cartilage degradation and invasion in rheumatoid arthritis.25,28 Enhanced plasminogen activation have also been revealed in pancreatitis.29,30 However, direct demonstration of enhanced level of plasminogen in pancreatitis juice has not been reported before. In addition to its role in inflammation, plasminogen activation also plays an important role in tumor cell invasiveness and metastasis.31,32 Plasmin generated at the surface of tumor cells is considered a key event in tumor invasion and metastasis in the pancreatic cancer.31,32
Neural Cell Adhesion Molecule L1
Another up-regulated protein in pancreatitis juice is neural cell adhesion molecule L1 (NCAM L1). Neural cell adhesion molecule L1 is a multidomain membrane glycoprotein of the immunoglobulin superfamily33–36 that is over-expressed by a variety of highly malignant tumors, including neuroblastomas,37 ovarian tumors,38 renal cell carcinoma,39 and endometrial tumors.40 Neural cell adhesion molecule L1 promotes many cellular activities by interacting through its extracellular domain with other cell adhesion molecules, extracellular matrix molecules, and signal receptors.41–43 In light of its role in cell proliferation44 and tumor progression,41 NCAM L1 has emerged as a promising marker for cancers.45 Expression of NCAM L1 has not been previously reported in pancreatic cancer or pancreatitis. However, another adhesion molecule, neural cell adhesion molecule, has expression in 66.7% of pancreatic cancer cases.46 These 2 cell adhesion proteins are generated from different genes on different chromosomes; however, both are membrane glycoproteins belonging to a superfamily of immunoglobulins that mediate cell-to-cell adhesion at the cell surface. The expression of neural cell adhesion molecule has been detected more frequently in lesions where neural invasion was excessive.46 It is not clear why NCAM L1 was up-regulated in the setting of pancreatitis and not in pancreatic cancer juice. It will be interesting in the future to determine the expression pattern of this protein in normal pancreas and pancreatic disease, such as cancer and pancreatitis.
Caldecrin
One of the down-regulated proteins in pancreatitis juice is caldecrin (2.5-fold decrease). Caldecrin, also known as serum calcium-decreasing factor or chymotrypsin C, is a novel-type serine protease expressed in the pancreas.47 Its association with pancreatic disease has not been reported before. Given the fact that several digestive enzymes were differentially regulated in pancreatitis juice presented in this study, it is not surprising to see caldecrin also differentially expressed in pancreatitis juice. Further work is needed to evaluate the role of caldecrin in various pancreatic diseases.
Comparison of Differentially Expressed Proteins in the Pancreatic Juices from Pancreatic Cancer and Pancreatitis
Comparison of the differentially expressed proteins in pancreatitis to those from pancreatic cancer juice and normal juice,14 revealed 9 proteins (hemoglobin, fibrinogen, trypsin I, trypsin II, chymotrypsinogen b, Ig-α1 chain c region, Ig-μ chain c region, ribonuclease, and human serum albumin) that were both up-regulated in pancreatic cancer juice and pancreatitis juice (Table 3). These proteins should be excluded in future biomarker study for pancreatic cancer. Interestingly, down-regulated proteins in pancreatic cancer do not overlap with those from pancreatitis. There are 21 proteins that were differentially expressed only in pancreatic cancer, whereas 18 proteins were differentially expressed only in the pancreatitis (Fig. 3 and Table 3). In addition, elastase 2b, which was variable in comparison of normal juice with pooled normal juice,14 was also down-regulated pancreatitis juice but not found in pancreatic cancer juice. These data thus provide useful basis for future development of biomarkers for pancreatic cancer and pancreatitis and reduce the chance of false-positive biomarkers.
TABLE 3.
Comparison of Proteins With at Least 2-fold Change in Pancreatic Juice from Pancreatitis and Cancer
| Database ID | Protein Name | Ratio (CP/nl) | Ratio (CA/nl) |
|---|---|---|---|
| At least 2-fold change in abundance in both of pancreatic cancer and pancreatitis | |||
| SW:CTRB_HUMAN | Chymotrypsinogen b precursor | 2.1 | 2.8 |
| SW:FIBB_HUMAN | Fibrinogen β chain precursor | 3.0 | 3.8 |
| SW:HBB_HUMAN | Hemoglobin β chain | 6.8 | 3.3 |
| GP:AF542069_1 | Human serum albumin | 3.9 | 3.7 |
| SW:ALC1_HUMAN | Ig-α1 chain C region | 4.7 | 4.0 |
| SW:MUC_HUMAN | Ig-μ chain C region | 2.5 | 3.9 |
| SW:RNP_HUMAN | Ribonuclease pancreatic precursor | 2.3 | 2.8 |
| SW:TRY1_HUMAN | Trypsin I precursor | 3.8 | 2.5 |
| SW:TRY2_HUMAN | Trypsin II precursor | 2.2 | 2.9 |
| At least 2-fold change in abundance in pancreatic cancer only | |||
| SW:A1AG_HUMAN | α1-Acid glycoprotein 1 precursor | 1.2 | 0.3 |
| SW:B2MG_HUMAN | β2-Microglobulin | 3.3 | |
| SW:CPB2_HUMAN | Carboxypeptidase a2 precursor | 0.7 | 0.3 |
| SW:CO3_HUMAN | Complement C3 precursor | 1.7 | 3.0 |
| SW:EL3B_HUMAN | Elastase 3b precursor | 1.7 | 2.2 |
| SW:FIBA_HUMAN | Fibrinogen α/αe chain precursor | 0.2 | |
| SW:FIBG_HUMAN | Fibrinogen γ chain precursor | 1.4 | 7.1 |
| SW:KAC_HUMAN | Ig-κ light chain VLJ region | 1.5 | 4.7 |
| SW:IBP2_HUMAN | Insulin-like growth factor binding protein 2 | 4.8 | |
| SW:KLK1_HUMAN | Kallikrein 1 precursor | 2.7 | |
| SW:LITA_HUMAN | Lithostathine 1α (pancreatic stone protein) | 0.72 | 2.3 |
| SW:LITB_HUMAN | Lithostathine 1β precursor | 4.8 | |
| SW:GP2_HUMAN | Pancreatic secretory granule membrane major glycoprotein | 0.72 | 2.4 |
| SW:PAP1_HUMAN | Pancreatitis-associated protein 1 precursor | 3.1 | |
| SW:SAP_HUMAN | Proactivator polypeptide precursor | 0.2 | |
| SW:CLPP_HUMAN | Putative ATP-dependent clp protease proteolytic subunit | 0.3 | |
| PIR2:S36262 | Rearranged Ig-κ region V-domain | 3.7 | |
| GP:U66061_9 | T-cell receptor β chain (human germline) | 10.2 | |
| GP:U88581_1 | Transferrin, C2 allele | 0.4 | |
| SW:TRY4_HUMAN | Trypsin IV precursor | 5.6 | |
| SW:IPK1_HUMAN | Tumor-associated trypsin inhibitor | 5.6 | |
| At least 2-fold change in abundance in pancreatitis only | |||
| 3D:1clyH | 1clyH IGG FAB (human IGG1) | 7.0 | |
| SW:A1BG_HUMAN | α1b-Glycoprotein precursor | 3.1 | 0.9 |
| SW:A2MG_HUMAN | α2-Macroglobulin precursor | 2.8 | 1.1 |
| SW:APOH_HUMAN | β2-glycoprotein I precursor (apolipoprotein H) | 6.5 | 1.1 |
| SW:BMP8_HUMAN | Bone morphogenetic protein 8 precursor | 0.2 | |
| SW:CLCR_HUMAN | Caldecrin precursor | 0.4 | 1.2 |
| SW:EL2A_HUMAN | Elastase 2a precursor | 0.5 | 1.0 |
| SW:EL2B_HUMAN | Elastase 2b precursor | 0.4 | |
| SW:SGCG_HUMAN | γ-Sarcoglycan | 0.1 | |
| SW:HPT2_HUMAN | Haptoglobin 2 precursor | 2.4 | 2.0 |
| GP:D84239_1 | IgG Fc binding protein | 0.3 | |
| GP:AJ294732_1 | Immunoglobulin heavy chain constant region γ3 | 11.1 | |
| GP:AB021510_1 | Immunoglobulin heavy chain variable region (IgM) | 9.6 | |
| SW:CAML_HUMAN | NCAM L1 precursor | 34.5 | |
| PIR2:A29934 | Pancreatic elastase IIIA | 0.3 | 1.7 |
| SW:PLMN_HUMAN | Plasminogen/plasmin | 2.7 | |
| SW:TRFE_HUMAN | Serotransferrin precursor (transferrin) | 2.7 | 1.7 |
| GP:AY190093_1 | T-cell receptor β chain (clone PAS.S.20) | 0.1 | |
GP indicates GenPept; PIR, Protein information resource; IPI, International Protein Index; SW, SWISS-PROT; CA, cancer; CP, chronic pancreatitis; nl, normal.
FIGURE 3.
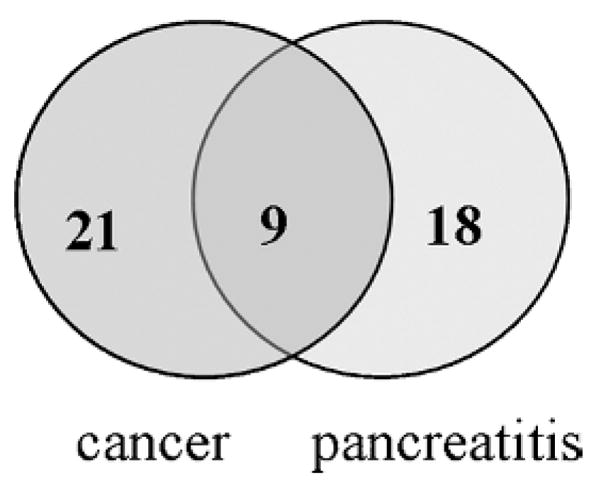
Comparison of protein abundance changes in pancreatic juices from pancreatic cancer and pancreatitis patients. Of the 27 differentially regulated proteins in pancreatitis juice, 9 proteins were also differentially regulated in pancreatic cancer, and they are all up-regulated. Eighteen and 21 proteins are only differentially regulated in pancreatitis and pancreatic cancer, respectively.
Summary
In this study, we performed a comparative and comprehensive proteomic analysis on pancreatic juice from pancreatitis versus pooled normal controls. In total, we identify and quantify 72 proteins in comparison of the pancreatic juice from pancreatitis and pooled normal control, of which 19 were overexpressed, and 8 were underexpressed in pancreatitis juice by at least 2-fold. In these 27 differentially expressed proteins, several proteins (including plasminogen, NCAM L1, and caldecrin) have not been previously associated with pancreatic cancer or pancreatitis, and pancreatic juice. Further study is needed to elucidate the roles of these newly discovered proteins in pancreatitis and pancreatic cancer. In addition, 9 proteins (hemoglobin, fibrinogen, trypsin I, trypsin II, chymotrypsinogen b, Ig-α1 chain c region, Ig-μ chain c region, ribonuclease, and human serum albumin) that were previously shown to be up-regulated in pancreatic cancer juice are also differentially expressed in pancreatitis juice. Thus, these proteins may create false-positive results as biomarkers for pancreatic cancer.
Acknowledgments
Supported by the National Cancer Institute NIH R01-CA-107209, Canary Foundation, the Concern Foundation, Gene and Mary Ann Walters Pancreatic Cancer Foundation, the AACR-PanCAN Career Development Award for Pancreatic Cancer Research, and Federal funds from the National Heart, Lung and Blood Institute NIH, under contract no. NOI-HV-28179.
References
- 1.Greenlee RT, Hill-Harmon MB, Murray T, et al. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Thomas A, Murray T, et al. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 4.Chen R, Yi EC, Donohoe D, et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Lu Z, Hu L, Evers S, et al. Differential expression profiling of human pancreatic adenocarcinoma and healthy pancreatic tissue. Proteomics. 2004;4:3975–3988. doi: 10.1002/pmic.200300863. [DOI] [PubMed] [Google Scholar]
- 6.Shen J, Person MD, Zhu J, et al. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 7.Chen R, Pan S, Brentnall TA, et al. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol Cell Proteomics. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Lohr M, Faissner R. Proteomics in pancreatic disease. Pancreatology. 2004;4:67–75. doi: 10.1159/000077212. [DOI] [PubMed] [Google Scholar]
- 9.Goke B, Keim V, Dagorn JC, et al. Resolution of human exocrine pancreatic juice proteins by reversed-phase high performance liquid chromatography (HPLC) Pancreas. 1990;5:261–266. doi: 10.1097/00006676-199005000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Keim V, Iovanna JL, Rohr G, et al. Characterization of a rat pancreatic secretory protein associated with pancreatitis. Gastroenterology. 1991;100:775–782. doi: 10.1016/0016-5085(91)80025-5. [DOI] [PubMed] [Google Scholar]
- 11.Scheele GA. Two-dimensional gel analysis of soluble proteins. Characterization of guinea pig exocrine pancreatic proteins. J Biol Chem. 1975;250:5375–5385. [PubMed] [Google Scholar]
- 12.Scheele GA, Palade GE. Studies on the guinea pig pancreas. Parallel discharge of exocrine enzyme activities. J Biol Chem. 1975;250:2660–2670. [PubMed] [Google Scholar]
- 13.Gronborg M, Bunkenborg J, Kristiansen TZ, et al. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Pan S, Donohoe S, et al. Quantitative Proteomic Profiling of Pancreatic Cancer Juice. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 15.Gygi SP, Rist B, Gerber SA, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 16.Han DK, Eng J, Zhou H, et al. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin DB, Gifford DR, Wright ME, et al. Quantitative proteomic analysis of proteins released by neoplastic prostate epithelium. Cancer Res. 2004;64:347–355. doi: 10.1158/0008-5472.can-03-2062. [DOI] [PubMed] [Google Scholar]
- 18.Eng J, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 19.Keller A, Nesvizhskii AI, Kolker E, et al. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI, Keller A, Kolker E, et al. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 21.Li XJ, Zhang H, Ranish JA, et al. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75:6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 22. [Accessed 2006];Gene Ontology Software and Databases. Available at: http://www.godatabase.org/dev.
- 23.Wojtukiewicz MZ, Rucinska M, Zacharski LR, et al. Localization of blood coagulation factors in situ in pancreatic carcinoma. Thromb Haemost. 2001;86:1416–1420. [PubMed] [Google Scholar]
- 24.Charlton LA, Sayed M, Clark-Lewis I, et al. Characterization of an activated ribosomal S6 kinase variant from maturing sea star oocytes: association with phosphatase 2A and substrate specificity. J Cell Biochem. 1999;75:310–326. [PubMed] [Google Scholar]
- 25.Syrovets T, Simmet T. Novel aspects and new roles for the serine protease plasmin. Cell Mol Life Sci. 2004;61:873–885. doi: 10.1007/s00018-003-3348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inman RD, Harpel PC. Alpha 2–plasmin inhibitor–plasmin complexes in synovial fluid. J Rheumatol. 1986;13:535–537. [PubMed] [Google Scholar]
- 27.Kawakami M, Kawagoe M, Harigai M, et al. Elevated plasma levels of alpha 2-plasmin inhibitor–plasmin complex in patients with rheumatic diseases. Possible role of fibrinolytic mechanism in vasculitis. Arthritis Rheum. 1989;32:1427–1433. doi: 10.1002/anr.1780321112. [DOI] [PubMed] [Google Scholar]
- 28.van der Laan WH, Pap T, Ronday HK, et al. Cartilage degradation and invasion by rheumatoid synovial fibroblasts is inhibited by gene transfer of a cell surface-targeted plasmin inhibitor. Arthritis Rheum. 2000;43:1710–1718. doi: 10.1002/1529-0131(200008)43:8<1710::AID-ANR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Friess H, Cantero D, Graber H, et al. Enhanced urokinase plasminogen activation in chronic pancreatitis suggests a role in its pathogenesis. Gastroenterology. 1997;113:904–913. doi: 10.1016/s0016-5085(97)70186-0. [DOI] [PubMed] [Google Scholar]
- 30.Friess H, Duarte R, Kleeff J, et al. The plasminogen activator/plasmin system is up-regulated after acute necrotizing pancreatitis in human beings. Surgery. 1998;124:79–86. [PubMed] [Google Scholar]
- 31.Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57:25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt M, Harbeck N, Thomssen C, et al. Clinical impact of the plasminogen activation system in tumor invasion and metastasis: prognostic relevance and target for therapy. Thromb Haemost. 1997;78:285–296. [PubMed] [Google Scholar]
- 33.Brummendorf T, Kenwrick S, Rathjen FG. Neural cell recognition molecule L1: from cell biology to human hereditary brain malformations. Curr Opin Neurobiol. 1998;8:87–97. doi: 10.1016/s0959-4388(98)80012-3. [DOI] [PubMed] [Google Scholar]
- 34.Kowitz A, Kadmon G, Eckert M, et al. Expression and function of the neural cell adhesion molecule L1 in mouse leukocytes. Eur J Immunol. 1992;22:1199–1205. doi: 10.1002/eji.1830220514. [DOI] [PubMed] [Google Scholar]
- 35.Nolte C, Moos M, Schachner M. Immunolocalization of the neural cell adhesion molecule L1 in epithelia of rodents. Cell Tissue Res. 1999;298:261–273. doi: 10.1007/s004419900063. [DOI] [PubMed] [Google Scholar]
- 36.Pancook JD, Reisfeld RA, Varki N, et al. Expression and regulation of the neural cell adhesion molecule L1 on human cells of myelomonocytic and lymphoid origin. J Immunol. 1997;158:4413–4421. [PubMed] [Google Scholar]
- 37.Deichmann M, Kurzen H, Egner U, et al. Adhesion molecules CD171 (L1CAM) and CD24 are expressed by primary neuroendocrine carcinomas of the skin (Merkel cell carcinomas) J Cutan Pathol. 2003;30:363–368. doi: 10.1034/j.1600-0560.2003.00073.x. [DOI] [PubMed] [Google Scholar]
- 38.Fogel M, Gutwein P, Mechtersheimer S, et al. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet. 2003;362:869–875. doi: 10.1016/S0140-6736(03)14342-5. [DOI] [PubMed] [Google Scholar]
- 39.Meli ML, Carrel F, Waibel R, et al. Anti-neuroblastoma antibody chCE7 binds to an isoform of L1-CAM present in renal carcinoma cells. Int J Cancer. 1999;83:401–408. doi: 10.1002/(sici)1097-0215(19991029)83:3<401::aid-ijc17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 40.Fogel M, Huszar M, Altevogt P, et al. L1 (CD171) as a novel biomarker for ovarian and endometrial carcinomas. Expert Rev Mol Diagn. 2004;4:455–462. doi: 10.1586/14737159.4.4.455. [DOI] [PubMed] [Google Scholar]
- 41.Allory Y, Matsuoka Y, Bazille C, et al. The L1 cell adhesion molecule is induced in renal cancer cells and correlates with metastasis in clear cell carcinomas. Clin Cancer Res. 2005;11:1190–1197. [PubMed] [Google Scholar]
- 42.Brummendorf T, Lemmon V. Immunoglobulin superfamily receptors: cis-interactions, intracellular adapters and alternative splicing regulate adhesion. Curr Opin Cell Biol. 2001;13:611–618. doi: 10.1016/s0955-0674(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 43.Kamiguchi H, Lemmon V. Neural cell adhesion molecule L1: signaling pathways and growth cone motility. J Neurosci Res. 1997;49:1–8. doi: 10.1002/(sici)1097-4547(19970701)49:1<1::aid-jnr1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 44.Primiano T, Baig M, Maliyekkel A, et al. Identification of potential anticancer drug targets through the selection of growth-inhibitory genetic suppressor elements. Cancer Cell. 2003;4:41–53. doi: 10.1016/s1535-6108(03)00169-7. [DOI] [PubMed] [Google Scholar]
- 45.Grunberg J, Novak-Hofer I, Honer M, et al. In vivo evaluation of 177Lu-and 67/64Cu-labeled recombinant fragments of antibody chCE7 for radioimmunotherapy and PET imaging of L1-CAM–positive tumors. Clin Cancer Res. 2005;11:5112–5120. doi: 10.1158/1078-0432.CCR-05-0227. [DOI] [PubMed] [Google Scholar]
- 46.Kameda K, Shimada H, Ishikawa T, et al. Expression of highly polysialylated neural cell adhesion molecule in pancreatic cancer neural invasive lesion. Cancer Lett. 1999;137:201–207. doi: 10.1016/s0304-3835(98)00359-0. [DOI] [PubMed] [Google Scholar]
- 47.Yoshino-Yasuda I, Kobayashi K, Akiyama M, et al. Caldecrin is a novel-type serine protease expressed in pancreas, but its homologue, elastase IV, is an artifact during cloning derived from caldecrin gene. J Biochem (Tokyo) 1998;123:546–554. doi: 10.1093/oxfordjournals.jbchem.a021971. [DOI] [PubMed] [Google Scholar]