Abstract
Serine hydrolase KIAA1363 is an acetyl monoalkylglycerol ether (AcMAGE) hydrolase involved in tumor cell invasiveness. It is also an organophosphate (OP) insecticide-detoxifying enzyme. The key to understanding these dual properties was the use of KIAA1363 +/+ (wildtype) and −/− (gene deficient) mice to define the role of this enzyme in brain and other tissues and its effectiveness in vivo in reducing OP toxicity. KIAA1363 was the primary AcMAGE hydrolase in brain, lung, heart and kidney and was highly sensitive to inactivation by chlorpyrifos oxon (CPO) (IC50 2 nM) [the bioactivated metabolite of the major insecticide chlorpyrifos (CPF)]. Although there was no difference in hydrolysis product monoalkylglycerol ether (MAGE) levels in +/+ and −/− mouse brains in vivo, isopropyl dodecylfluorophosphonate (30 mg/kg) and CPF (100 mg/kg) resulted in 23-51 % decrease in brain MAGE levels consistent with inhibition of AcMAGE hydrolase activity. On incubating +/+ and −/− brain membranes with AcMAGE and cytidine-5’-diphosphocholine, the absence of KIAA1363 activity dramatically increased de novo formation of platelet-activating factor (PAF) and lyso-PAF, signifying that metabolically-stabilized AcMAGE can be converted to this bioactive lipid in brain. On considering detoxification, KIAA1363 −/− mice were significantly more sensitive than +/+ mice to ip-administered CPF (100 mg/kg) and parathion (10 mg/kg) with increased tremoring and mortality that correlated for CPF with greater brain acetylcholinesterase inhibition. Docking AcMAGE and CPO in a KIAA1363 active site model showed similar positioning of their acetyl and trichloropyridinyl moieties, respectively. This study establishes the relevance of KIAA1363 in ether lipid metabolism and OP detoxification.
Keywords: acetyl monoalkylglycerol ether, chlorpyrifos, KIAA1363, organophosphate, platelet activating factor
Introduction
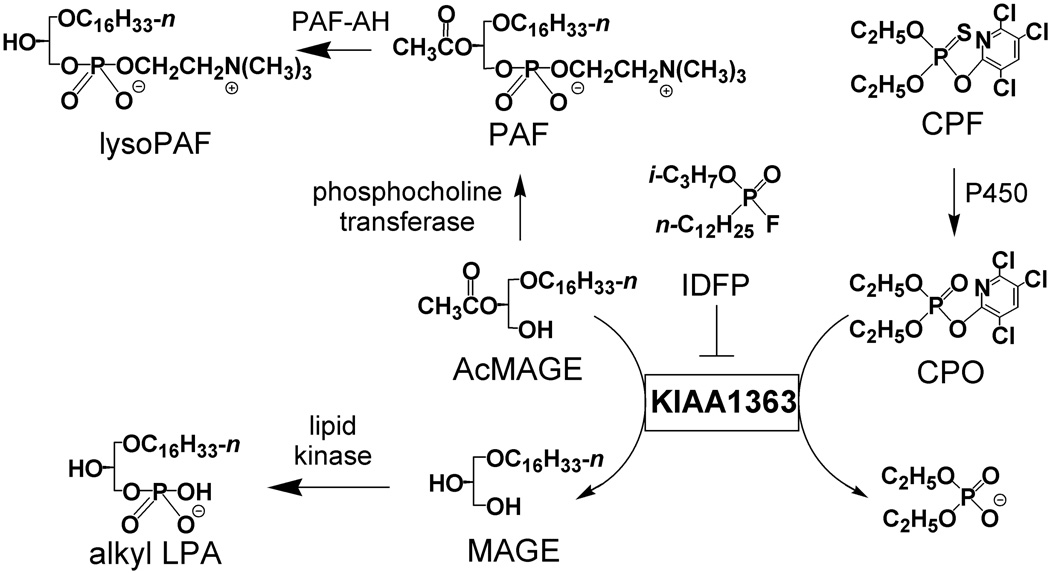
Many enzymes not only catalyze reactions of their endogenous substrates but also degrade xenobiotics. A recent example of this duality is serine hydrolase KIAA1363. It cleaves both acetyl monoalkylglycerol ether (AcMAGE) (Chiang et al., 2006) [the penultimate precursor in the de novo biosynthesis of platelet activating factor (PAF) (Lee et al., 1990; Snyder, 1995)] and organophosphate (OP) insecticides (Nomura et al., 2005, 2006) such as chlorpyrifos oxon (CPO) [the bioactivated metabolite of the major insecticide chlorpyrifos (CPF)] (Fig. 1).
Fig. 1.
Role of KIAA1363 in de novo PAF biosynthesis and OP metabolism. Another presumed KIAA1363 substrate is paraoxon (the 4-nitrophenyl analog of CPO) formed on P450 oxidative desulfuration of parathion.
KIAA1363 was initially recognized as a marker for aggressive tumors with higher activity in invasive breast, melanoma and ovarian cancer cell lines (Jessani et al., 2002) and human breast tumors (Jessani et al., 2005) compared to their noninvasive counterparts. This enzyme in cancer cells was characterized as a hydrolase converting AcMAGE to monoalkylglycerol ether (MAGE) (Chiang et al., 2006). Short-hairpin RNA knockdown of KIAA1363 selectivity decreased cancer cell migration in vitro and tumor growth in vivo possibly through lowering the level of alkyl lysophosphatidic acid (alkyl LPA), a downstream metabolite of MAGE (Chiang et al., 2006). On the other hand, KIAA1363 was also identified as the predominant enzyme for hydrolyzing CPO to diethyl phosphate and possibly also for detoxifying some other OP toxicants in brain possessing the unusual ability to spontaneously reactivate following phosphorylation (Nomura et al., 2005, 2006).
Although a metabolic function for KIAA1363 is proposed in cancer cells, the physiological role is not established in vivo. Therefore, this investigation uses knockout (−/−) mice, ether lipids and OP probes to consider the physiological function of KIAA1363 in brain and other tissues and its potential relevance and effectiveness in vivo as an OP detoxification enzyme.
Materials and methods
Chemicals
[3H-acetyl]AcMAGE was prepared from 10 µM unlabeled PAF (Sigma, St. Louis, MO) supplemented with tracer levels of [3H-acetyl]PAF (Perkin Elmer, Boston, MA) by incubation with phospholipase C from Bacillus cereus (20 units) (Sigma) in 100 mM phosphate buffer pH 7.4 (500 µl) for 30 min at 25°C (Blank et al., 1988). The identity of AcMAGE as the product was confirmed by liquid chromatography/mass spectrometry (LC/MS) analysis of corresponding reactions without added [3H]PAF. Analytical standards of 1-O-hexadecyl-2-acetylglycerol (C16:0 AcMAGE) and 1-O-dodecylglycerol, 1-O-hexadecylglycerol and 1-O-octadecylglycerol (C12:0, C16:0 and C18:0 MAGE, respectively) were from Alexis Biochemicals (San Diego, CA). OP insecticides (CPO, CPF and parathion) were purchased from ChemService (West Chester, PA) and isopropyl dodecylfluorophosphonate (IDFP) was prepared according to Segall et al. (2003).
Animal studies
KIAA1363 −/− and wild-type (+/+) mice on a mixed genetic background (129S6/SvEv and C57BL/6) (Nomura et al., 2005) were backcrossed for four generations. Age-and sex-matched mice were used for toxicology studies with test compounds administered ip using dimethyl sulfoxide (DMSO) (1 µl/g body weight) as the vehicle or DMSO alone as a control. Females were employed for CPF studies and males for parathion experiments, based on availability at the time. There is no evidence of gender-related differences between KIAA1363 +/+ and −/− mice. The degree of tremoring was scored as described later. Statistical analysis for mortality and tremor studies were performed with the Fisher’s exact test and the Mann-Whitney U-test, respectively. Brains were removed and immediately placed on powdered dry ice and held at - 80°C until analyzed.
AcMAGE hydrolase and acetylcholinesterase (AChE) activity assays
Frozen brain homogenized in 100 mM pH 7.4 sodium phosphate (phosphate buffer) was centrifuged at 1000 × g and the supernatant was spun at 100,000 × g to separate the membranes (pellet) from the cytosol (supernatant). AcMAGE hydrolase activity was measured in two ways: radiometric with [3H]AcMAGE for [3H]acetate formation (Blank et al., 1988); LC/MS with unlabeled AcMAGE for MAGE formation. Thus, [3H]AcMAGE (10 µM) was incubated with brain membranes (1.25 µg protein) in buffer (500 µl) for 2 h at 37°C. Reactions were terminated by addition of 2.5 ml of chloroform:methanol:hexane (1.25:1.4:1) followed by 0.83 ml of 100 mM potassium carbonate. After extraction, the upper aqueous layer containing potassium [3H]acetate was removed for scintillation counting. KIAA1363 activity by LC/MS was determined by incubation of 10 µM unlabeled AcMAGE as above, then sequential addition of 10 µM C18:0 MAGE (as an internal standard), ethyl acetate (0.6 ml) and ammonium sulfate (0.1 g). Following extraction, the upper ethyl acetate phase was removed and analyzed by LC/MS quantifying MAGE and AcMAGE. The Hewlett-Packard model 1100 liquid chromatograph equipped with a mass selective detector and a Kromasil C4 5 µm column (250 mm × 4.6 mm, Supelco, Sigma-Aldrich, St. Louis, MO) used a gradient of acetonitrile/water containing 0.05% formic acid beginning with 80% acetonitrile at 0.1 ml/min (to relieve the backpressure associated with injecting ethyl acetate) and increasing to 100% acetonitrile over 20 min at 1 ml/min. Analysis involved retention time (tR) and total positive ion scanning of 200–800 m/z and single ion monitoring (SIM) of C16:0 (m/z 317 parent, tR 14.1 min) and C18:0 (m/z 345 parent, tR 16.9 min) MAGE and C16:0 AcMAGE (m/z 341 dehydro, tR 16.8 min). AChE activity was monitored colorimetrically using 1 mM acetylthiocholine and 50 µg brain membrane protein in phosphate buffer (250 µl) (Nomura et al., 2006) and was the same for KIAA1363 +/+ and −/− mice. Significance of changes in activity or % inhibition was assessed using the Student’s unpaired t-test.
AcMAGE and MAGE analysis
AcMAGE and MAGE levels in brain were analyzed by gas chromatography/mass spectrometry (GC/MS). The frozen brain (420–480 mg) (see above) was homogenized in ethyl acetate (3 ml) and phosphate buffer (3 ml) with 10 nmol of internal standard (C12:0 MAGE). Following centrifugation for complete phase separation, the upper ethyl acetate layer was recovered, dried with anhydrous magnesium sulfate (0.5 g), passed through a nylon syringe filter (13 mm, 0.2 µm) (Pall Life Sciences, East Hills, NY) and evaporated to dryness under nitrogen. The residue was treated with N,O-bis(trimethylsilyl)trifluoroacetamide (Sigma) (200 µl) for 30 min at room temperature with sonication. An aliquot (1 µl) of the trimethylsilyl (TMS) derivatives (Panikashvili et al., 2001) was injected in the splitless mode into an Agilent Technologies model 6890N GC equipped with a DB-XLB fused-silica capillary column (30 m × 0.25 mm × 25 µm). The injector temperature was 250 °C and the helium carrier gas flow rate was 1.2 ml/min. The GC oven temperature was held at 100 °C for 2 min and then programmed to 280 °C at 10 °C /min and held for 10 min. The mass spectra were obtained by electron impact ionization at 70 eV and an ion source temperature of 250 °C. An Agilent 5973A mass selective detector was used for total scan of m/z 35 to 550 for characterization and SIM for quantitation of individual lipids. Normalization was based on brain weight and internal standard. tR values for the TMS derivatives and mass fragmentation patterns [m/z (abundance)] were as follows: C12:0 MAGE; 15.31 min; 389 (3), 314 (1), 257 (9), 205 (100), 147 (60), 133 (44), 117 (66), 73 (84); C16:0 MAGE (Kaneshiro et al., 1998); 18.63 min; 445 (2), 313 (7), 205 (100), 147 (50), 133 (39), 117 (48), 73 (58); C16:0 AcMAGE; 19.25 min; 415 (3), 355 (6), 313 (16), 145 (12), 130 (100), 117 (62), 73 (50). The quantitation limit for both MAGE and AcMAGE was 30 pmol/g brain. Significance of metabolite changes was determined with the Student’s unpaired t-test.
Potential in vitro conversion of AcMAGE to PAF and other products
The possible conversion of AcMAGE to PAF in brain was examined by determining if the relevant enzymes are present using a coupled in vitro system (Goracci and Francescangeli, 1991) (Fig. 1). Cytidine 5’-diphosphocholine (CDP-choline) (the cofactor for phosphocholine transferase to convert AcMAGE to PAF) was added since this endogenous cofactor would not be retained in membrane preparations. Thus, AcMAGE (10 µM) and CDP-choline (100 µM) were incubated with 100 µg of KIAA1363 +/+ or −/− mouse brain membrane protein for 2 h at 37 °C in 0.5 ml of 50 mM Tris pH 8.0 with 20 mM MgCl2 and 20 mM DTT. The metabolites were then extracted by addition of 1 ml 2:1 chloroform:methanol. The mixture was centrifuged and the bottom organic layer was removed. A 10 µl aliquot was then injected in LC/MS. A Finnigan SpectraSYSTEM P4000 Quaternary Gradient pump coupled with a Finnigan LCQ Classic mass spectrometer detector (Thermo Electron, San Jose, CA) and a HAISIL 300 C18 5 µm column (100 mm × 4.6 mm, Higgins Analytical, Mountain View, CA) used a gradient of methanol/10 mM ammonium acetate containing 3% methanol beginning with 80% methanol at 0.1 ml/min (to relieve the backpressure associated with injecting chloroform) and increasing to 100% methanol over 20 min at 0.8 ml/min. Analysis involved tR and SIM of C12:0 (m/z 278.1 ammonia adduct, tR 6.0 min) and C16:0 (m/z 334.1 ammonia adduct, tR 13.0 min) MAGE, C16:0 AcMAGE (m/z 341.1 dehydro, tR 14.4 min), PAF (m/z 524.3 parent, tR 12.2 min) and lyso-PAF (m/z 482.3 parent, tR 11.4 min). C16:0 MAGE, C16:0 AcMAGE and C16:0 PAF levels were quantified using standard curves for authentic samples with C12:0 MAGE as the internal standard. LysoPAF levels were quantified using the standard curve for PAF and expressed as PAF equivalent. Quantitation limits were 1.0 µM for MAGE and AcMAGE and 0.1 µM for lysoPAF and PAF. Significance of metabolite changes was assessed using the Student’s unpaired t-test.
Modeling of enzyme-substrate interactions
The CPO and AcMAGE molecules were built in Maestro 7.5 and imported into AutoDock 3.0 and docked into the KIAA1363 structural model (Nomura et al., 2006) using the Lamarckian Genetic Algorithm.
Results
Tissue distribution of AcMAGE hydrolase activity
KIAA1363 is the primary AcMAGE hydrolase in tumor cells (Chiang et al., 2006) but the distribution of activity is not established in mammalian tissues. AcMAGE hydrolase activity was measured here by radiometric and LC/MS methods with very similar results (Fig. 2), thereby validating the analytical procedures. The contribution of KIAA1363 to AcMAGE hydrolysis was then examined with +/+ and −/− tissues. The major AcMAGE hydrolase was KIAA1363 not only in brain but also in heart, lung and kidney. Brain, heart, lung and kidney membranes from −/− mice analyzed by the two methods had 5–6-fold, 24–28-fold, 6–9-fold, and 2–4-fold less AcMAGE hydrolase activity, respectively, compared to +/+ mice but there was no difference in liver. The liver AcMAGE hydrolase activity is therefore not due to KIAA1363. In vitro loss of AcMAGE with all enzyme preparations corresponded with formation of the hydrolysis product MAGE determined by LC/MS (data not shown).
Fig. 2.
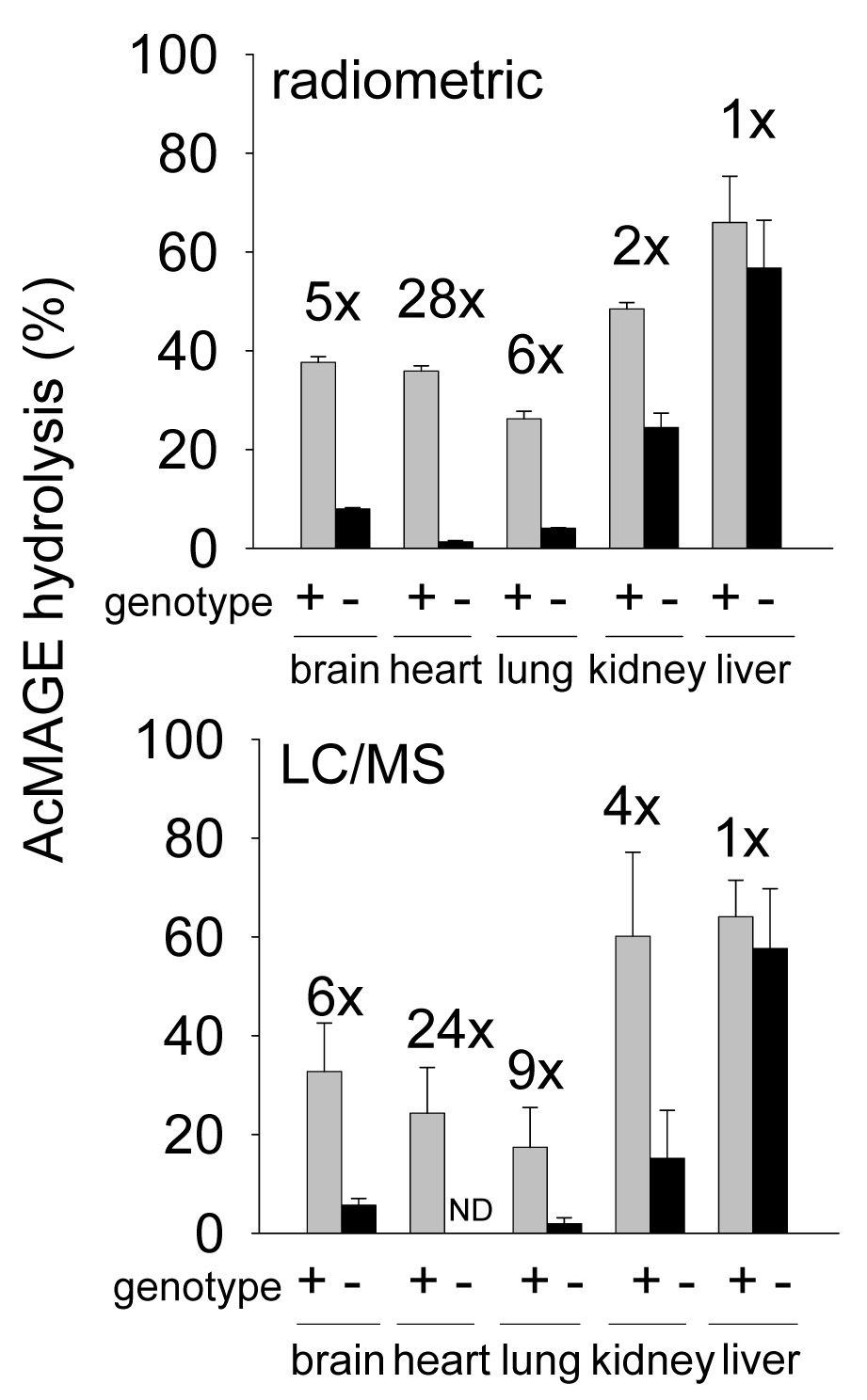
Tissue distribution of AcMAGE hydrolase activity compared by radiometric and LC/MS methods in KIAA1363 +/+ and −/− mice. Membrane protein (1.25 µg) was incubated with 10 µM [3H]AcMAGE or unlabeled AcMAGE for 2 h at 37°C and [3H]acetate or MAGE production determined by radiometric analysis or LC/MS, respectively. All differences in brain, heart, lung and kidney are expressed as fold-change (x) and are significant (p<0.001) between +/+ and −/− membranes. Mean ± SD (n=3). ND= not detected.
Evidence for distinct AcMAGE hydrolases other than KIAA1363
Although KIAA1363 is the predominant AcMAGE hydrolase in brain and heart, there is significant residual activity in −/− tissue membranes not associated with this enzyme (Fig. 2). Therefore, this alternate AcMAGE hydrolase activity in −/− mouse brain and heart was characterized in terms of substrate kinetics and CPO inhibition (Fig. 3). The Km values were higher and the Bmax values were lower in −/− compared to +/+ membranes and the −/− AcMAGE hydrolase activity was 90–2500-fold less sensitive to CPO compared to +/+, thus allowing differentiation of at least two hydrolases. In addition, brain cytosol protein had ~4-fold less AcMAGE hydrolase activity compared to membrane protein in both +/+ and −/− mice with a CPO IC50 of 1300 nM (data not shown).
Fig. 3.
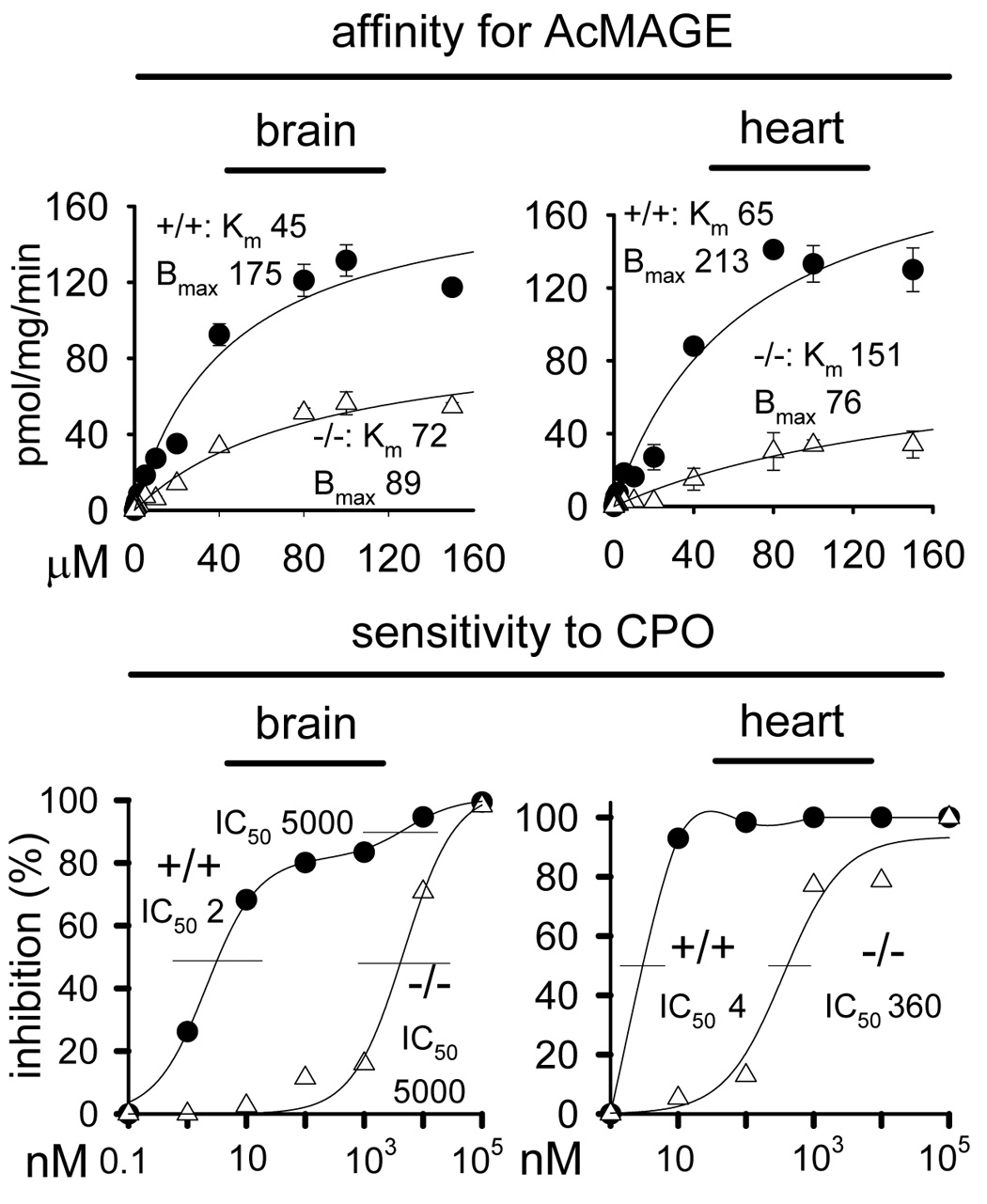
Affinity for AcMAGE and sensitivity to CPO for [3H]AcMAGE hydrolase activity of brain and heart membranes from KIAA1363 +/+ and −/− mice. Assays with −/− preparations have low activity relative to those with +/+ membranes (see Fig. 2). Mean ± SD (n=3). Km is given as µM, Bmax as pmol/mg/min and IC50 as nM.
Inhibition in vivo of brain AcMAGE hydrolase activity
IDFP and CPO are potent in vivo inhibitors of KIAA1363 assayed as [3H]CPO hydrolysis and labeling by a fluorophosphonate-rhodamine probe (Nomura et al., 2005, 2006). They were therefore used in this study to determine possible in vivo inhibition of brain AcMAGE hydrolase activity (Table 1). IDFP at 10 and 30 mg/kg inhibited total [3H]AcMAGE hydrolysis by 28 and 70 %, respectively. CPF at 100 mg/kg also inhibited AcMAGE hydrolysis by 32 % in KIAA1363 +/+ brain and 17 % of the residual activity in −/− brain. Although not detailed here, IDFP at 10 mg/kg almost completely blocked fluorophosphonate-rhodamine labeling of brain KIAA1363.
Table 1.
OP-Induced in Vivo Changes in Brain [3H]AcMAGE Hydrolase Activities and MAGE Levels in KIAA1363 +/+ and −/− Mice
| Genotype and treatment |
|||
|---|---|---|---|
| Compound | Mg/kg | [3H]AcMAGE hydrolysis relative to control [%, mean ± SD (n=3)] | MAGE level a [nmol/g, mean ± SD (n=5–12)] (% relative to control) |
| KIAA1363 +/+ | |||
| Control | 100 | 0.52 ± 0.04 (100) | |
| IDFP | 10 | 72 ± 16 ** | 0.39 ± 0.03 (75) *** |
| 30 | 30 ± 5 ** | 0.36 ± 0.05 (69) *** | |
| CPF | 100 | 68 ± 5 ** | 0.40 ± 0.05 (77) *** |
| KIAA1363 −/− | |||
| Control | 100 | 0.59 ± 0.06 (100) | |
| IDFP | 30 | -- | 0.29 ± 0.05 (49) *** |
| CPF | 100 | 83 ± 5 ** | 0.43 ± 0.05 (73) ** |
AcMAGE is not detected in any case at its quantitation limit of 0.03 nmol/g brain.
p<0.01
p<0.001 for significance of differences from the corresponding controls.
Endogenous MAGE levels
To further investigate the physiological role of KIAA1363, endogenous MAGE levels were compared in KIAA1363 +/+ versus −/− brain by GC/MS. C16:0 MAGE was identified by identical tR and mass fragmentation pattern to the standard (Fig. 4). There was no difference in MAGE levels (0.52–0.59 nmol/g) between +/+ and −/− brains (Table 1). AcMAGE levels were below the quantitation limits in all studies.
Fig. 4.
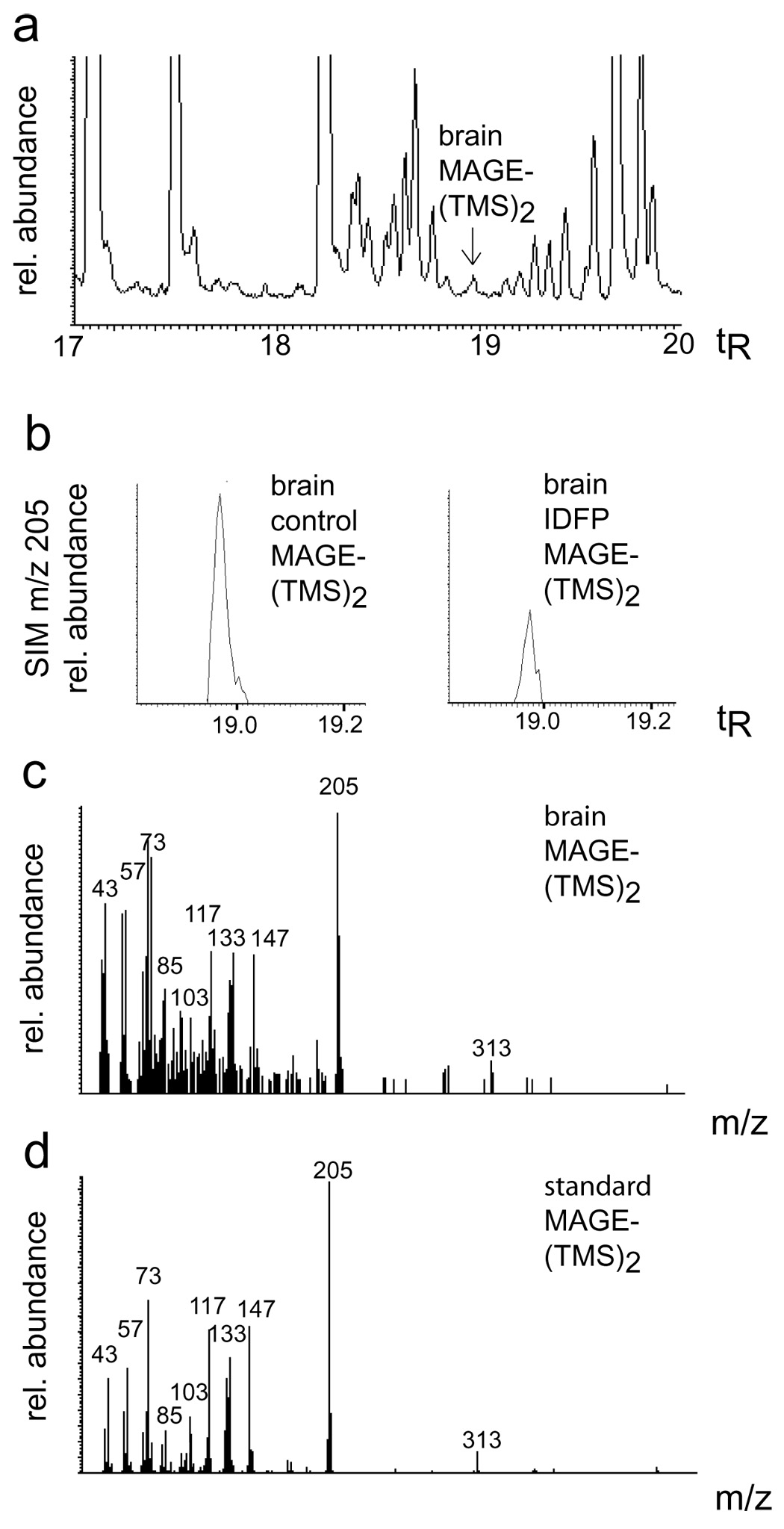
Identification of C16:0 MAGE as bis(trimethylsilyl)[(TMS)2] derivative (tR 18.97 min) in mouse brain showing (a) total positive ion monitoring (200–800 m/z), (b) representative SIM m/z 205 relative abundance for brain control and brain IDFP (30 mg/kg), and fragmentation pattern from brain (c) compared with a standard (d).
OP-induced in vivo changes in brain MAGE levels
IDFP and CPF at ip doses largely inhibiting AcMAGE hydrolase activity were examined for potential effects on MAGE levels (Table 1). MAGE in brain was significantly decreased to 49–77 % in the IDFP- and CPF-treated +/+ and −/− mice (Table 1; Fig. 4b) whereas AcMAGE levels, even with the OP inhibitors, remained below the quantitation limit.
KIAA1363 modulation of in vitro conversion of AcMAGE to PAF
Possible involvement of KIAA1363 as a limiting factor in PAF biosynthesis via the de novo pathway was examined in vitro by coincubation of AcMAGE and CDP-choline with +/+ or −/− brain membranes (Table 2). In the absence of KIAA1363 there was significantly more AcMAGE and less MAGE establishing that this enzyme in vitro is a limiting factor in maintaining AcMAGE levels. Metabolites of the alternative pathway for AcMAGE yielding PAF plus lysoPAF were correspondingly increased in the −/− membranes. When CPO (100 µM) was used to almost completely inhibit KIAA1363, other AcMAGE hydrolases, and PAF-acetylhydrolase (PAF-AH), the conversion of AcMAGE to PAF was the only major pathway.
Table 2.
KIAA1363 Modulation of in Vitro Conversion by Brain Membranes of AcMAGE with CDP-Choline to MAGE, PAF and Lyso-PAF
| Variables |
Analytes [µM, mean ± SD (n=3)] |
||||
|---|---|---|---|---|---|
| Products |
|||||
| KIAA1363 genotype | CPO (µM) | Substrate AcMAGE | MAGE | PAF | Lyso-PAF |
| +/+ | 0 | 1.2 ± 0.2 | 6.3 ± 1.2 | 0.24 ± 0.03 | 0.27 ± 0.02 |
| −/− | 0 | 3.0 ± 0.1 ** | 2.8 ± 0.9 * | 1.3 ± 0.1 ** | 0.74 ± 0.09 ** |
| +/+ | 100 | 4.9 ± 0.9 ** | < 1.0 a | 4.9 ± 0.4 ** | <0.1 a |
| −/− | 100 | 5.8 ± 1.3 * | < 1.0 a | 3.9 ± 0.9 ** | <0.1 a |
All data were below the quantitation limit with 100 µM CPO and above this limit with 0 µM CPO.
p<0.05
p<0.01 for significance of differences from the corresponding controls (−/− versus +/+ with 0 µM CPO; 100 versus 0 µM CPO with +/+ and −/−)
OP toxicity to KIAA1363 +/+ and −/− mice
Two historically important and widely used diethyl phosphorothionate insecticides, CPF and parathion, were examined to determine the effect, if any, of KIAA1363 gene deficiency on sensitivity to poisoning (Table 3). CPF at 100 mg/kg in +/+ mice induced mild cholinergic symptoms including tremoring and lacrimation consistent with the observed 38% AChE inhibition, whereas in −/− mice there was significantly increased tremoring, AChE inhibition and 48 h mortality. Parathion at 10 mg/kg caused dramatically increased tremoring and acute mortality (30 min) in −/− mice compared to +/+ mice with high AChE inhibition in both cases.
Table 3.
CPF and Parathion Toxicity in KIAA1363 +/+ and −/− Mice
| Treatment and genotype | Tremor scorea [mean ± SD (n=13–16)] | Mortality b [% (n=10–12)] | AChE inhibition [%, mean ± SD (n=3–4)] |
|---|---|---|---|
| CPF (100 mg/kg) | |||
| +/+ | 0.69 ± 0.87 | 18 | 38 ± 31 |
| −/− | 4.0 ± 0.6 *** | 92 *** | 92 ± 6 * |
| Parathion (10 mg/kg) | |||
| +/+ | 1.5 ± 1.3 | 0 | 68 ± 45 c |
| −/− | 4.4 ± 0.9 *** | 70 ** | 91 ± 9 c |
Post-treatment time 4 h for CPF and 30 min or prior to death for parathion. Tremors were scored using the following scale: 0= none; 1=infrequent slight tremor; 2=infrequent mild tremor; 3=intermittent to constant moderate tremor; 4=frequent to constant severe tremor; 5=death.
Post-treatment time 48 h for CPF and 30 min for parathion. CPF data at 24 h was also significantly different with 0 and 42 % mortality for +/+ and −/−, respectively.
The difference between −/− and +/+ mice was not significant (p=0.42).
p<0.05
p<0.01
p<0.001 for significance of differences for −/− versus +/+.
Model of KIAA1363-AcMAGE interaction
The KIAA1363-CPO active site model (Nomura et al., 2006) provides the basis for predicting the KIAA1363-AcMAGE interaction (Fig. 5). The acetyl moiety of AcMAGE fits similarly to the trichloropyridinyl moiety of CPO. The acetyl carbonyl carbon and the phosphorus are 4.0 and 2.7 Å, respectively, from the S191 hydroxyl oxygen. D348 and H378 of the catalytic triad are proximally positioned to S191. The diethylphosphoryl group of CPO and alkyl chain of AcMAGE fit well in the “entry gorge.”
Fig. 5.
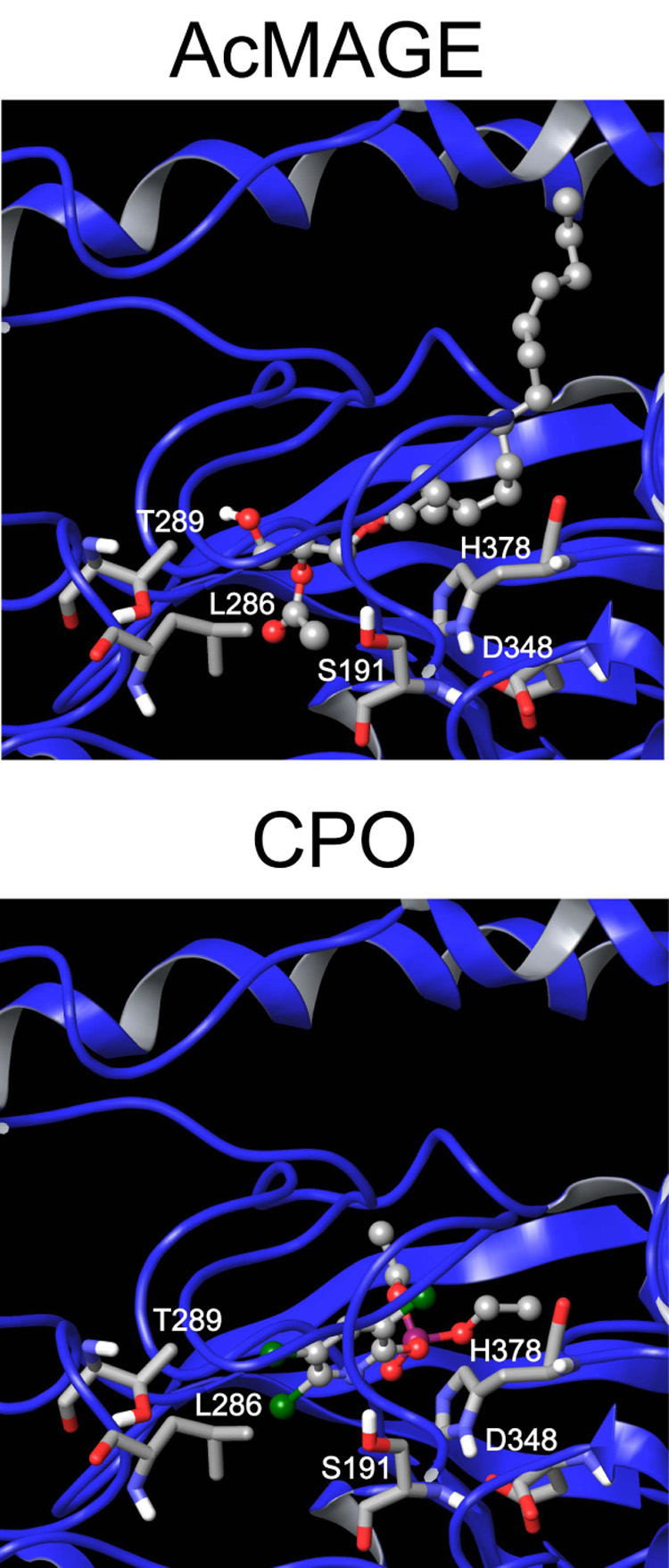
KIAA1363 docked with AcMAGE and CPO showing S191, D348 and H378 of the catalytic triad and T289 and L286 at the inner edge of the leaving group pocket.
Discussion
KIAA1363 is the principal AcMAGE and CPO hydrolase in some but not all tissues
KIAA1363 was first studied in tumor cells (Jessani et al., 2002) with subsequent identification as an AcMAGE hydrolase (Chiang et al., 2006) and then in brain resulting in assignment as the most important CPO hydrolase (Nomura et al., 2005). The present study shows that KIAA1363 is the principal but not the only AcMAGE hydrolase in brain, heart, lung and kidney. The non-KIAA1363 AcMAGE hydrolase activity in brain and heart of −/− mice with lower sensitivity to CPO may partially compensate for the loss of KIAA1363 function. The liver and kidney AcMAGE hydrolase activities are possibly due to the homologous enzyme arylacetamide deacetylase (Nomura et al., 2006) which is highly expressed in these tissues but not in brain (GNF SymAtlas v1.2 as cited by Su et al., 2004). The similar and modest changes in brain MAGE levels on treatment with IDFP and CPF (coupled with the lack of change between KIAA1363 +/+ versus −/− mice) may be due to other less OP-sensitive AcMAGE hydrolases or alternate sources of MAGE [e.g. alkyl LPA (Snyder, 1995)].
KIAA1363 modulation of brain ether lipid levels
AcMAGE levels in vivo were below detection limits so the significance of KIAA1363 can only be evaluated for now by in vitro studies on components of the ether lipid signaling network. This study shows that mouse brain membranes in vitro convert AcMAGE to PAF through the de novo route and that KIAA1363 limits PAF levels by diverting AcMAGE to MAGE (Fig. 1). Thus, there is dramatically increased PAF and lysoPAF formation in KIAA1363 −/− versus +/+ brain membranes coincubated with AcMAGE and CDP-choline. Further, with CPO at 100 µM inhibiting both AcMAGE hydrolases and PAF-AH, the major pathway for AcMAGE in this system is direct conversion to PAF. PAF levels must be tightly regulated in brain due to its highly neuroactive nature in modulating glutamate release and upregulating memory formation at moderate concentrations and causing neuroinflammation and neurotoxicity at higher levels (Bazan, 2003). Therefore, it is not surprising that there are multiple mechanisms for limiting PAF levels from both biosynthetic (KIAA1363 and redundant AcMAGE hydrolases) and degradation (PAF-AH) pathways. Consistent with their roles in neurotransmission and PAF biosynthesis and regulation, three components are highly clustered in the hippocampal formation particularly in CA1-3, i.e. KIAA1363 and PAF-AH isoform 1b, beta 1 in mouse brain (Lein et al., 2007) (Supplemental Figure) and PAF receptor in rat brain (Mori et al., 1996).
The contribution of KIAA1363 to regulating steady-state levels of ether lipid metabolites may be most relevant in cancer cells (Chiang et al., 2006), since they have dramatically increased ether lipid content (Snyder and Wood, 1969). This is supported by carbamate-induced inhibition and short-hairpin RNA knockdown of KIAA1363 shown to drastically reduce MAGE and alkyl LPA levels in cancer cells in a KIAA1363-dependent manner, thereby impairing cancer cell migration and tumor growth in vivo (Chiang et al., 2006).
KIAA1363 contribution to OP insecticide detoxification
Toxicity and AChE inhibition studies with +/+ and −/− mice played a major role in evaluating the contribution of KIAA1363 to OP detoxification. Cholinergic symptoms, mortality and AChE inhibition are less in KIAA1363 +/+ than −/− mice treated with CPF or parathion. Detoxification by KIAA1363 is less evident on direct administration of CPO (Nomura et al., 2006) most likely because of rapidity of action and the steep dose-response curve. The importance of KIAA1363 in detoxification therefore depends on its OP substrate specificity, the pharmacokinetic properties of phosphorothionates and phosphates allowing adequate time for detoxification before AChE inactivation and the activity of other metabolizing enzymes (Tang et al., 2006). Paraoxonase, with a his-his dyad and calcium at its active site, is of particular interest since, along with KIAA1363, it contributes to detoxifying both CPO and paraoxon and also plays a role in lipid metabolism (Harel et al., 2004; Costa et al., 2006).
Dual roles of KIAA1363
KIAA1363 is a proposed therapeutic target for tumor invasiveness since inhibiting this enzyme leads to decreased growth in tumor xenograft models for SKOV-3 and MDA-MB-231 cells (Chiang et al., 2006). Toxicity associated with inhibition of KIAA1363 in non-tumor cells is of potential concern since this enzyme is highly expressed in brain as well as other tissues. The present study indicates that therapeutics specifically targeting KIAA1363 in cancer may not cause adverse effects since −/− mice are normal and their endogenous MAGE levels are not disrupted compared to +/+ mice. KIAA1363 is also the primary CPO-detoxifying enzyme in nervous tissue thereby providing a defense against this OP and possibly a few others in the brain both in vitro (Nomura et al., 2005) and in vivo (this study). The two substrates and dual functions of KIAA1363 are made visually apparent with the similar docking of the seemingly disparate AcMAGE and CPO structures in the active site of the enzyme. KIAA1363 is unusual in its ability to spontaneously reactivate from phosphorylation and it can potentially be produced in soluble form and serve as a structural scaffold for directed evolution to increase its catalytic efficiency for OP detoxification.
Supplementary Material
Acknowledgments
This work was supported by Grants ES008762 (J.E.C.) and CA087660 (B.F.C.) from the National Institutes of Health. We thank our University of California at Berkeley colleagues Kathleen Durkin for help in molecular modeling and Rita Nichiporuk and Ulla Andersen for advice in the mass spectrometry studies.
Abbreviations
- AcMAGE
acetyl monoalkylglycerol ether
- alkyl LPA
alkyl lysophosphatidic acid
- CDP-choline
cytidine-5’-diphosphocholine
- CPF
chlorpyrifos
- CPO
chlorpyrifos oxon
- DMSO
dimethyl sulfoxide
- GC/MS
gas chromatography/mass spectrometry
- IDFP
isopropyl dodecylfluorophosphonate
- LC/MS
liquid chromatography/mass spectrometry
- MAGE
monoalkylglycerol ether
- OP
organophosphate
- PAF
platelet activating factor
- PAF-AH
platelet activating factor-acetylhydrolase
- SIM
single ion monitoring
- TMS
trimethylsilyl
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bazan NG. Synaptic lipid signaling: significance of polyunsaturated fatty acids and platelet-activating factor. J. Lipid Res. 2003;44:2221–2233. doi: 10.1194/jlr.R300013-JLR200. [DOI] [PubMed] [Google Scholar]
- Blank ML, Lee YJ, Cress EA, Snyder F. Stimulation of the de novo pathway for biosynthesis of platelet-activating factor (PAF) via cytidylyltransferase activation in cells with minimal endogenous PAF production. J. Biol. Chem. 1988;263:5656–5661. [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Vitalone A, Furlong CE. Paraoxonase polymorphisms and toxicity of organophosphates. In: Gupta RC, editor. Toxicology of Organophosphate and Carbamate Compounds. London: Elsevier Academic Press; 2006. pp. 247–255. [Google Scholar]
- Goracci G, Francescangeli E. Properties of PAF-synthesizing phosphocholinetransferase and evidence for lysoPAF acetyltransferase activity in rat brain. Lipids. 1991;26:986–991. doi: 10.1007/BF02536489. [DOI] [PubMed] [Google Scholar]
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RBG, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh D-Y, Yates JR, III, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat. Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Kaneshiro ES, Guo Z, Sul D, Kallam KA, Jayasimhulu K, Beach DH. Characterizations of Pneumocystis carinii and rat lung lipids: glyceryl ethers and fatty alcohols. J. Lipid Res. 1998;39:1907–1917. [PubMed] [Google Scholar]
- Lee T-c, Malone B, Blank ML, Fitzgerald V, Snyder F. Regulation of the synthesis of platelet-activating factor and its inactive storage precursor (1-alkyl-2-acyl-sn-glycero-3-phosphocholine) from 1-alkyl-2-acetyl-sn-glycerol by rabbit platelets. J. Biol.Chem. 1990;265:9181–9187. [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Mori M, Aihara M, Kume K, Hamanoue M, Kohsaka S, Shumizu T. Predominant expression of platelet-activating factor receptor in the rat brain microglia. J Neurosci. 1996;16:3590–3600. doi: 10.1523/JNEUROSCI.16-11-03590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Leung D, Chiang KP, Quistad GB, Cravatt BF, Casida JE. A brain detoxifying enzyme for organophosphorus nerve poisons. Proc. Natl. Acad Sci, U.S.A. 2005;102:6195–6200. doi: 10.1073/pnas.0501915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Durkin KA, Chiang KP, Quistad GB, Cravatt BF, Casida JE. Serine hydrolase KIAA1363: toxicological and structural features with emphasis on organophosphate interactions. Chem. Res. Tox. 2006;19:1142–1150. doi: 10.1021/tx060117m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Simeonidou C, Ben-Shabat S, Hanuš L, Breuer A, Mechoulam R, Shohami E. An endogenous cannabinoid (2-AG) is neuroprotective after brain injury. Nature. 2001;413:527–531. doi: 10.1038/35097089. [DOI] [PubMed] [Google Scholar]
- Segall Y, Quistad GB, Casida JE. Cannabinoid CB1 receptor chemical affinity probes: methods suitable for preparation of isopropyl [11,12-3H]dodecylfluorophosphonate and [11,12-3H]dodecanesulfonyl fluoride. Synth. Commun. 2003;33:2151–2159. [Google Scholar]
- Snyder F. Platelet-activating factor: the biosynthetic and catabolic enzymes. Biochem. J. 1995;305:689–705. doi: 10.1042/bj3050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder F, Wood R. Alkyl and alk-1-enyl ethers of glycerol in lipids from normal andneoplastic human tissues. Cancer Res. 1969;29:251–257. [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Rose RL, Chambers JE. Metabolism of organophosphorus and carbamate pesticides. In: Gupta RC, editor. Toxicology of Organophosphate and Carbamate Compounds. London: Elsevier Academic Press; 2006. pp. 127–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.