Abstract
Although it is advantageous for hypoxic cells to inhibit protein synthesis and conserve energy, it is also important to translate mRNAs critical for adaptive responses to hypoxic stress. Because internal ribosome entry sites (IRES) have been postulated to mediate this preferential synthesis, we analyzed the 5 ′-untranslated regions from a panel of stress-regulated mRNAs for m7GTP cap-independent translation and identified putative IRES elements in encephalomyocarditis virus, vascular endothelial growth factor, hypoxia-inducible factors (HIFs) 1α and 2α, glucose transporter-like protein 1, p57Kip2, La, BiP, and triose phosphate isomerase transcripts. However, when capped and polyadenylated dicistronic RNAs were synthesized in vitro and transfected into cells, cellular IRES-mediated translation accounted for less than 1% that of the level of cap-dependent translation. Moreover, hypoxic stress failed to activate cap-independent synthesis, indicating that it is unlikely that this is the primary mechanism for the maintenance of the translation of these mRNAs under low O2. Furthermore, although HIF-1α is frequently cited as an example of an mRNA that is preferentially translated, we demonstrate that under different levels and durations of hypoxic stress, changes in newly synthesized HIF-1α and β-actin protein levels mirror alterations in corresponding mRNA abundance. In addition, our data suggest that cyclin-dependent kinase inhibitor p57Kip2 and vascular endothelial growth factor mRNAs are selectively translated by an IRES-independent mechanism under hypoxic stress.
Hypoxic stress is a central component of normal development and physiology as well as the pathology of multiple human diseases (1–4). Cells adapt to low oxygen (O2) levels by altering their mRNA expression and translation profiles (5, 6). Most of the transcriptional effects are mediated by a family of hypoxia inducible factors (HIFs)2 that transactivate genes by binding to hypoxia response elements in their promoters, introns, and/or enhancers (4, 7). In addition to the transcriptional effects mediated by HIF, hypoxic stress is associated with energy starvation (8, 9) and alterations in cell cycle progression (10, 11). Furthermore, whereas global protein synthesis is attenuated under low O2, select mRNAs believed to be involved in the adaptive response to hypoxia are preferentially translated.
Hypoxia-mediated inhibition of general protein synthesis (12) is regulated primarily by the modification of eukaryotic translation initiation factors (eIFs) at two steps; that is, regeneration of the “ternary complex” (eIF2-GTP and met-tRNA) and regulation of the m7GTP cap binding complex eIF4F (eIF4E/eIF4A/eIF4G (13, 14)). The small (40 S) ribosomal subunit is recruited to the mRNA 5′ end by eIF4F. This complex is modulated by mTOR, a conserved Ser/Thr kinase that integrates environmental stimuli such as nutrient availability, energy status, and cellular stress to regulate growth and cell cycle progression (15). Activated mTOR binds to the eIF3 translation initiation complex and phosphorylates ribosomal kinase S61 (S6K1), which then dissociates and phosphorylates downstream targets such as eIF4B and ribosomal protein S6 (rpS6 (16)). Under limited O2, mTOR signaling is inhibited; 4E-BP is hypophosphorylated and binds eIF4E to hinder the assembly of the cap binding complex. The level and duration of O2 deprivation influences a variety of mechanisms of translation inhibition. For example, a biphasic decrease in global translation is observed during anoxic (0.0% O2) stress (14). Decreased translation under acute (1–4 h) anoxia is mediated by phosphorylation of eIF2α, whereas under prolonged anoxia (16 h), translational repression is maintained by eIF4E inhibition via 4E-BP and eIF4E sequestration by 4E-T. In contrast, under hypoxia (0.5–1.5% O2), a delay between rapid phosphorylation changes detected for elongation factor eEF2, S6K1, rpS6, and 4E-BP and decreased protein synthesis is observed (8).
In most cells O2 deprivation causes a delay in cell cycle progression (10, 11) in a HIF-1α- and c-Myc-dependent manner (17, 18). HIF-1α induces cell cycle arrest by functionally counteracting c-Myc and regulating the expression of G1 checkpoint genes. In addition, mTOR signaling has also been linked to cell cycle progression (19, 20). Rapamycin treatment or mTOR depletion affects the cell cycle, and both S6K1 and 4E-BP1 are believed to be mediators of reduced G1 phase progression.
Many genes exhibit distinct patterns of translation under hypoxia (6, 21, 22). Although hypoxia is known to reduce global protein synthesis, a select subset of mRNAs encoding proteins with adaptive functions continues to be translated under low O2. For example, translation of an endothelium-specific receptor tyrosine kinase for angiopoietins (Tie2) has been shown to increase under prolonged hypoxia as assessed by metabolic labeling of human umbilical vein endothelial cells followed by immunoprecipitation with anti-Tie2 antibodies (23). In addition, multiple transcripts such as eIF5, Tax-1-binding protein (TXBP151), activating transcription factor 4 (ATF4), and ATF6 have been shown by microarray analysis to remain associated with high molecular weight polysomes during anoxic stress (6).
Although the majority of translation is cap-dependent, 3–5% of cellular mRNAs are believed to be translated by internal initiation (24–26). In this mechanism the 40 S ribosome is recruited to mRNA in the vicinity of the initiation codon by sequences generally located in the 5′-untranslated region (UTR) called internal ribosome entry sites (IRES). Multiple hypoxia-regulated mRNAs have been reported to contain IRES, and it was proposed that these elements allow mRNAs to continue to be efficiently translated in O2-starved cells when cap-dependent translation is diminished (27–30). Both HIF-1α and vascular endothelial growth factor (VEGF) contain functional IRES based on dicistronic vector assays (27). However, monocistronic transcripts can be generated from cryptic promoters or cryptic splice sites within the 5′-UTR in this assay, confounding the accurate assessment of IRES activity. This was highlighted when a promoter-less dicistronic vector was developed to test the claim of cellular IRES activity in the 5′-UTR of eIF4G (31). Consequently, the existence of cellular IRES and their biological role in cell adaptation to stress remains a controversial field (32).
We have analyzed the influence of hypoxic stress on the translational regulation of multiple mRNAs encoding proteins involved in hypoxic adaptations. In particular, we wished to determine whether IRES elements provided a mechanism for the selective translation of hypoxia-regulated mRNAs. When capped and polyadenylated in vitro synthesized RNAs were analyzed in in vitro and in vivo translation assays, only the encephalomyocarditis virus (EMCV) 5′-UTR exhibited significant IRES-mediated translation. Moreover, hypoxic conditions did not activate VEGF, HIF-1α, or HIF-2α cellular IRES-mediated translation. We directly measured levels of endogenous VEGF, HIF-1α, β-actin, and cyclin-dependent kinase inhibitor p57Kip2 protein in O2-starved HeLa and Hep3B cells and compared the results to analysis of the distribution of these mRNAs along polysomes isolated from HeLa cells cultured under normoxic and hypoxic conditions. Unexpectedly, when decoupled from post-translational degradation, levels of newly synthesized HIF-1α, like β-actin, decreased under low O2 in parallel with the down-regulation of global protein synthesis and decreased HIF-1α mRNA. In contrast, under low O2 increases in newly synthesized VEGF protein mirror significant increases in VEGF mRNA abundance as well as a hypoxia-mediated increase in the translation efficiency of VEGF mRNA. Under low O2, despite attenuated global translation, the synthesis of p57Kip2 appears to be maintained in an IRES-independent manner, and moreover, p57Kip2 may compete more effectively for binding to translating ribosomes.
EXPERIMENTAL PROCEDURES
Cell Culture—NIH3T3, HeLa, and Hep3B cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 4.5 g/ml glucose, l -glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Dicistronic vector constructs and in vitro synthesized dicistronic RNA were transiently transfected into NIH3T3 cells using Lipofectamine 2000 (Invitrogen). NIH3T3 cells were plated at 5 × 104 cells/well of a 24-well plate (Costar). The next day 1 μg of bicistronic vector or 1 μg of in vitro synthesized RNA was transfected/well, and after 4 h cells were split into 2 wells and cultured under normoxic or hypoxic conditions. Upon completion, 20 μl of whole cell lysate was analyzed for Rluc and Fluc activity using the Dual Luciferase Reporter Assay System (Promega).
Metabolic Labeling, Immunoprecipitation, and Western Blot Analysis—Before metabolic labeling cells were starved for methionine and cysteine for 1 h (HeLa cells O2 < 0.1%; Hep3B cells O2 0.5%) or 5 h (Hep3B cellsO2 < 0.1%). Labeling was carried out on exponentially growing cells for 1 h in Dulbecco's modified Eagle's medium without cold methionine or cysteine, 10% dialyzed fetal calf serum, 200 μm deferoxamine mesylate (DFX), and 100 μCi of [35S]methionine and [35S]cysteine per ml. Cells were washed twice with phosphate-buffered saline containing 200 μm DFX and lysed in cell lysis buffer (1% Triton X-100, 25 mm Tris, pH 8, 300 mm NaCl, 200 μm DFX, and 1 tablet of protease inhibitor (Roche Applied Science) per 10 ml at 4 °C. Lysates were clarified by centrifugation for 15 min at 120,000 rpm. Specific activity was determined by trichloroacetic acid precipitation onto Whatman No. 3MM filter paper and liquid scintillation counting. Lysate protein concentration was determined, and 500 μg of protein was used for each immunoprecipitated sample. Lysates were precleared overnight at 4 °C with 30 μg of washed protein G-Sepharose beads (Amersham Biosciences). Beads were pelleted by spinning the lysates at 6000 rpm for 1 min, and supernatants were transferred to Eppendorf tubes containing 30 μg of pre-washed protein G-Sepharose beads and 1.5 μg of antibody (anti-p57Kip2 (BD Pharmingen), anti-HIF-1α antibody (BIOMOL), and anti-β-actin antibody (Abcam)) and incubated on a rocker for 3 h at 4 °C. Beads were pelleted and washed 3× with cell lysis buffer, 20 μl of 2× SDS loading dye were added, and samples were heated to 100 °C for 10 min, resolved on a 10% SDS-polyacrylamide gel, and visualized by phosphorimaging densitometry. Whole cell lysates were prepared, and Western blot analysis was carried out as previously described (33).
Hypoxic Treatments—Exponentially growing cells were cultured under moderate hypoxic (0.5% O2 42 h), acute severe hypoxic (O2 < 0.1% 5 h), and prolonged severe hypoxic (O2 < 0.1% 16 h) conditions with normoxic (21% O2) controls. Moderate hypoxic conditions were achieved in a Ruskinn In vivo 400 work station, and severe hypoxic conditions were achieved in a Bio-Bag Environmental Chamber (BD Biosciences). Media was preconditioned to the correct O2 level in hypoxic experiments.
Plasmid Constructs—The control dicistronic vector construct pRF and pRF vectors with either the VEGF or HIF-1α 5′-UTR were gifts from Dr. G. Goodall, University of Adelaide (27). Primers at the 5′ and 3′ ends of the 5′-UTRs from murine HIF-2α, Glut-1, La, BiP, HMG-box-containing protein 1 (HMGB1), calreticulin, aldolase A, and triose phosphate isomerase (TPI) were used to PCR the respective 5′-UTRs from ES cell RNA (sequences available upon request). A Spe I site was included in the 5′ primer, and an EcoRI site was included in the 3′ primer. PCR products were digested with SpeI and EcoRI and directionally cloned into the SpeI-EcoRI sites in the pRF and promoter-less pRF linker region between the Renilla luciferase (Rluc) and firefly luciferase (Fluc) coding regions.
Dicistronic pSP64 plasmids were constructed as follows. pSP64 (Promega) was HindIII-digested, blunt-ended, digested with XbaI, and then ligated to the EcoRV-XbaI Rluc fragment from pRF. Subsequently, the XbaI IRES Fluc fragments isolated from the corresponding 5′-UTR pRF constructs were cloned into the XbaI-digested pSP64 Rluc plasmid.
In Vitro Synthesis of Dicistronic RNA—Dicistronic pSP64 constructs were linearized by digestion with EcoRI. Capped and polyadenylated transcripts were synthesized, DNase-treated, and precipitated using the mMessage mMachine large scale RNA protocol (Ambion). A 4:1 ratio of cap analog m7G(5′)ppp(5′)G:GTP was used.
In Vitro Translation—EMCV, VEGF, HIF-1α, and HIF-2α in vitro synthesized dicistronic RNA was translated in rabbit reticulocyte lysates (RRL; Promega) according to the protocol supplied by the manufacturer. 0.1 μg of in vitro synthesized transcript was added to a 50-μl reaction, and upon completion 2.5 μl of the lysate was then analyzed for Rluc and Fluc activity using the Dual Luciferase Reporter Assay System (Promega).
VEGF ELISA—The concentration of human VEGF protein in cell culture supernatants was determined by ELISA assay (R&D Systems). One hour before collecting the supernatant the media was changed so that only newly synthesized VEGF would be assayed. Preconditioned media was used for hypoxic experiments.
Quantitative RT-PCR—Total RNA was isolated from cells using the Trizol reagent protocol (Invitrogen). Polysome RNA was extracted with acidic phenol from sucrose fractions and ethanol-precipitated with 15 μg of glycogen. cDNA was prepared using the Superscript First-strand Synthesis System for RT-PCR (Invitrogen). Quantitative RT-PCR (QRT-PCR) analysis was performed in an Applied Biosystems 7900HT Sequence Detection System with amplification quantified by SYBR green (VEGF, HIF-1α, and β-actin) and Taqman (p57Kip2). Sequences are available upon request.
Polysome Analysis—Polysome analysis was carried out as described previously (34). Ten to 50% linear sucrose gradients containing 100 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, and 20 mm HEPES, pH 7.4, were prepared in 12-ml Beckman ultracentrifuge tubes with a two-chamber gradient mixer. Normoxic control and hypoxic HeLa cells were incubated with cycloheximide (100 μg/ml, freshly prepared in ethanol) for 15 min before harvesting. Cells were washed and then lysed on ice by treatment with 500 μl of ice-cold TMK100 lysis buffer (10 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 100 mm KCl, 2 mm dithiothreitol, 1% Triton X-100, and 100 units of RNase inhibitor (Promega) per ml in diethyl pyrocarbonate-treated water) for 5 min. The nuclei were cleared at 10,000 × g for 10 min at 4 °C, and the supernatants were loaded over the top of sucrose gradients. These gradients were ultracentrifuged at 38,000 rpm for 90 min at 4 °C (Beckman SW41 rotor). Thirteen fractions (750 μl per fraction) were collected into 1.5-ml microcentrifuge tubes containing 70 μl of 10% SDS, and the gradient profile was monitored via UV absorbance at 254 nm with a UA-5 detector (ISCO, Lincoln, NE). Each sample was digested with 8 μl of protease K (20 mg/ml) solution at 37 °C for 30 min and stored at –80 °C before RNA extraction.
RESULTS
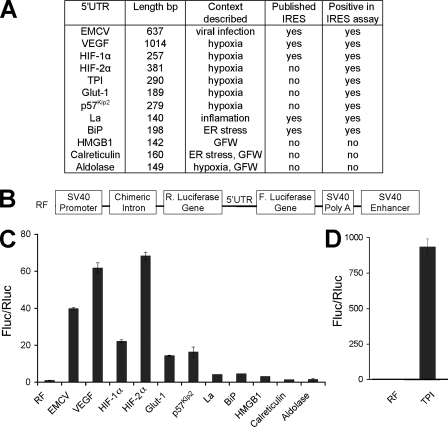
Analysis of the 5′-UTRs of Stress-regulated mRNAs for IRES-mediated Translation—To confirm and extend the observation that hypoxia-regulated mRNAs contain IRES sequences, we cloned murine 5′-UTRs of multiple stress-regulated mRNAs (Fig. 1A) into the dicistronic vector pRF (Fig. 1B) upstream of the second cistron encoding firefly luciferase activity (Fluc). In this vector a bicistronic RNA is transcribed from the SV40 promoter, and Renilla luciferase activity (Rluc) indicates the level of cap-dependent translation, whereas Fluc activity depicts cap-independent translation. The 5′-UTR of EMCV was chosen as a positive control for viral IRES activity, and the immunoglobulin heavy chain-binding protein (BiP) 5′-UTR was used as a positive control for cellular IRES elements (35) because it was the first cellular IRES identified. The La autoantigen 5′-UTR was also selected for analysis because La has been reported to be translated by an IRES-mediated mechanism and acts as an IRES trans-acting factor (36). We analyzed VEGF and HIF-1α 5′-UTRs because these mRNAs are important in the response to hypoxia, and functional IRES elements have been described in their 5′-UTRs based on similar assays (27). In addition, we assayed HIF-2α, glucose transporter-like protein 1 (Glut-1), p57Kip2, HMGB1, calreticulin, aldolase A, and TPI transcripts for IRES elements, because the level of these proteins is elevated upon growth factor withdrawal or hypoxic stress (6, 37). Constructs containing murine 5′-UTRs were transfected into murine NIH 3T3 cells because the original studies characterizing hypoxia-mediated IRES translation were performed in these cells (27). Cell lysate luciferase activity was assayed and expressed as a ratio of Fluc/Rluc activity indicating the relative levels of IRES to cap-dependent translation. The value of Fluc/Rluc activity was normalized to the control vector, pRF, arbitrarily set at 1. The BiP 5′-UTR in this dicistronic vector assay resulted in a 5-fold increase in relative Fluc activity over the pRF control (Fig. 1C). Because BiP was previously reported to contain an IRES element (35), we chose to further characterize 5′-UTR exhibiting Fluc/Rluc activity at this level or higher. We determined that EMCV, VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, La, and TPI appeared to support cap-independent translation at a level equal to or greater than BiP, whereas HMGB1, calreticulin, and aldolase A did not (Fig. 1, C and D).
FIGURE 1.
Analysis of the 5 ′-UTRs of stress-regulated mRNAs for IRES-mediated translation. A, a list of the 5′-UTRs from stress-regulated mRNAs that were analyzed in the dicistronic vector assay for IRES-mediated translation and the results, shown in table form. EMCV was chosen as a positive control for viral IRES activity and BiP and La as positive controls for cellular IRES activity. VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, HMGB1, calreticulin, aldolase A, and TPI transcripts were analyzed for IRES elements because the level of these proteins is elevated under growth factor withdrawal (GFW) or hypoxic stress. ER, endoplasmic reticulum. B, schematic representation of the dicistronic vector RF. C and D, NIH3T3 cells were transiently transfected with dicistronic vector constructs with the listed 5′-UTRs cloned in the polylinker region between the Rluc and Fluc codons and analyzed 42 h later for luciferase activity. Luciferase activity was expressed as the ratio of Fluc/Rluc to indicate the relative level of IRES to cap-dependent translation. The value of Fluc/Rluc was normalized to the control pRF arbitrarily set at 1. TPI is graphed separately due to extremely high Fluc/Rluc levels.
To test the role of IRES as a mechanism for cap-independent translation of select mRNAs involved in the cellular response to hypoxic stress, dicistronic vector constructs containing the 5′-UTRs of EMCV, VEGF, HIF-1α, and HIF-2α mRNAs were transfected into NIH3T3 cells cultured under moderate hypoxic (0.5% O2 42 h; supplemental Fig. 1A) or severe hypoxic (O2 < 0.1% 16 h; supplemental Fig. 1B) conditions with a normoxic control. Levels of Rluc activity dropped under low O2 and served as a control for hypoxia-mediated decreases in global translation. Because IRES elements are postulated to maintain the translation of mRNAs involved in the adaptive response, we expected to observe an increase in cap-independent relative to cap-dependent translation. However, we found no significant differences in the ratio of Fluc/Rluc between normoxic and hypoxic conditions, indicating that IRES activity of the mRNAs analyzed was not regulated by hypoxia (supplemental Fig. 1, A and B). Therefore, we hypothesized that the maintenance of IRES-mediated translation of select mRNAs under low O2 was sufficient for the adaptive response.
Fluc Activity from the VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, La, BiP, or TPI 5′-UTR Is Not the Result of Cellular IRES-mediated Translation—Although the dicistronic vector assay is a standard approach for characterizing cellular IRES activity, cryptic promoters or aberrant splice sites have been observed in potential IRES sequences, and these can generate monocistronic transcripts that encode Fluc activity. To test for the existence of cryptic promoters in the VEGF, HIF-1α HIF-2α, Glut-1, p57Kip2, La, BiP, and TPI 5′-UTRs, we employed a promoter-less dicistronic vector (Fig. 2A) as described by Han and Zhang (31). The SV40 promoter and intron sequence were removed from the dicistronic vector constructs, and these modified pRF plasmids were subsequently transfected into NIH3T3 cells and assayed for Rluc and Fluc activity (Fig. 2C). Results were compared with those obtained from the intact dicistronic vector constructs (Fig. 2B). Without the SV40 promoter, a dicistronic RNA is not synthesized, and only background levels of Rluc and Fluc activity should be observed. As expected, when control promoter-less pRF and EMCV constructs were analyzed, only base-line levels of Fluc and Rluc activity were detected (Fig. 2C). In addition, as predicted, when the VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, La, BiP, and TPI promoterless constructs were analyzed, only base-line levels of Rluc activity were observed. However, similar levels of Fluc activity were obtained in the promoter-less versus the corresponding intact dicistronic vectors (Fig. 2, C versus B). This indicates that the Fluc activities for the VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, La, BiP, and TPI constructs are due to a cryptic promoter in each construct rather than cellular IRES sequences.
FIGURE 2.
Firefly luciferase activity from the VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, La, BiP, or TPI 5 ′-UTR is not the result of cellular IRES-mediated translation. A, schematic representation of dicistronic (RF) and promoter-less constructs (RF–). Dicistronic vector constructs (B) and promoter-less constructs (C) were transiently transfected into NIH3T3 cells and assayed 16 h later for Fluc and Rluc activity.
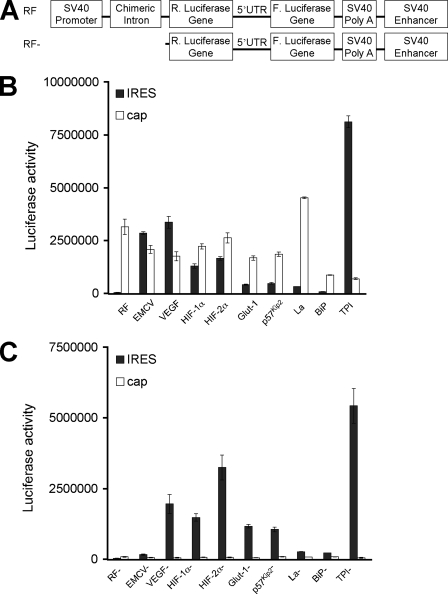
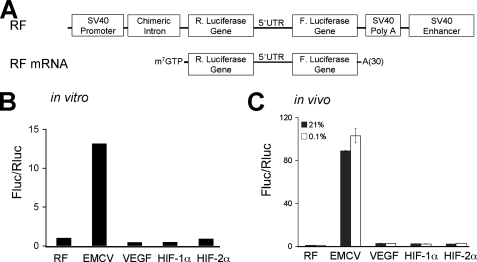
Only the 5′-UTR from EMCV Exhibits Significant IRES-mediated Translation—Although the data in Fig. 2 demonstrate that the 5′-UTRs of VEGF, HIF-1α, and HIF-2α contain cryptic promoters, it remained possible that these 5′-UTRs nevertheless contain IRES elements. To determine whether cellular IRES were present in VEGF, HIF-1α, and HIF-2α 5′-UTRs, we analyzed the translation of in vitro synthesized dicistronic RNAs. This method circumvents the aberrant transcriptional regulation observed with dicistronic vector assays (Fig. 2). Capped and polyadenylated RNAs from the dicistronic vector constructs were synthesized from the SP6 promoter (Fig. 3A) and in vitro translated in RRL (Fig. 3B) or introduced into NIH3T3 cells and cultured under normoxic or hypoxic conditions (O2 < 0.1% 6 h; Fig. 3C). Lysates and RRL translation extracts were assayed for luciferase activity, and results are expressed as the ratio of Fluc/Rluc activity and normalized to RF values. Of note, the EMCV IRES directed Fluc activity at 14-fold greater levels than control in RRL and 90-fold greater levels than control in NIH3T3 lysates, and no increase in EMCV IRES translation was observed when cells were cultured under hypoxia. In contrast, the level of VEGF, HIF-1α, and HIF-2α cellular IRES translation was only 1–3-fold over that of control and less than 1% that of the level of cap-dependent translation. Cellular IRES are theorized to allow the efficient translation of select mRNAs under low O2 (38–40). However, we did not observe a significant increase in cellular IRES activity under hypoxic conditions (Fig. 3C). This result demonstrates that the 5′-UTRs of VEGF, HIF-1α, and HIF-2α mRNAs do not direct efficient cap-independent translation under normoxic or hypoxic conditions.
FIGURE 3.
Only the 5 ′-UTR from EMCV exhibits significant IRES-mediated translation. A, schematic representation of dicistronic vector constructs and in vitro transcribed dicistronic RNAs. Dicistronic in vitro transcribed RNAs were translated in RRL (B) and transiently transfected into NIH3T3 cells cultured under normoxic and hypoxic conditions (C). Luciferase activity was expressed as the ratio of Fluc/Rluc to indicate the relative level of IRES to cap-dependent translation. The value of Fluc/Rluc was normalized to the control vector, pRF, that was set at 1.
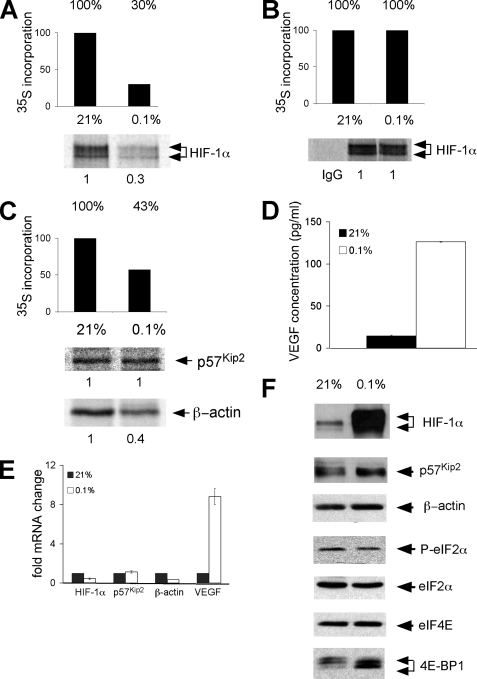
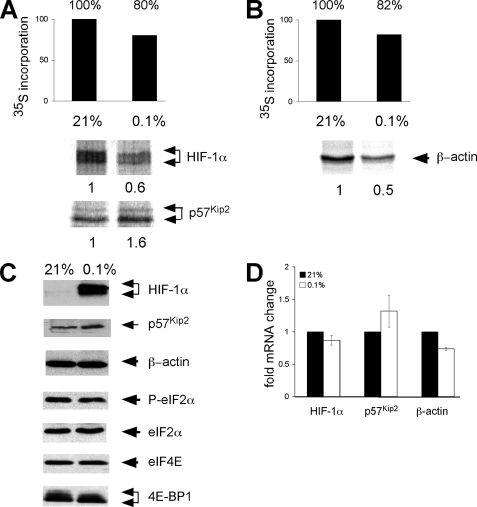
Translational Regulation of Endogenous mRNAs Involved in the Adaptive Response to Low O2—Our results indicate that cap-independent translation is not the primary mechanism for hypoxia-mediated synthesis of the mRNAs examined. As an initial step toward understanding what mechanisms might be important, we assayed levels of global protein synthesis under severe hypoxia (O2 < 0.1% for 16 h). For these experiments HeLa cells were used because translational regulation is well studied in this system (6, 13, 41). In addition, we have performed many of the dicistronic assays in HeLa cells as well as NIH3T3 cells and obtained identical results (data not shown). Cells were cultured under prolonged normoxic and severe hypoxic conditions and pulse-labeled for 1 h with [35S]methionine and [35S]cysteine. The rate of protein synthesis was determined by measuring the incorporation of trichloroacetic acid-precipitable counts (Fig. 4, A–C). Prolonged and severe hypoxia reduced total protein synthesis in HeLa cells by 57–70% and served as an important control for hypoxic treatment.
FIGURE 4.
Translational regulation of endogenous mRNAs involved in the adaptive response to low O2. HeLa cells were cultured under normoxic, moderate hypoxic (B; O2 < 0.5% 2 h) or severe hypoxic (A and C; O2 < 0.1% 16 h) conditions and then pulse-labeled for the last hour with [35S]methionine and [35S]cysteine. The rate of protein synthesis was determined by the incorporation of trichloroacetic acid-precipitable counts. Metabolically labeled HeLa cells were immunoprecipitated with anti-HIF-1α, anti-p57Kip2, and anti-β-actin antibodies. Cells were pretreated and labeled in the presence of DFX to eliminate the post-translational normoxic degradation of HIF-1α subunits. D, HeLa cells were cultured under prolonged normoxia or hypoxia, and then 1 h before analysis the medium was changed so that only newly synthesized VEGF protein was measured. The concentration of VEGF protein in the media was determined by ELISA assay. E, the levels of HIF-1α, p57Kip2, β-actin, and VEGF mRNA in HeLa cells cultured under normoxia or prolonged hypoxia was assessed by QRT-PCR. Results are the average of three experiments, and error bars represent the S.E. F, Western blot analysis showing the accumulation of HIF-1α, p57Kip2, β-actin, phosphorylate (P)-eIF2α, eIF2α, eIF4E, and 4E-BP1 protein under normoxia and prolonged hypoxia. Cells for HIF-1α Western blot analysis were not treated with DFX during culture.
To determine how severe hypoxic stress affects the translation of endogenous HIF-1α mRNA, we pulse-labeled HeLa cells under normoxic and prolonged severe hypoxic culture conditions (O2< 0.1% for 16 h) and immunoprecipitated cell lysates with anti-HIF-1α antibodies (Fig. 4A). Cells were pretreated and labeled in the presence of DFX, an iron-chelating agent, which inhibits the post-translational degradation of HIF-1α. Others have reported that HIF-1α is preferentially translated under hypoxic stress (38, 42–44); consequently, we were surprised to observe that levels of newly synthesized HIF-1α were not maintained under prolonged, severe hypoxia and, instead, decreased in parallel to attenuated global protein synthesis (Fig. 4A). The specificity of the HIF-1α antibody was confirmed by the absence of nonspecific bands in immunoprecipitations performed with the pre-immune control (Fig. 4B). When cells were instead cultured for 2 h under moderate hypoxia (0.5% O2) with DFX in the culture media, we did not observe a decrease in global protein synthesis or change in the translation rate of HIF-1α (Fig. 4B). This control demonstrates that reduced levels of HIF-1α protein (Fig. 4A) correlate with a decrease in global protein synthesis and is not the result of potential off target effects of DFX.
QRT-PCR analysis of polysome gradients from HeLa cells cultured under normoxic and anoxic conditions indicated p57Kip2 mRNA remained associated with translating polysomes under stress, whereas β-actin did not (6, 23). Therefore, we hypothesized that p57Kip2 might be preferentially translated under hypoxia. We analyzed the translation of β-actin as a negative control because its synthesis has been shown to decrease under hypoxic stress (23). Concordantly, we observed that the synthesis of p57Kip2 protein was maintained under prolonged, severe hypoxic stress, but levels of newly synthesized β-actin decreased 60% (Fig. 4C).
To determine whether the synthesis of another critical adaptive protein, VEGF, is maintained under severe hypoxic stress, we examined the levels of VEGF by ELISA assay because multiple anti-VEGF antibodies failed to immunoprecipitate VEGF protein (data not shown). HeLa cells were cultured under normoxic or prolonged, severe hypoxic conditions (O2 < 0.1% for 16 h), and the medium was changed 1 h before analysis so that only newly released VEGF was measured. A 9-fold increase in VEGF protein was observed under prolonged, severe hypoxic versus normoxic conditions (Fig. 4D).
Changes in the level and phosphorylation status of factors critical for the regulation of protein translation initiation were examined by Western blot analysis (Fig. 4F). Under prolonged and severe hypoxic treatment, we did not detect a change in the phosphorylation status of eIF2α. This is consistent with previously published data indicating that the phosphorylation of eIF2α peaks between 1 and 2 h in HeLa cells under anoxic conditions and remains unaltered under severe hypoxia (41). Although we did not see a change in eIF4E protein levels, we did observe a shift in 4E-BP1 to the hypophosphorylated form, consistent with published studies (8, 14). Cells cultured for Western blot analyses were not treated with DFX so that the post-translational stabilization of HIF-1α under low O2 could be observed. This also served as an additional control for hypoxic treatment. As expected, HIF-1α protein levels increased dramatically under hypoxia; in contrast, p57Kip2 protein levels exhibited a small increase under prolonged, severe hypoxia (Fig. 4F). HeLa cells were treated with cycloheximide, and the stability of p57Kip2 protein was measured. We did not detect significant differences between the rate of normoxic and hypoxic p57Kip2 degradation (supplemental Fig. 2). Although β-actin mRNA and newly synthesized protein decreased 60% under severe hypoxic stress (Fig. 4C), total β-actin protein levels remained constant (Fig. 4F), most likely due to the long half-life of β-actin protein.
The distinct expression patterns of HIF-1α, VEGF, p57Kip2, and β-actin proteins could be caused by differential mRNA accumulation, selective translation, or altered protein stability. To further explore the underlying mechanism for these observations, we analyzed mRNA abundance in HeLa cells cultured under normoxic and prolonged, severe hypoxic conditions by QRT-PCR. In cells exposed to O2 < 0.1% for 16 h, HIF-1α and β-actin mRNA levels decreased 55 and 64%, respectively, whereas VEGF mRNA levels increased 8–9-fold (Fig. 4E). We concluded that the increase in newly synthesized VEGF protein under severe hypoxic stress (9-fold; Fig. 4D) correlates with increased VEGF mRNA levels (8–9-fold), whereas decreases in newly synthesized HIF-1α and β-actin protein (70%, 60%) correlate with decreased mRNA levels (55%, 64%). In contrast, the accumulation of p57Kip2 mRNA (15% increase) and the translation of p57Kip2 protein (1:1) is maintained under prolonged, severe hypoxia.
Translational Regulation of Select mRNAs under Acute Hypoxic Stress—In the previous section we characterized the translational regulation of select mRNAs under prolonged, severe hypoxia. However, because translational regulation provides a mechanism for rapid changes in select gene expression, we were interested in testing whether the translation of mRNAs involved in the adaptive response to low O2 was different under acute versus prolonged severe hypoxia. To accomplish this, human hepatoma cells (Hep3B) were cultured under normoxia and acute, severe hypoxia (O2 < 0.1% for 5 h) and analyzed using the same experimental design described in Fig. 4. In cells cultured at O2 < 0.1% for 5 h, global protein synthesis was reduced 18–20% (Fig. 5, A and B), whereas the level of newly synthesized HIF-1α and β-actin protein decreased 40 and 50%, respectively (Fig. 5, A and B). In contrast, the level of newly synthesized p57Kip2 increased 60% (Fig. 5A). Similar results were obtained in HeLa cells cultured under acute, severe hypoxia (data not shown).
FIGURE 5.
Translational regulation of select mRNAs under acute hypoxic stress. A and B, Hep3B cells were cultured under normoxic or hypoxic (O2 < 0.1% 5 h) conditions and then pulse-labeled for the last hour with [35S]methionine and [35S]cysteine. The rate of protein synthesis was determined by the incorporation of trichloroacetic acid-precipitable counts. Pulse-labeled Hep3B cells were immunoprecipitated with anti-HIF-1α, anti-p57Kip2, and anti-β-actin antibodies. Cells were pretreated and labeled in the presence of DFX to eliminate the post-translational normoxic degradation of HIF-1α subunits. C, Western blot analysis showing the accumulation of HIF-1α, p57Kip2, β-actin, phosphorylated (P)-eIF-2α, eIF2α, eIF4E, and 4E-BP1 protein under normoxia and acute hypoxia. Cells for Western blot analysis were not treated with DFX during culture. D, the levels of HIF-1α, p57Kip2, and β-actin mRNA in Hep3B cells cultured under normoxia or acute hypoxia were assessed by QRT-PCR. Results are the average of three experiments, and error bars represent the S.E.
The levels and phosphorylation status of factors critical for translation initiation were examined by Western blot analysis (Fig. 5C). No significant changes in eIF-2α phosphorylation or levels of eIF4E total protein were observed, and as predicted, 4E-BP1 shifted to a more hypophosphorylated form. HIF-1α protein levels increased significantly due to post-translational stabilization, as expected. p57Kip2 and β-actin protein levels remained the same under these conditions. We measured p57Kip2, HIF-1α, and β-actin mRNA levels in Hep3B cells by QRT-PCR (Fig. 5D). HIF-1α and β-actin mRNA levels decreased under acute hypoxia by 13 and 36%, whereas p57Kip2 mRNA levels increased 32%. Again we observed that hypoxia-mediated changes in newly synthesized protein correlated with changes in mRNA levels. In conclusion, the translational regulation of HIF-1α, β-actin, and p57Kip2 mRNAs is the same under acute versus prolonged hypoxia.
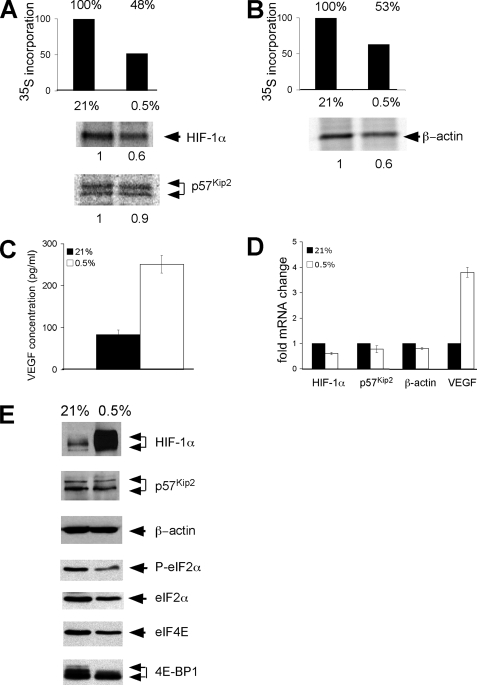
Hypoxia-mediated Changes in the Translation of HIF-1α, p57Kip2, β-Actin, and VEGF Correlate with Changes in mRNA Levels under Both Moderate and Severe Hypoxia—As stated previously, the mechanisms of inhibition of global protein synthesis depend on the level of O2. We, therefore, determined whether different degrees of O2 deprivation influence the synthesis of a subset of mRNAs encoding proteins critical for cellular responses to hypoxic stress. This question was of particular importance to us because in development various disease states and the tumor microenvironment cells are exposed to a wide range of O2 levels. Therefore, we compared the translation of HIF-1α, β-actin, VEGF, and p57Kip2 mRNAs under moderate hypoxia (0.5% O2 42 h) to our previous results obtained under severe hypoxia (O2 < 0.1% 5 and 16 h: Figs. 4 and 5) using the same experimental design (Fig. 6, A and B). After moderate hypoxic stress total protein synthesis was reduced by 47–52% (Fig. 6, A and B). The level of both newly synthesized HIF-1α and β-actin protein decreased 40%, whereas the level of newly synthesized p57Kip2 was maintained (90%; Fig. 6, A and B). Under these conditions newly synthesized VEGF protein increased 2.5-fold (Fig. 6C).
FIGURE 6.
Hypoxia-mediated changes in the translation of HIF-1α, VEGF, and β-actin correlate with changes in mRNA levels under both moderate and severe hypoxia. A and B, Hep3B cells were cultured under normoxic or moderate hypoxic conditions (0.5% O2 42 h) and then metabolically labeled for the last hour with [35S]methionine and [35S]cysteine. The rate of protein synthesis was determined by the incorporation of trichloroacetic acid-precipitable counts. Pulse-labeled Hep3B cells were immunoprecipitated with anti-HIF-1α, anti-p57Kip2, and anti-β-actin antibodies. Cells were pretreated and labeled in the presence of DFX to eliminate the post-translational normoxic degradation of HIF-1α subunits. C, Hep3B cells were cultured under normoxia and prolonged moderate hypoxia, and then 1 h before analysis the media was changed so that only newly synthesized VEFG protein was measured. The concentration of VEGF protein in the media was determined by ELISA assay. D, the levels of HIF-1α, p57Kip2, β-actin, and VEGF mRNA in Hep3B cells cultured under normoxia or prolonged moderate hypoxia were assessed by QRT-PCR. Results are the average of three experiments, and error bars represent the S.E. E, Western blot analysis showing the accumulation of HIF-1α, p57Kip2, β-actin, phosphorylated (P)-eIF-2α, eIF2α, eIF4E, and 4E-BP1 protein under normoxia and prolonged moderate hypoxia. Cells for HIF-1α Western blot analysis were not treated with DFX during culture.
When we analyzed total protein levels and the phosphorylation status of key translation initiation factors upon moderate hypoxic treatment, we observed that the level of total eIF-2α and eIF4E protein dropped with 42 h of hypoxic treatment (Fig. 6E). eIF2α phosphorylation decreased in parallel with total eIF2α protein levels, and 4E-BP1 protein exhibited a shift to a hypophosphorylated form. As expected, HIF-1α protein levels increased under prolonged hypoxic stress, whereas p57Kip2 and β-actin protein levels were unchanged.
We quantitated p57Kip2, VEGF, HIF-1α, and β-actin mRNA levels in Hep3B cells cultured under normoxia or prolonged hypoxia by QRT-PCR (0.5% O2 42 h; Fig. 6D). HIF-1α, p57Kip2, and β-actin mRNA levels decreased 39, 22, and 23% and were similar to the decreases in newly synthesized HIF-1α (40%), β-actin (40%), and p57Kip2 protein (10%) under prolonged hypoxia. Moreover, the change in VEGF protein paralleled the changes in VEGF mRNA (2.5:3.6) under prolonged hypoxia.
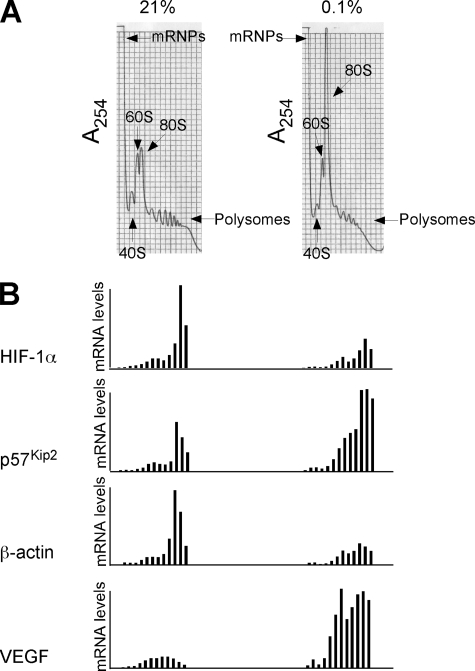
VEGF and p57Kip2 mRNAs Preferentially Associate with Polysomes under Low O2—To complete our investigation of selective translation under low O2 we analyzed the polysome profile of HeLa cells cultured under normoxic and hypoxic (<0.1% O2 16 h) conditions (Fig. 7). When we compared the bulk A254 of sedimenting ribosomes under normoxic and hypoxic conditions, we observed an increase in the 80 S peak and a corresponding collapse in polysome fractions (Fig. 7A). This is consistent with the hypoxia-mediated decrease in global protein synthesis measured by metabolic labeling (Fig. 4, A and C). QRT-PCR analysis of the distribution of HIF-1α and β-actin mRNAs in fractions isolated from normoxic and hypoxic polysome gradients reveals that their sedimentation rate does not change with hypoxic stress. However, there is less overall HIF-1α and β-actin RNA associated with the gradient under low O2 (Fig. 7B). This is in agreement with our previous data that demonstrate decreases in the accumulation of newly synthesized HIF-1α and β-actin proteins are matched by decreases in corresponding mRNA abundance (Fig. 4, HIF-1α 70%:55%; β-actin 60%:64%). In addition, the levels of VEGF mRNA associated with polysomes increased significantly with hypoxic stress, and this result strongly correlates with significant hypoxia-mediated increases in VEGF mRNA and accumulation of newly synthesized VEGF protein (8–9-fold, 9-fold; see Figs. 4 and 7). Furthermore, VEGF mRNA displays a shift toward heavier polysomes under hypoxia. From these data we conclude that hypoxia-mediated VEGF gene regulation includes both a significant increase in mRNA levels as well as an increase in translation efficiency. Under low O2, the accumulation of newly synthesized p57Kip2 protein parallels the levels of p57Kip2 mRNA (100%:115%; Fig. 4); however, p57Kip2 mRNA exhibits a shift toward heavier polysomes. This suggests that p57Kip2 mRNA may compete more effectively for binding to translating ribosomes in hypoxic cells.
FIGURE 7.
Distribution of HIF-1α, p57Kip2, β-actin, and VEGF mRNAs across HeLa cell polysome gradient. A, sucrose gradient profiles of cytosolic extracts from normoxic and hypoxic HeLa cells. The absorbance profile (A254 nm) of the gradient is shown. The top of the gradient is to the left; the positions of absorbance peaks corresponding to pre-ribosomal ribonucleoprotein particles (RNPs), 40 S, 60 S, and 80 S, and polysomes are indicated. B, detection of mRNAs across the HeLa cell polysome gradient. RNAs were extracted from the collected fractions, and the level of HIF-1α, p57Kip2, β-actin, and VEGF mRNAs in each fraction were assessed by QRT-PCR. Values in each fraction were normalized to the first normoxic fraction that was set at 1. Analysis is representative of three independent normoxic and hypoxic polysome gradients.
We began this study with the hypothesis that the translation of a subset of mRNAs critical for hypoxic adaptation was maintained under low O2 by an IRES-mediated mechanism. Instead, our data indicate that under different levels and durations of hypoxic stress, changes in the synthesis of HIF-1α, p57Kip2, β-actin, and VEGF protein mirror alterations in the corresponding mRNA levels. Furthermore, our data suggest that p57Kip2 and VEGF mRNAs are translated more competitively under low O2 in an IRES-independent manner. In conclusion, this study clarifies the translational regulation of a subset of mRNAs important in the response to low O2.
DISCUSSION
To test the theory that mRNAs important in the adaptive response to low O2 were selectively translated in an IRES-dependent manner, we analyzed a panel of hypoxia-regulated mRNAs for IRES-mediated translation using a dicistronic vector assay system. Although viral IRES elements have been well characterized, the existence of cellular IRES has been debated (31, 45). In keeping with previous reports, we found that VEGF, HIF-1α, HIF-2α, Glut-1, p57Kip2, and TPI 5′-UTRs conferred “IRES-mediated translation” in the dicistronic vector assay; however, results obtained from promoter-less dicistronic experiments and direct mRNA transfection of dicistronic transcripts demonstrated that this was due to cryptic promoter activity rather than IRES-mediated translation. During the preparation of this manuscript, Bert et al. (45) reported that 5′-UTRs from VEGF receptor-1, early growth response 1, HIF-1α, VEGF, c-Myc, and X-linked inhibitor of apoptosis protein mRNAs exhibit cryptic promoter activity. To circumvent the aberrant transcriptional regulation observed with dicistronic vector assays, we transfected capped and polyadenylated in vitro synthesized dicistronic mRNAs into NIH3T3 cells. When VEGF, HIF-1α, and HIF-2α 5′-UTRs were analyzed, they displayed low levels of IRES activity (<1% the level of cap-dependent translation). However, because cellular IRES have been postulated to play a central role in the adaptation of the cell to hypoxic stress, it was critical to analyze levels of IRES-mediated translation under low O2. We find that low levels of IRES-mediated activity observed with VEGF, HIF-1α, and HIF-2α 5′-UTRs did not increase under acute, severe stress (<0.1% O2 6 h). IRES-mediated translation is an attractive model for the maintenance of hypoxia-regulated mRNAs under stress. However, given that cap-independent translation levels for VEGF, HIF-1α, and HIF-2α mRNAs are less than 1% that of the level of cap-mediated translation (which drops 20–70% during O2 deprivation), it is unlikely that IRES play a primary role in the translation of these mRNAs under hypoxic stress.
Although it is advantageous for cells to inhibit protein synthesis to conserve energy, it is also important to translate mRNAs involved in the adaptive response to hypoxia. Because mRNAs can have intrinsically different initiation rates under conditions where translation components are limiting, (46) we examined whether this level of regulation was important for a subset of mRNAs important in O2 adaptation. Microarray analysis of polysome profiles from normoxic and anoxic cells has yielded important information regarding selectively translated mRNAs under anoxic stress (6, 14). However, the recruitment of mRNAs onto polysomes could either increase or decrease protein production depending on subsequent post-translational processes, such as translation elongation, peptide chain termination, and protein folding (47). Therefore, we chose to directly study the translation of endogenous HIF-1α, p57Kip2, β-actin, and VEGF mRNAs by metabolic labeling and immunoprecipitation or ELISA assays and compare these results to their distribution on normoxic and hypoxic polysome gradients. Although HIF-1α is frequently cited as an mRNA that is preferentially translated, our studies indicate that, when decoupled from post-translational degradation, the accumulation of newly synthesized HIF-1α protein, like β-actin, decreases under moderate and severe hypoxia. This is supported by the observation that under low O2 less HIF-1α and β-actin mRNA is associated with translating polysomes, and this parallels decreased HIF-1α and β-actin mRNA abundance. In contrast, the sedimentation rate of HIF-1α and β-actin mRNA under normoxic and hypoxic conditions along the polysome gradient appears unchanged. This suggests that the translation efficiency of these mRNAs does not change with hypoxic stress.
Likewise, the translational regulation of VEGF has been widely investigated. We observed that increases in newly synthesized VEGF protein mirrors increases in VEGF mRNA. Under hypoxic stress, more VEGF mRNA is transcribed, its stability increases, and these significant increases in VEGF mRNA levels are consistent with the biologically important increases in VEGF protein levels under low O2. Furthermore, we observed that the level of p57Kip2 and VEGF mRNA associated with translating polysomes increases and shifts to heavier weight fractions under low O2 despite attenuated global protein synthesis. This is in agreement with previous studies which have shown that p57Kip2 and VEGF mRNA remain associated with the translating polysome peak in microarray analysis of normoxic- and anoxic-bound polysomes (6). We suggest that p57Kip2 and VEGF mRNAs may be more competitive in their ability to bind a limiting translation initiation factor(s) in hypoxic cells.
In this manuscript we examined the hypoxia-mediated regulation of a subset of mRNAs involved in the adaptive response to low O2. Two classes of mRNAs were observed. We found evidence of genes that maintain their rate of protein synthesis (HIF-1α and β-actin) and, in addition, genes whose rate of protein synthesis was increased (p57Kip2 and VEGF) under low O2, as accessed by their distribution along normoxic and hypoxic polysome gradients. There are also mRNAs such as glyceraldehyde-3-phosphate dehydrogenase and 5′-terminal oligopyrimidine-RNAs, whose rate of protein synthesis decreases with hypoxic stress (48, 49). These mRNAs display a shift toward free ribosomes along hypoxic compared with normoxic polysome gradients, suggesting that they are less competitive in binding critical translation initiation factors and contribute to decreases in global protein synthesis observed with hypoxic stress (Figs. 4, 5, 6). However, our data suggest that decreased transcription may also contribute to hypoxia-mediated global translation attenuation. This likely contributes to the delay between the rapid phosphorylation changes detected for eEF2, S6K1, rpS6, and 4E-BP and the down-regulation of protein synthesis during hypoxic stress (8). In accordance with these results, inactive RNA polymerase II preinitiation complexes have been observed on a subset of promoters in hypoxic cell extracts (50).
Interestingly, under all the hypoxic conditions studied, we observed hypophosphorylation of 4E-BP1. Significant increases in eIF2α phosphorylation were not detected; however, these studies were not performed under acute anoxic conditions where this level of translational control is prominent (13, 14). Limiting eIF4E activity caused by increased 4E-BP1 binding may be associated with decreased cell proliferation (20), and alterations in eIF4E activity may act as a sensor for cell cycle regulation. Finally, the cyclin-dependent kinase inhibitor p57Kip2 promotes G1 arrest in cell cycle progression (51), and its translation may contribute to adaptive, energy conserving responses to hypoxic stress.
In conclusion, these studies clarify the hypoxia-mediated translational regulation of a subset of mRNAs believed to be important in the adaptive response to low O2. For the mRNAs examined, hypoxia-mediated changes in newly synthesized protein parallel changes in the corresponding mRNA levels. Moreover, we observe this mechanism of regulation irrespective of the cell line or level and duration of hypoxic stress examined. In addition, the distribution ofp57Kip2 and VEGF mRNAs shifts along the polysome gradient to higher weight fractions, which may also contribute to the adaptive response to low O2.
Supplementary Material
Acknowledgments
We thank Dr. Greg Goodall for the gift of the pRF, pRF-VEGF, and pRF-HIF-1α plasmids and Doug Lin, Brian Keith, and all members of the Simon laboratory for thoughtful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 CA104838 (NCI, to M. S. C.), CA1048387-03S1 (to R. M. Y.), and R37 HL65449 (MERIT; to S. A. L.). This work was also supported by the Howard Hughes Medical Institute and the Abramson Family Cancer Research Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: HIF, hypoxia-inducible factor; UTR, untranslated region; IRES, internal ribosome entry sites; eIF, eukaryotic translation initiation factor; RRL, rabbit reticulocyte lysate; QRT-PCR, quantitative reverse phase-PCR; Glut-1, glucose transporter-like protein 1; VEGF, vascular endothelial growth factor; EMCV, encephalomyocarditis virus; DFX, deferoxamine mesylate; TPI, triose phosphate isomerase; ELISA, enzyme-linked immunosorbent assay; HMGB1, high mobility group-box-containing protein 1.
References
- 1.Covello, K. L., Kehler, J., Yu, H., Gordan, J. D., Arsham, A. M., Hu, C. J., Labosky, P. A., Simon, M. C., and Keith, B. (2006) Genes Dev. 20557 –570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaufman, B., Scharf, O., Arbeit, J., Ashcroft, M., Brown, J. M., Bruick, R. K., Chapman, J. D., Evans, S. M., Giaccia, A. J., Harris, A. L., Huang, E., Johnson, R., Kaelin, W., Jr., Koch, C. J., Maxwell, P., Mitchell, J., Neckers, L., Powis, G., Rajendran, J., Semenza, G. L., Simons, J., Storkebaum, E., Welch, M. J., Whitelaw, M., Melillo, G., and Ivy, S. P. (2004) Cancer Res. 643350 –3356 [DOI] [PubMed] [Google Scholar]
- 3.Wouters, B. G., van den Beucken, T., Magagnin, M. G., Lambin, P., and Koumenis, C. (2004) Drug Resist. Updat. 725 –40 [DOI] [PubMed] [Google Scholar]
- 4.Semenza, G. L. (2001) Curr. Opin. Cell Biol. 13167 –171 [DOI] [PubMed] [Google Scholar]
- 5.Wouters, B. G., van den Beucken, T., Magagnin, M. G., Koritzinsky, M., Fels, D., and Koumenis, C. (2005) Semin. Cell Dev. Biol. 16487 –501 [DOI] [PubMed] [Google Scholar]
- 6.Blais, J. D., Filipenko, V., Bi, M., Harding, H. P., Ron, D., Koumenis, C., Wouters, B. G., and Bell, J. C. (2004) Mol. Cell. Biol. 247469 –7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin, W. G., Jr. (2002) Nat. Rev. Cancer 2673 –682 [DOI] [PubMed] [Google Scholar]
- 8.Liu, L., Cash, T. P., Jones, R. G., Keith, B., Thompson, C. B., and Simon, M. C. (2006) Mol. Cell 21521 –531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arsham, A. M., Howell, J. J., and Simon, M. C. (2003) J. Biol. Chem. 27829655 –29660 [DOI] [PubMed] [Google Scholar]
- 10.Gardner, L. B., Li, Q., Park, M. S., Flanagan, W. M., Semenza, G. L., and Dang, C. V. (2001) J. Biol. Chem. 2767919 –7926 [DOI] [PubMed] [Google Scholar]
- 11.Green, S. L., Freiberg, R. A., and Giaccia, A. J. (2001) Mol. Cell. Biol. 211196 –1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pettersen, E. O., Juul, N. O., and Ronning, O. W. (1986) Cancer Res. 464346 –4351 [PubMed] [Google Scholar]
- 13.Koumenis, C., Naczki, C., Koritzinsky, M., Rastani, S., Diehl, A., Sonenberg, N., Koromilas, A., and Wouters, B. G. (2002) Mol. Cell. Biol. 227405 –7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koritzinsky, M., Magagnin, M. G., van den Beucken, T., Seigneuric, R., Savelkouls, K., Dostie, J., Pyronnet, S., Kaufman, R. J., Weppler, S. A., Voncken, J. W., Lambin, P., Koumenis, C., Sonenberg, N., and Wouters, B. G. (2006) EMBO J. 251114 –1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wullschleger, S., Loewith, R., and Hall, M. N. (2006) Cell 124471 –484 [DOI] [PubMed] [Google Scholar]
- 16.Holz, M. K., Ballif, B. A., Gygi, S. P., and Blenis, J. (2005) Cell 123569 –580 [DOI] [PubMed] [Google Scholar]
- 17.Koshiji, M., Kageyama, Y., Pete, E. A., Horikawa, I., Barrett, J. C., and Huang, L. E. (2004) EMBO J. 231949 –1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordan, J. D., Bertout, J. A., Hu, C. J., Diehl, J. A., and Simon, M. C. (2007) Cancer Cell 11335 –347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham, R. T., and Wiederrecht, G. J. (1996) Annu. Rev. Immunol. 14483 –510 [DOI] [PubMed] [Google Scholar]
- 20.Fingar, D. C., Richardson, C. J., Tee, A. R., Cheatham, L., Tsou, C., and Blenis, J. (2004) Mol. Cell. Biol. 24200 –216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koritzinsky, M., Seigneuric, R., Magagnin, M. G., van den Beucken, T., Lambin, P., and Wouters, B. G. (2005) Radiother. Oncol. 76177 –186 [DOI] [PubMed] [Google Scholar]
- 22.Bi, M., Naczki, C., Koritzinsky, M., Fels, D., Blais, J., Hu, N., Harding, H., Novoa, I., Varia, M., Raleigh, J., Scheuner, D., Kaufman, R. J., Bell, J., Ron, D., Wouters, B. G., and Koumenis, C. (2005) EMBO J. 243470 –3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, E. H., Lee, J. M., Blais, J. D., Bell, J. C., and Pelletier, J. (2005) J. Biol. Chem. 28020945 –20953 [DOI] [PubMed] [Google Scholar]
- 24.Mokrejs, M., Vopalensky, V., Kolenaty, O., Masek, T., Feketova, Z., Sekyrova, P., Skaloudova, B., Kriz, V., and Pospisek, M. (2006) Nucleic Acids Res. 34125 –130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holcik, M., and Sonenberg, N. (2005) Nat. Rev. Mol. Cell Biol. 6318 –327 [DOI] [PubMed] [Google Scholar]
- 26.Bonnal, S., Boutonnet, C., Prado-Lourenco, L., and Vagner, S. (2003) Nucleic Acids Res. 31427 –428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang, K. J., Kappel, A., and Goodall, G. J. (2002) Mol. Biol. Cell 131792 –1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huez, I., Bornes, S., Bresson, D., Creancier, L., and Prats, H. (2001) Mol. Endocrinol. 152197 –2210 [DOI] [PubMed] [Google Scholar]
- 29.Bornes, S., Boulard, M., Hieblot, C., Zanibellato, C., Iacovoni, J. S., Prats, H., and Touriol, C. (2004) J. Biol. Chem. 27918717 –18726 [DOI] [PubMed] [Google Scholar]
- 30.Akiri, G., Nahari, D., Finkelstein, Y., Le, S. Y., Elroy-Stein, O., and Levi, B. Z. (1998) Oncogene 17227 –236 [DOI] [PubMed] [Google Scholar]
- 31.Han, B., and Zhang, J. T. (2002) Mol. Cell. Biol. 227372 –7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak, M. (2005) Nucleic Acids Res. 336593 –6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansfield, K. D., Guzy, R. D., Pan, Y., Young, R. M., Cash, T. P., Schumacker, P. T., and Simon, M. C. (2005) Cell Metab. 1393 –399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji, X., Kong, J., and Liebhaber, S. A. (2003) Mol. Cell. Biol. 23899 –907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, Q., and Sarnow, P. (1997) Nucleic Acids Res. 252800 –2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter, M. S., and Sarnow, P. (2000) J. Biol. Chem. 27528301 –28307 [DOI] [PubMed] [Google Scholar]
- 37.Lum, J. J., Bui, T., Gruber, M., Gordan, J. D., DeBerardinis, R. J., Covello, K. L., Simon, M. C., and Thompson, C. B. (2007) Genes Dev. 211037 –1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galban, S., Kuwano, Y., Pullmann, R., Jr., Martindale, J. L., Kim, H. H., Lal, A., Abdelmohsen, K., Yang, X., Dang, Y., Liu, J. O., Lewis, S. M., Holcik, M., and Gorospe, M. (2008) Mol. Cell. Biol. 2893 –107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bornes, S., Prado-Lourenco, L., Bastide, A., Zanibellato, C., Iacovoni, J. S., Lacazette, E., Prats, A. C., Touriol, C., and Prats, H. (2007) Circ. Res. 100305 –308 [DOI] [PubMed] [Google Scholar]
- 40.Schepens, B., Tinton, S. A., Bruynooghe, Y., Beyaert, R., and Cornelis, S. (2005) Nucleic Acids Res. 336884 –6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koritzinsky, M., Rouschop, K. M., van den Beucken, T., Magagnin, M. G., Savelkouls, K., Lambin, P., and Wouters, B. G. (2007) Radiother. Oncol. 83353 –361 [DOI] [PubMed] [Google Scholar]
- 42.Bernardi, R., Guernah, I., Jin, D., Grisendi, S., Alimonti, A., Teruya-Feldstein, J., Cordon-Cardo, C., Simon, M. C., Rafii, S., and Pandolfi, P. P. (2006) Nature 442779 –785 [DOI] [PubMed] [Google Scholar]
- 43.van den Beucken, T., Koritzinsky, M., and Wouters, B. G. (2006) Cancer Biol. Ther. 5749 –755 [DOI] [PubMed] [Google Scholar]
- 44.Hui, A. S., Bauer, A. L., Striet, J. B., Schnell, P. O., and Czyzyk-Krzeska, M. F. (2006) FASEB J. 20466 –475 [DOI] [PubMed] [Google Scholar]
- 45.Bert, A. G., Grepin, R., Vadas, M. A., and Goodall, G. J. (2006) RNA 121074 –1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walden, W. E., Godefroy-Colburn, T., and Thach, R. E. (1981) J. Biol. Chem. 25611739 –11746 [PubMed] [Google Scholar]
- 47.Gebauer, F., and Hentze, M. W. (2004) Nat. Rev. Mol. Cell Biol. 5827 –835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, J. D., and Johannes, G. J. (2007) RNA 131116 –1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hornstein, E., Tang, H., and Meyuhas, O. (2001) Cold Spring Harbor Symp. Quant. Biol. 66477 –484 [DOI] [PubMed] [Google Scholar]
- 50.Denko, N., Wernke-Dollries, K., Johnson, A. B., Hammond, E., Chiang, C. M., and Barton, M. C. (2003) J. Biol. Chem. 2785744 –5749 [DOI] [PubMed] [Google Scholar]
- 51.Matsuoka, S., Edwards, M. C., Bai, C., Parker, S., Zhang, P., Baldini, A., Harper, J. W., and Elledge, S. J. (1995) Genes Dev. 9650 –662 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.