Abstract
Factor VIIIa is inactivated by a combination of two mechanisms. Activation of factor VIII by thrombin results in a heterotrimeric factor VIIIa that spontaneously inactivates due to dissociation of the A2 subunit. Additionally, factor VIIIa is cleaved by the anticoagulant serine protease, activated protein C, at two cleavage sites, Arg336 in the A1 subunit and Arg562 in the A2 subunit. We previously characterized an engineered variant of factor VIII which contains a disulfide bond between the A2 and the A3 subunits that prevents the spontaneous dissociation of the A2 subunit following thrombin activation. Thus, in the absence of activated protein C, this variant has stable activity following activation by thrombin. To isolate the effects of the individual activated protein C cleavage sites on factor VIIIa, we engineered mutations of the activated protein C cleavage sites into the disulfide bond-cross-linked factor VIII variant. Arg336 cleavage is 6-fold faster than Arg562 cleavage, and the Arg336 cleavage does not fully inactivate factor VIIIa when A2 subunit dissociation is blocked. Protein S enhances both cleavage rates but enhances Arg562 cleavage more than Arg336 cleavage. Factor V also enhances both cleavage rates when protein S is present. Factor V enhances Arg562 cleavage more than Arg336 cleavage as well. As a result, in the presence of both activated protein C cofactors, Arg336 cleavage is only twice as fast as Arg562 cleavage. Therefore, both cleavages contribute significantly to factor VIIIa inactivation.
Hemophilia A is an X-linked bleeding disorder, affecting 1 in 5000 males. It is caused by deficiency of blood coagulation factor (F) VIII and can cause severe bleeding after e.g. surgery and trauma or chronic bleeding into muscles and joints (1). Upon initial production of small amounts of thrombin, the intrinsic coagulation pathway is initiated by a positive feedback loop (2) in which thrombin activates factors V, VIII, and XI. Activated FXI (FXIa) activates FIX to FIXa, which in turn activates FX to FXa. FXa then converts prothrombin to thrombin, leading to fibrin clot formation. FVa and FVIIIa are cofactors for FXa and FIXa, respectively, and enhance the proteolytic function of these enzymes considerably, resulting in rapid production of thrombin.
FVIII is a 2332-amino-acid protein with the domain structure A1-A2-B-A3-C1-C2 (3), which is cleaved within the B domain during secretion and circulates in the blood as a heterodimer bound to von Willebrand factor. Thrombin converts FVIII to its activated form (FVIIIa) by cleavages at positions Arg372, Arg740, and Arg1689 (4). Cleavage at Arg372 and Arg1689 are both required for complete activation of FVIII (5, 6). The cleavage at Arg740 leads to dissociation of the B domain from the rest of the molecule, but this cleavage is not required for FVIIIa activation (5). After cleavage and dissociation of the B domain, the remaining subunits form a non-covalently linked heterotrimer, consisting of the A1 subunit (1–372), the A2 subunit (373–740), and a light chain containing the A3, C1, and C2 domains (1690–2332) (8, 9).
In FVIIIa, the A2 subunit spontaneously dissociates, which inactivates FVIIIa with a half-life of about 2 min (10–12). FVIIIa can also be inactivated through proteolysis by activated protein C (APC),2 which cleaves FVIIIa at Arg336 in the A1 domain and at Arg562 in the A2 domain. The proteolytic activity of APC is enhanced by its non-enzymatic cofactors protein S (13) and factor V (FV) (14). Protein S enhances both FVa (15, 16) and FVIIIa (13, 15) inactivation by APC. FV enhances cleavage of FVIIIa by APC in the presence of protein S in purified systems and prolongs clotting time in plasma-based clotting assays measuring FVIIIa activity (14, 17). This APC cofactor effect of FV was discovered as a result of the thrombosis risk factor FVLeiden (18, 19). FVLeiden has Arg506 mutated to Gln and therefore cannot be cleaved by APC at this position (20, 21). This cleavage is required for APC to fully inactivate FVa (22). However, it is also required for the APC cofactor activity of FV (23).
Cleavage of FVIIIa at Arg336 has been correlated with the inactivation of FVIIIa (9) and is kinetically favored over cleavage at Arg562 in APC-mediated inactivation of FVIIIa (17, 24). As cleavage at Arg336 also induces A2 subunit dissociation, which fully inactivates FVIIIa, this is considered the primary inactivation site (24). The cofactors of APC likely influence the cleavage rates of these individual sites differently. O'Brien et al. (25) monitored FVIIIa proteolysis by APC via Western blot using antibodies specific for the A1 or A2 domains and determined that protein S enhanced both cleavages but had a greater effect on cleavage at Arg562.
In this study, we characterize the individual cleavages in FVIIIa by APC that led to the inactivation of FVIIIa and the effect of the APC cofactors, protein S and factor V, on each cleavage. Rapid inactivation of FVIIIa due to A2-subunit dissociation makes proteolytic inactivation of FVIIIa hard to study. This problem has been addressed in the past by either using very high concentrations of FVIIIa (24) or studying proteolysis of the procofactor FVIII rather than FVIIIa (4). To circumvent this problem, we used a FVIII variant that has a disulfide bond engineered between the A2 and A3 domains (M662C/D1828C FVIII). This disulfide bond prevents dissociation of the A2 domain and stabilizes the molecule (26). This allowed us to determine the effects of APC proteolysis on FVIIIa activity independently of the effects of A2 subunit dissociation. A similar disulfide bond introduced in FVa allowed for characterization of individual APC cleavage sites in FVa, independent of A2 subunit dissociation (27).
After activation by thrombin, M662C/D1828C FVIIIa has FIXa cofactor activity similar to that of wild type FVIIIa (26). As has been reported (24), cleavage at Arg336 was faster than cleavage at Arg562. However, we found that in the absence of A2 subunit dissociation, cleavage at Arg336 did not fully inactivate FVIIIa, whereas cleavage at Arg562 did. We also found that protein S stimulated both FVIIIa cleavages but stimulated Arg562 cleavage somewhat more. FV also enhanced cleavage at Arg562 more than cleavage at Arg336. As a result, in the presence of both protein S and FV, APC cleavage at Arg336 was only 2-fold faster than APC cleavage at Arg562.
MATERIALS AND METHODS
FVIII-deficient plasma was from George King Biomedical Inc. (Overland Park, KS). Recombinant FVIII and the plasmid p25D were from Bayer Corp. (Berkeley, CA). Refacto® B domain-deleted FVIII was from Wyeth Corp. (Collegeville, PA). Thrombin, protein S, and factor IXaβ were purchased from Enzyme Research Laboratories (South Bend, IN). Activated protein C and factor X were purchased from Hematologic Technologies (Essex Junction, VT). Hirudin was purchased from Calbiochem/EMD Biosciences. Bovine brain phosphatidyl serine and bovine brain phosphatidyl ethanolamine were purchased from Sigma. Bovine liver phosphatidyl choline was purchased from Avanti Polar Lipids (Alabaster, AL).
Characterization of FVIII Variants—M662C/D1828C FVIII and other variants were produced essentially as described previously (26, 28). The new FVIII variants were also created in plasmid p25D expressing B domain-deleted FVIII (with amino acids 744–1637 deleted) that already contained the M662C and D1828C mutations (p25D-M662C/D1828C), using the QuikChange mutagenesis kit from Stratagene (La Jolla, CA). Arg336 was mutated to Gln by changing codon CGA to CAG. Arg562 was mutated to Gln by changing codon AGA to CAG. Sequence was verified after subcloning the mutated FVIII sequence into p25D plasmid that did not go through mutagenesis. Plasmids were transfected into HEK293 cells and selected for stable expression with Geneticin, and the FVIII gene was purified from the selected HEK293 clones and sequenced to confirm mutant sequence in the clones expressing FVIII. After purification as described (28), proteins were stored in HEPES-buffered saline (50 mm HEPES, 150 mm NaCl, 4 mm CaCl2, pH 7.4).
FVIII antigen concentration (mg/ml) was determined using the Asserachrom factor VIIIC:Ag enzyme-linked immunosorbent assay kit from Diagnostica Stago (Parsippany, NJ) and the Immubind FVIII enzyme-linked immunosorbent assay kit from American Diagnostica (Greenwich, CT) using the standard protocols with Refacto® FVIII as the standard. FVIII activity concentration (units/ml) was determined using the Chromogenix Coamatic VIII kit (Diapharma Group, Inc., West Chester, OH) with Refacto® FVIII as the standard (13,350 units/mg). Phospholipid vesicles (40% phosphatidyl choline, 20% phosphatidyl serine, 40% phosphatidyl ethanolamine) were prepared as described (29).
FVIII purity and proteolysis by thrombin and APC were monitored with silver-stained gels. For purity analysis, about 25 ng FVIII was loaded per lane. Samples were either reduced with dithiothreitol followed by treatment with iodoacetamide to prevent reoxidation or just treated with iodoacetamide to block all free cysteines and prevent reduction and disulfide shuffling. For proteolysis, FVIII at 5 nm with 25 μm phospholipid vesicles was incubated with 1.6 units/ml IIa for 1 min followed by the addition of 4.8 units/ml hirudin to inactivate the thrombin before adding 25 nm APC/50 nm protein S for a 1-h incubation at room temperature. The buffers used were the same as for the FXase assays (see below) except that no bovine serum albumin was added. Samples were electrophoresed on a 4–12% Bis/Tris gel with MOPS-SDS buffer (Invitrogen), and gels were stained with the SilverXpress kit (Invitrogen).
Functional Assays of FVIIIa Inactivation and Kinetic Properties—The functional properties of FVIIIa were monitored in assays of the FIXa-FVIIIa complex (FXase). We first determined the functional concentration of FVIIIa binding sites for FIXa in FXase assays as described (26). Using the FVIIIa concentrations for each FVIIIa variant determined in this manner, FIXa titrations were performed as described with the exception that FVIII was activated with 0.8 units/ml thrombin for 1 min (four times as much as used previously) followed by 4.8 units/ml hirudin to ensure complete inactivation of the thrombin. Data were fit to a hyperbolic curve to derive K½ values for formation of the FVIIIa-FIXa complex (26). FX titrations were performed as described and fit to the Michaelis-Menten equation to derive kcat and Km (26). The actual FXase complex concentration value used was the concentration of the limiting component (3–5 pm FVIIIa) times the fraction of this component that should be bound, based on the apparent Kd (K½) of the interaction of that respective FVIIIa variant with FIXa at that concentration (4.9 nm).
Assays of FVIIIa inactivation by APC/protein S/FV were performed as follows. FVIII at 5 nm was incubated in FXase buffer (150 mm NaCl, 50 mm HEPES, 5 mm CaCl2, 0.1 mm MnCl2, 5 mg/ml bovine serum albumin, pH 7.4) with 25 μm phospholipid vesicles. Thrombin was added to this mix at 0.8 units/ml (5 nm) to activate the FVIII, and then at 1 min, 4.8 units/ml hirudin was added to inactivate the thrombin. An aliquot was removed for assay in the FXase assay. Then APC and/or protein S and/or FV were added immediately (time 0 in graph). Aliquots were removed over time and added to FIXa and 25 μm phospholipid vesicles. Then FX was added to initiate the FXase reaction as described (26). Final concentrations in the FXase reaction were 150 pm FVIIIa, 2 nm FIXa, and 0.5 μm FX. The reaction was stopped with EDTA, and FXa was measured with substrate S2765 (Chromogenix, Lexington, MA).
The variant M662C/D1828C FVIIIa has two APC cleavage sites, and we assume that FVIIIa inactivation can proceed via two pathways
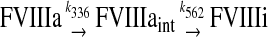 |
REACTION 1 |
and
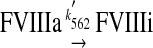 |
REACTION 2 |
In the first pathway, FVIIIa is cleaved at Arg336, resulting in a reaction intermediate with partial activity (FVIIIaint); this cleavage is followed by cleavage at Arg562, which results in inactive FVIIIa (FVIIIi). In the second, alternate pathway, FVIIIa is cleaved first at Arg562, and this cleavage fully inactivates FVIIIa.
These data for M662C/D1828C-FVIIIa inactivation can be fit to a double exponential decay (22, 30). However, we determined that we could not achieve first order conditions in this assay setup; thus, the rate of inactivating cleavages was dependent upon substrate concentration. Therefore, we did not determine cleavage rates from curve fits to M662C/D1828C-FVIIIa inactivation.
M662C/D1828C/R336Q FVIII and M662C/D1828C/R562Q FVIII inactivation are both the result of a single cleavage; thus, inactivation profiles should fit an exponential decay. However, to account for the lack of first order conditions, only the initial rate of activity loss was determined. Thus, for M662C/D1828C/R336Q FVIII and M662C/D1828C/R562Q FVIII, the initial time points that appeared linear on a log scale were fit to a single exponential decay using non-linear least squares regression analysis with the following equation
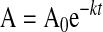 |
(Eq. 1) |
where A is activity of FVIIIa (in percentage), A0 is the initial activity of FVIIIa, k is the rate constant in min–1, and t is time in minutes (24). The data for M662C/D1828C/R562Q (cleavage at Arg336) were fit to an exponential decay to a plateau of 25%, and we then assumed that 25% was equivalent to complete cleavage at Arg336. Data in Table 1 are presented as rate constants of FVIIIa cleavage assuming that loss of activity is proportional to cleavage rate. Initial concentration of FVIIIa was 5 nm, and APC concentration varied as indicated in the figures.
TABLE 1.
Inactivation cleavage rates for cleavages at Arg336 and Arg562 in FVIIIa
Rates are derived from data that were averaged together to generate Figs. 3, 4, 5 according to curve fits described in the methods. Standard errors are shown, and the number of experiments used to determine mean and S.E. is indicated are parentheses. In some cases, values were determined at two APC concentrations. For example, second order rate constants derived from Fig. 4B with 2.5 nm APC, 100 nm PS and from Fig. 5A with 1 nm APC, 100 nm PS were averaged together to determine values in the APC/PS row. ND = not determined.
| CC662-562Q FVIIIa Arg336 cleavage | CC662-336Q FVIIIa Arg562 cleavage | |
|---|---|---|
| nmol FVIIIa/min/nmol APC | nmol FVIIIa/min/nmol APC | |
| APC | 0.10 ± 0.03 (5) | 0.019 ± 0.004 (5) |
| APC/FV | 0.10 ± 0.04 (2) | 0.030 ± 0.007 (2) |
| APC/PS | 0.61 ± 0.30 (6) | 0.13 ± 0.03 (6) |
| APC/PS/FV | 2.6 ± 0.6 (3) | 1.1 ± 0.2 (3) |
FVIIIa inactivation by APC and protein S was also monitored with an APTT assay essentially as described previously, except that FVIII was activated with 0.8 units/ml IIa for 1 min followed by 3.2 units/ml hirudin (28). Time courses were started with the addition of APC and protein S to final concentrations of 1 and 100 nm, respectively. Clotting time was converted to units FVIII activity with a standard curve of Refacto® FVIII corrected for the hirudin in the FVIII sample.
RESULTS
Preparation and Characterization of FVIII Variants—M662C/D1828C FVIII, which contains an engineered disulfide bond between residues 662 and 1828, has been characterized and described (26). This variant was made in the background of B domain-deleted FVIII. Within the construct containing M662C/D1828C FVIII, we introduced individual mutations of the APC cleavage sites. Both sites were mutated from Arg to Gln, generating the constructs, p25D-M662C/D1828C-R336Q and p25D-M662C-D1828C-R562Q. The constructs were expressed in HEK293 cells, and all three proteins were purified as described (26, 28). FVIII concentration and activity were measured as described under “Materials and Methods,” and M662C/D1828C FVIII had specific activity of 7640 units/mg of FVIII, M662C/D1828C/R336Q FVIII had specific activity of 9510 units/mg of FVIII, and M662C/D1828C/R562Q FVIII had specific activity of 14,500 units/mg of FVIII.
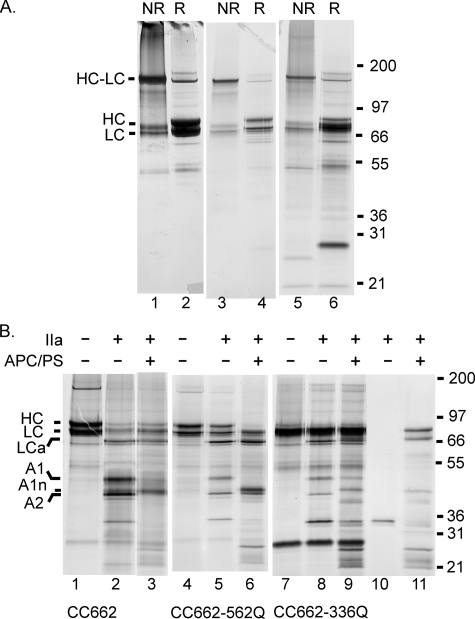
To evaluate the purity of these preparations and to confirm the presence of the disulfide bond, we ran silver-stained gels of M662C/D1828C FVIII, M662C/D1828C/R336Q FVIII, and M662C/D1828C/R562Q FVIII (Fig. 1A). Each protein was electrophoresed in non-reduced (lanes 1, 3, and 5) and reduced form (lanes 2, 4, and 6). Some free heavy chain and light chain were present in the non-reduced lanes. However, a majority of the FVIII was present as a cross-linked heavy chain and light chain band at about 170 kDa at least for M662C/D1828C FVIII and M662C/D1828C/R562Q FVIII. M662C/D1828C/R336Q FVIII was less pure (see below), so we could not estimate the degree of cross-linking of M662C/D1828C/R336Q FVIII. When the proteins were reduced, most of the protein density shifted from cross-linked heavy chain/light chain to separate heavy chain and light chain.
FIGURE 1.
Gel analysis of disulfide bond formation and proteolysis of FVIII variants. A, purified FVIII variants were analyzed by SDS-PAGE with or without reduction by dithiothreitol. Non-reduced lanes (NR) were treated with iodoacetamide before the addition of SDS to prevent disulfide shuffling. Reduced lanes (R) were treated with dithiothreitol while boiling in SDS sample buffer and then with iodoacetamide to prevent reformation of disulfide bonds. Lanes 1 and 2, M662C/D1828C FVIII, lanes 3 and 4, M662C/D1828C/R562Q FVIII, lanes 5 and 6, M662C/D1828C/R336Q FVIII. B, gel analysis of thrombin proteolysis of FVIII variants and APC proteolysis of thrombin-activated FVIIIa. Lanes 1–3, M662C/D1828C, lanes 4–6, M662C/D1828C/R562Q, lanes 7–9, M662C/D1828C/R336Q, lane 10, thrombin alone, and lane 11, thrombin, APC, and protein S. Each variant is shown with no proteolysis (lanes 1, 4, and 7), proteolysis by 1.6 units/ml thrombin (13.5 nm) for 10 min at room temperature (lanes 2, 5, and 8), and proteolysis first by thrombin and then followed by 25 nm APC/50 nm protein S (APC/PS) for 60 min at room temperature (lanes 3, 6, and 9). In panel B, samples were all reduced with dithiothreitol, and all gels were silver-stained. In both panels, molecular weight markers are indicated on the right, and FVIII fragments are indicated on the left. HC-LC, heavy chain cross-linked to light chain; HC, heavy chain; LC, light chain; LCa, activated light chain; A1, A1 subunit; A1n, APC cleaved A1 (residues 1–336); A2, A2 subunit.
M662C/D1828C FVIII and M662C/D1828C/R562Q FVIII were reasonably pure according to silver stain. However, M662C/D1828C/R336Q FVIII did have significant contaminants present. In particular, in the reduced lane (lane 6), there was a band below 31 kDa, and the apparent light chain band was much more intense than the heavy chain band. This was unlike the other two proteins, which suggested that there may be a contaminating protein in M662C/D1828C/R336Q FVIII that ran at about the same mobility as the light chain in the reduced sample. This was possibly because M662C/D1828C/R336Q FVIII was expressed at significantly lower levels in the stably expressing cell line, so purification was less effective. We were not able to improve this purity in two attempts at producing this variant.
To confirm that mutations at Arg336 and Arg562 prevented cleavage by APC, we electrophoresed FVIIIa variants in a reducing gel after proteolysis first by thrombin to activate to FVIIIa and then by APC with protein S (Fig. 1B). Thrombin alone was in lane 10, and thrombin with APC and protein S were in lane 11 to identify bands derived from thrombin, APC, and protein S in the FVIII lanes. M662C/D1828C FVIIIa (lane 3) was cleaved at both Arg336 and Arg562 by APC. Both the A1 subunit was lost (due to Arg336 cleavage) and the A2 subunit was lost (due to Arg562 cleavage). The A1 subunit (see lane 2, 48 kDa, residues 1–372) was replaced by the A1 N-terminal fragment directly above the A2 subunit (43 kDa, residues 1–336). The A2 subunit (41 kDa, residues 373–740) was a sharp band (see lane 2) that was lost completely. The resulting fragments would be around 20 kDa and were not visualized in this gel.
Thrombin-activated M662C/D1828C/R562Q FVIIIa (lane 5) had the typical A1 and A2 subunit bands. Following APC cleavage, the A1 band was gone and was replaced by the A1 N-terminal fragment (lane 6), but the A2 band seen in lane 5 remained in lane 6. Thus, there was no cleavage at Gln562.
M662C/D1828C/R336Q FVIII was not as pure as the others, so the gel was not as clear. In lane 7, in the absence of thrombin or APC proteolysis, the heavy chain and light chain bands were not clearly separated, as was seen in lanes 1 and 4. As stated, this may have been due to a contaminating protein. This hypothesis was supported by the fact that the major band intensity did not decrease upon thrombin or APC reaction in lanes 8 and 9. Additionally, other contaminating bands were seen, and the FVIIIa bands were not as intense as the other FVIIIa variants. However, the kinetic properties of all three variants were similar (Table 2), suggesting that this contaminant did not affect the functional properties of this FVIII preparation.
TABLE 2.
Kinetic constants of the FXase complex containing variant FVIIIa before and after digest with APC
Rates derived from hyperbolic curve fits to titrations of FIXa or FX into the FXase complex as described in the methods, 4–8 experiments averaged together. ND = not detectable.
|
Factor IXa affinity (K½) |
FX activation |
||
|---|---|---|---|
| Km | kcat | ||
| nm | nm | s-1 | |
| CC662 | 0.62 ± 0.16 | 7.8 ± 1.3 | 6.1 ± 2.2 |
| CC662 + APC | ND | ND | ND |
| CC662-336Q | 0.86 ± 0.28 | 4.5 ± 1.0 | 6.4 ± 2.7 |
| CC662-336Q + APC | ND | ND | ND |
| CC662-562Q | 0.55 ± 0.15 | 5.0 ± 0.9 | 4.4 ± 0.9 |
| CC662-562Q + APC | 2.10 ± 0.93 | 26 ± 4 | 2.5 ± 1.2 |
Nevertheless, the A1 and A2 subunit bands could clearly be seen in lane 8 at 48 and 41 kDa. After proteolysis by APC (lane 9), the A1 subunit band at 48 kDa was still present. A band visible at 43 kDa was equivalent to the A1 subunit cleaved at Arg336. However, this was probably a protein present in the mix of thrombin, APC, and protein S (see lane 11). In contrast, the A2 subunit band at 41 kDa was completely gone, indicating complete cleavage at Arg562.
Note that the thrombin, which was visible in this gel at about 35 kDa apparent molecular mass (lanes 2, 5, 8, and 10), decreased in intensity in the lanes containing APC and protein S, although these samples also contain thrombin (lanes 3, 6, 9, and 11). This result was seen consistently in many gels (not shown) and suggests a possible cleavage of thrombin in these reactions. However, we do not have a definite explanation for this phenomenon.
Inactivation of FVIIIa Variants by APC and Protein S—We first tested the inactivation of the FVIII variants in a time course using the APTT clotting assay to monitor FVIIIa activity (Fig. 2). FVIII variants were activated with thrombin. In the absence of APC and protein S, all three variants were fairly stable. M662C/D1828C FVIIIa and M662C/D1828C/R336Q FVIIIa lost about 25% activity, and M662C/D1828C/R562Q lost only 5% activity. This stability was presumably due to the disulfide bond, which prevents A2 subunit dissociation in FVIIIa, as was previously confirmed with M662C/D1828C FVIIIa (26). However, in this experiment, M662C/D1828C FVIIIa was more stable than previously observed (26). We now use more thrombin to activate FVIII than used previously (0.8 units/ml versus 0.2 units/ml) and also use a greater excess of hirudin to assure full inactivation of IIa. Control experiments confirmed that this explains the difference in these current results (see also Fig. 3A). We believe that the FVIII is more fully activated with 0.8 units/ml thrombin.
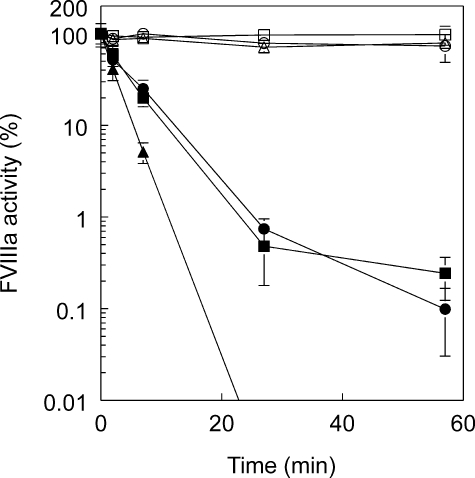
FIGURE 2.
M662C/D1828C FVIIIa variants containing APC cleavage site mutants are partially resistant to APC inactivation. FVIII variants were first activated by reaction with 5.4 nm thrombin followed by excess hirudin, and then at time 0, either 1 nm APC and 100 nm protein S or an equivalent volume of buffer was added. At time points, aliquots were removed and assayed for FVIIIa activity in an APTT coagulation assay with FVIII-deficient plasma. ▵, ▴, M662C/D1828C; ○, •, M662C/D1828C/R336Q; □, ▪, M662C/D1828C/R562Q; open symbols, without APC/protein S, closed symbols, with APC/protein S. Four experiments were averaged together, and standard errors are shown as error bars.
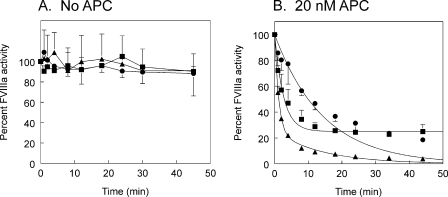
FIGURE 3.
Time courses of inactivation of M662C/D1828C FVIIIa variants by APC. Thrombin-activated FVIIIa activity was monitored over time by FXase assay, a purified system containing FIXa, FX, and phospholipid vesicles. A, FVIIIa alone. B, FVIIIa with 20 nm APC. ▴, M662C/D1828C; •, M662C/D1828C/R336Q; ▪, M662C/D1828C/R562Q. Two to three experiments were averaged together. M662C/D1828C inactivation was fit to a double exponential decay. M662C/D1828C/R336Q and M662C/D1828C/R562Q were fit to single exponential decay curves. Standard deviations are shown as error bars.
When APC and protein S were added to the thrombin-activated FVIIIa variants, M662C/D1828C FVIIIa was readily inactivated even in the absence of A2 subunit dissociation (less than 0.001% activity at 60 min). Both APC cleavage site variants were inactivated more slowly but to similar extents.
Rates of Inactivation by APC Alone—To clarify the profiles of inactivation of M662C/D1828C/R336Q FVIIIa and M662C/D1828C/R562Q FVIIIa, we monitored inactivation of thrombin-activated FVIIIa variants using a FXase assay with purified components. In this assay, all three FVIIIa variants lost only about 10% activity after 45 min in the absence of APC (Fig. 3A). However, in the presence of 20 nm APC without any cofactor, they were all inactivated (Fig. 3B). M662C/D1828C FVIIIa was fully inactivated in the time course. M662C/D1828C/R336Q FVIIIa was inactivated much more slowly, but activity continued to decrease throughout the time course. In contrast to this, M662C/D1828C/R562Q FVIIIa quickly lost about 75% of its activity, but then the activity leveled off.
The inactivation profile of M662C/D1828C-FVIIIa was fit to a double exponential decay (Fig. 3B). However, first order rate conditions did not apply in these experiments. Specifically, the apparent rate of inactivation was dependent upon substrate concentration (data not shown). Therefore, we did not calculate rate constants from the double exponential decay. Furthermore, inactivation profiles of M662C/D1828C/R336Q and M662C/D1828C/R562Q were fit to single exponential decay equations using only the initial time points that appeared linear on a log scale. These curve fits for the two cleavage site variants were used to derive initial cleavage rates for the Arg336 cleavage alone and the Arg562 cleavage alone (Table 1). In M662C/D1828C/R562Q FVIIIa, APC cleaved Arg336 at a rate of 0.1 nmol FVIIIa/min per nmol of APC. APC cleaved Arg562 in M662C/D1828C/R336Q FVIIIa at a rate of 0.019 nmol FVIIIa/min per nmol of APC.
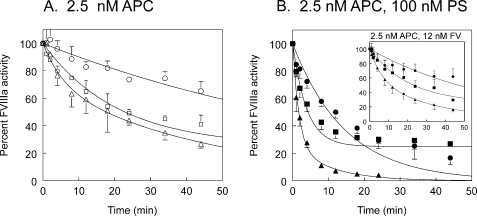
Effect of APC Cofactors on Inactivation Rates—Protein S and FV are both cofactors for APC in the proteolysis of FVIIIa (14, 15). APC alone at a concentration of 2.5 nm did not fully inactivate the FVIIIa variants during a 45-min time course (Fig. 4A). However, protein S significantly enhanced the rate of inactivation of all three FVIIIa variants by APC (Fig. 4B). Protein S increased the rate of Arg336 cleavage about 5-fold in M662C/D1828C/R562Q FVIIIa. Furthermore, protein S increased the rate of Arg562 cleavage 7-fold in M662C/D1828C/R336Q FVIIIa (Table 1). FV had very little APC cofactor effect in the absence of protein S (Fig. 4B, inset). A slight increase in cleavage at Arg562 may or may not be significant. This was consistent with published results (14, 17).
FIGURE 4.
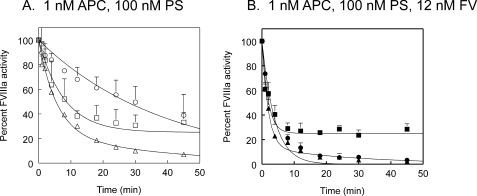
Time courses of inactivation of M662C/D1828C FVIIIa variants by APC and protein S (PS) or FV. Activity of thrombin-activated FVIIIa variants was monitored over time by FXase assay. A, FVIIIa with 2.5 nm APC. B, FVIIIa with 2.5 nm APC and 100 nm protein S. Inset, FVIIIa with 2.5 nm APC and 12 nm FV. ▵, ▴, M662C/D1828C; ○, •, M662C/D1828C/R336Q; □, ▪, M662C/D1828C/R562Q. Two to three experiments were averaged together. M662C/D1828C inactivation was fit to a double exponential decay. M662C/D1828C/R336Q and M662C/D1828C/R562Q were fit to single exponential decay curves. Standard deviations are shown as error bars.
However, in the presence of protein S, FV was a potent cofactor for APC proteolysis of FVIIIa (Fig. 5). With APC concentration further decreased to 1 nm and with 100 nm protein S, 12 nm FV increased the cleavage rate of Arg336 in M662C/D1828C/R562Q FVIIIa by 4-fold over the APC/protein S rate, and it increased the cleavage rate of Arg562 in M662C/D1828C/R336Q FVIIIa by 9-fold over the APC/protein S rate (Table 1). Arg336 was cleaved at a rate of 2.6 nmol FVIIIa/min per nmol of APC, and Arg562 was cleaved at a rate of 1.1 nmol FVIIIa/min per nmol of APC. So although protein S and FV enhanced both cleavage rates, together they served to increase the Arg562 cleavage rate about 60-fold and to increase the Arg336 cleavage rate about 24-fold such that the Arg562 cleavage rate was close to 50% of the Arg336 cleavage rate in the presence of both cofactors. Cleavage rates could be determined at two different APC concentrations for APC alone (Figs. 3B and 4A) and for APC plus protein S (Figs. 4B and 5A). These numbers were not significantly different; thus, the values presented in Table 1 were averaged together over both experiments where possible.
FIGURE 5.
Time courses of inactivation of M662C/D1828C FVIIIa variants by APC, protein S and FV. Thrombin-activated FVIIIa variants were monitored over time by FXase assay. A, FVIIIa with 1 nm APC and 100 nm protein S. B, FVIIIa with 1 nm APC, 100 nm protein S, and 12 nm FV. ▵, ▴, M662C/D1828C; ○, •, M662C/D1828C/R336Q; □, ▪, M662C/D1828C/R562Q. Two to three experiments were averaged together. M662C/D1828C inactivation was fit to a double exponential decay. M662C/D1828C/R336Q and M662C/D1828C/R562Q were fit to single exponential decay curves. Standard deviations are shown as error bars.
Effect of APC Proteolysis on Kinetic Parameters of FVIIIa Cofactor Activity—We determined a functional dissociation constant (K½) for binding of FVIIIa to FIXa with a titration of FIXa into the FXase complex assay with limiting FVIIIa and excess substrate (FX). We determined Km and kcat for the FXase complex with a titration of the substrate, FX, into the FXase complex. M662C/D1828C FVIIIa had a Kd for FIXa of 0.62 nm, Km for FX of 7.8 nm, and kcat of 5.5 s–1 (Table 2). After complete cleavage by APC, no FVIIIa cofactor activity was detectable. M662C/D1828C/R336Q FVIIIa had similar kinetic constants with a Kd of 0.86 nm, Km of 4.5 nm, and kcat of 5.4 s–1. This variant also had no detectable activity in the FXase complex after cleavage by APC (Table 2). In all these inactivation time courses, M662C/D1828C/R562Q FVIIIa maintained a plateau of about 25% activity after apparent complete cleavage at Arg336. APC proteolysis at Arg336 increased the Kd for FIXa from 0.55 to 2.1 nm and increased the Km for FX activation from 5.0 to 26 nm, whereas decreasing the kcat from 3.9 to 1.8 s–11. Therefore, cleavage at Arg562 (in the variant M662C/D1828C/R336Q) fully inactivated FVIIIa, whereas cleavage at Arg336 (in the variant M662C/D1828C/R562Q) only partially inactivated FVIIIa in the absence of dissociation of the A2 subunit.
DISCUSSION
We previously demonstrated that M662C/D1828C FVIII formed a disulfide bond between Cys662 in the A2 domain and Cys1828 in the A3 domain (28). We further determined that M662C/D1828C FVIII had normal functional properties in the FXase complex but was not inactivated via the normal mechanism of spontaneous dissociation of the A2 subunit following activation by thrombin (26). This established that although the disulfide bond prevented A2 subunit dissociation as expected, it did not otherwise distort the FVIIIa protein in any way that impacted the extensive interactions that are involved in the normal cofactor function of FVIIIa in the FXase complex. Additionally, the fact that the addition of this disulfide bond does not change the functional properties of FVIIIa confirms that inactivation mechanisms due to A2 subunit dissociation versus inactivation mechanisms due to proteolysis could be isolated (26).
These kinetic constants were significantly different from our previously published results for M662C/D1828C FVIIIa (26). This was due to a variety of factors. Most notably, we activated FVIII with more thrombin in these experiments than previously (see “Materials and Methods”). We also had to change our sources for phospholipids and factor X, both of which are critical for this assay. We believe that these numbers more accurately reflect the functional properties of the FXase complex. However, this does not negate any earlier conclusions since our previously published data were all internally consistent.
We used a similar disulfide-cross-linked FVa variant to isolate the inactivating effects of APC proteolysis from A2 subunit dissociation that follows APC proteolysis in wild type FVa (27). Therefore, this disulfide bond-stabilized FVIIIa was a valid tool for isolating the effects of the individual APC cleavages on FVIIIa function. Furthermore, because A2 subunit dissociation does not occur, it allows accurate determination of the effects of the APC cofactors, protein S, and factor V at near physiological concentrations.
We confirmed that APC alone cleaves Arg336 faster than it cleaves Arg562. However, the difference in rates that we saw (Arg336 cleavage was 6-fold faster than Arg562 cleavage) was less than that seen in a study of APC cleavage site mutants without a stabilizing disulfide bond. Varfaj et al. (24) determined that Arg336 cleavage by APC took place around 25-fold faster than Arg562 cleavage. This difference may be due to different assay conditions, in particular the extremely high concentration of FVIIIa that was used in previous studies. In general, however, our results confirm these previous results.
In the absence of A2 subunit dissociation, the Arg562 cleavage within the A2 subunit still fully inactivates FVIIIa. This has been suggested in other work and likely happens because Arg562 is located in a FIXa interactive site within residues 558–565 (24, 31). Arg336 cleavage within the A1 subunit does not fully inactivate FVIIIa. The partial loss of activity is attributable to a 4-fold increase in Kd for FIXa as well as a 5-fold increase in Km for FX and a minor decrease in kcat for FX (Table 2). This partial loss of activity due to cleavage at Arg336 was also measured in experiments where cleaved or uncleaved FVIIIa subunits were reconstituted at very high concentrations to recover activity (32). The effect of cleavage at Arg336 on Km is expected because this cleavage releases peptides 337–372, which bind to FX (33), and cleavages by FXa at both Lys36 and Arg336 together result in a 5-fold increase in Km (32). However, this is the first instance where the complete functional effects (Kd, Km and kcat) of cleavage at Arg336 were measured in isolation from other cleavages or from subunit dissociation.
Despite the high level of homology between FVIII and FV, there are significant differences in the mechanisms of inactivation of FVIIIa and FVa as a result of APC cleavages. As was previously established, Arg 336 cleavage is faster than Arg562 cleavage (24). This is in contrast to FVa inactivation, where APC cleavage at Arg506 (homologous to Arg562) is faster than cleavage at Arg306 (homologous to Arg336).
The APC cofactor protein S (and FV in the case of FVIIIa inactivation) enhances the cleavage rate of the slower APC cleavage more than the faster APC cleavage. Thus, protein S primarily enhances Arg306 cleavage in FVa (7), whereas protein S and FV together preferentially enhance Arg562 cleavage in FVIIIa, although in FVIIIa, both cleavage rates are enhanced. The effect for protein S alone that we observed was similar to previous measurements of protein S effect (25). Therefore, as is the case for FVa, in the presence of both endogenous cofactors for APC, the APC cleavages take place at similar rates.
There are similarities in the inactivation of FVa and FVIIIa. In both FVa and FVIIIa, the homologous cleavages in the A1 domain (Arg306 and Arg336) result in dissociation (in the case of FVa) or enhanced dissociation (in the case of FVIIIa) of the A2 subunit, which fully inactivates the cofactor. However, when A2 subunit dissociation is prevented, both cofactors retain significant activity. In both FVa and FVIIIa, the homologous cleavages in the A2 domain (Arg506 or Arg562) result in a greater loss of activity than does the A1 subunit cleavage when A2 subunit dissociation is prevented. In FVa, Arg506 cleavage results in a 40-fold decrease in FXa affinity, whereas Arg306 cleavage only results in a 7-fold decrease in FXa affinity (22, 27). In FVIIIa, Arg562 cleavage fully inactivates FVIIIa independent of subunit dissociation, whereas Arg336 cleavage only partially inactivates FVIIIa independent of subunit dissociation (Table 2).
In summary, these FVIIIa variants have allowed us to isolate and therefore precisely characterize the functional effects of each APC cleavage on FVIIIa independent of FVIIIa inherent instability. In addition, this has allowed a new level of detail in characterization of the role of the APC cofactors, protein S and FV, in these reactions. In the presence of the APC cofactors, which is the normal state in vivo, both cleavages should play a significant role in FVIIIa inactivation.
This work was supported, in whole or in part, by National Institutes of Health Grant HL82588 (to A. J. G.). This work was also supported by grants from the Sam and Rose Stein Endowment Fund, the Factor Foundation, and the Armstrong McDonald Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: APC, activated protein C; MOPS, 4-morpholinepropanesulfonic acid; Bis/Tris, 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol; APTT, activated partial thromboplastin time.
References
- 1.Lenting, P. J., van Mourik, J. A., and Mertens, K. (1998) Blood 92 3983–3996 [PubMed] [Google Scholar]
- 2.Gailani, D., and Broze, G. J., Jr. (1991) Science 253 909–912 [DOI] [PubMed] [Google Scholar]
- 3.Kane, W. H., and Davie, E. W. (1988) Blood 71 539–555 [PubMed] [Google Scholar]
- 4.Amano, K., Michnick, D. A., Moussalli, M., and Kaufman, R. J. (1998) Thromb. Haemostasis 79 557–563 [PubMed] [Google Scholar]
- 5.Pittman, D. D., and Kaufman, R. J. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 2429–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Brien, D. P., Pattinson, J. K., and Tuddenham, E. G. (1990) Blood 75 1664–1672 [PubMed] [Google Scholar]
- 7.Rosing, J., Hoekema, L., Nicolaes, G. A. F., Thomassen, M. C. L. G. D., Hemker, H. C., Varadi, K., Schwarz, H. P., and Tans, G. (1995) J. Biol. Chem. 270 27852–27858 [DOI] [PubMed] [Google Scholar]
- 8.Lollar, P., Knutson, G. J., and Fass, D. N. (1985) Biochemistry 24 8056–8064 [DOI] [PubMed] [Google Scholar]
- 9.Eaton, D., Rodriquez, H., and Vehar, G. A. (1986) Biochemistry 25 505–512 [DOI] [PubMed] [Google Scholar]
- 10.Fay, P. J., Haidaris, P. J., and Smudzin, T. M. (1991) J. Biol. Chem. 266 8957–8962 [PubMed] [Google Scholar]
- 11.Lollar, P., Parker, E. T., and Fay, P. J. (1992) J. Biol. Chem. 267 23652–23657 [PubMed] [Google Scholar]
- 12.Pipe, S. W., Eickhorst, A. N., McKinley, S. H., Saenko, E. L., and Kaufman, R. J. (1999) Blood 93 176–183 [PubMed] [Google Scholar]
- 13.Koedam, J. A., Meijers, J. C. M., Sixma, J. J., and Bouma, B. N. (1988) J. Clin. Investig. 82 1236–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen, L., and Dahlbäck, B. (1994) J. Biol. Chem. 269 18735–18738 [PubMed] [Google Scholar]
- 15.Walker, F. J. (1980) J. Biol. Chem. 255 5521–5524 [PubMed] [Google Scholar]
- 16.Walker, F. J. (1981) J. Biol. Chem. 256 11128–11131 [PubMed] [Google Scholar]
- 17.Lu, D., Kalafatis, M., Mann, K. G., and Long, G. L. (1996) Blood 87 4708–4717 [PubMed] [Google Scholar]
- 18.Dahlbäck, B., Carlsson, M., and Svensson, P. J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahlbäck, B., and Hildebrand, B. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1396–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertina, R. M., Koeleman, B. P. C., Koster, T., Rosendaal, F. R., Dirven, R. J., de Ronde, H., van der Velden, P. A., and Reitsma, P. H. (1994) Nature 369 64–67 [DOI] [PubMed] [Google Scholar]
- 21.Greengard, J. S., Sun, X., Xu, X., Fernández, J. A., Griffin, J. H., and Evatt, B. (1994) Lancet 343 1361–1362 [DOI] [PubMed] [Google Scholar]
- 22.Nicolaes, G. A. F., Tans, G., Thomassen, M. C. L. G. D., Hemker, H. C., Pabinger, I., Varadi, K., Schwarz, H. P., and Rosing, J. (1995) J. Biol. Chem. 270 21158–21166 [DOI] [PubMed] [Google Scholar]
- 23.Thorelli, E., Kaufman, R. J., and Dahlback, B. (1999) Blood 93 2552–2558 [PubMed] [Google Scholar]
- 24.Varfaj, F., Neuberg, J., Jenkins, P. V., Wakabayashi, H., and Fay, P. J. (2006) Biochem. J. 396 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien, L. M., Mastri, M., and Fay, P. J. (2000) Blood 95 1714–1720 [PubMed] [Google Scholar]
- 26.Gale, A. J., Radtke, K. P., Cunningham, M. A., Chamberlain, D., Pellequer, J. L., and Griffin, J. H. (2006) J. Thromb. Haemostasis 4 1315–1322 [DOI] [PubMed] [Google Scholar]
- 27.Gale, A. J., Xu, X., Pellequer, J. L., Getzoff, E. D., and Griffin, J. H. (2002) Protein Sci. 11 2091–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gale, A. J., and Pellequer, J. L. (2003) J. Thromb. Haemostasis 1 1966–1971 [DOI] [PubMed] [Google Scholar]
- 29.Mesters, R. M., Houghten, R. A., and Griffin, J. H. (1991) J. Biol. Chem. 266 24514–24519 [PubMed] [Google Scholar]
- 30.Gale, A. J., Heeb, M. J., and Griffin, J. H. (2000) Blood 96 585–593 [PubMed] [Google Scholar]
- 31.Fay, P. J., Beattie, T., Huggins, C. F., and Regan, L. M. (1994) J. Biol. Chem. 269 20522–20527 [PubMed] [Google Scholar]
- 32.Nogami, K., Wakabayashi, H., Schmidt, K., and Fay, P. J. (2003) J. Biol. Chem. 278 1634–1641 [DOI] [PubMed] [Google Scholar]
- 33.Lapan, K. A., and Fay, P. J. (1997) J. Biol. Chem. 272 2082–2088 [DOI] [PubMed] [Google Scholar]