Abstract
Previous studies have shown that the acute stimulation of endothelial nitric-oxide synthase (eNOS) mRNA transcription by laminar shear stress is dependent on nuclear factor κ B (NFκB) subunits p50 and p65 binding to a shear stress response element (SSRE) in the human eNOS promoter and that mutation of the SSRE abrogates the shear-stimulated increase in eNOS promoter activity. In the present study, we found that although shear markedly increased eNOS mRNA, the increase in nuclear translocation of p50 and p65 caused by shear was only 2-fold, suggesting that shear has additional effects on NFκB cofactor activity beyond nuclear translocation. Chromatin immunoprecipitation assays showed that virtually no p50 or p65 was bound to the eNOS promoter at base line but that shear increased the binding of these subunits to the human eNOS SSRE by 10- to 20-fold. Co-immunoprecipitation studies demonstrated during the first 30 min of shear p300 bound to p65. Shear also increased p300 histone acetyltransferase (HAT) activity by 2.5-fold and increased acetylation of p65. The increase in eNOS mRNA caused by shear was completely blocked by pharmacological inhibition of p300/HAT activity with curcumin or by p300 small interfering RNA. Chromatin immunoprecipitation assays also showed that shear stimulated acetylation of histones 3 and 4 at the region of the eNOS promoter SSRE and extended 3′ toward the eNOS coding region. This was associated with opening of chromatin at the SSRE. In conclusion, these studies reveal a previously unknown role of p300/HAT activation as a very early response to shear that is essential for increasing eNOS mRNA levels.
The endothelial lining of blood vessels is continually subjected to laminar shear stress by virtue of its contact with flowing blood. Exposure of endothelial cells in culture to laminar shear stress causes both acute and long term effects on their function and morphology (1). Acute effects include changes in intracellular calcium and activation of several protein kinases (2, 3). Over the long term, laminar shear alters endothelial cell shape and gene expression (4, 5). One of the most physiologically important adaptations to shear is an increase in production of nitric oxide. Acutely, shear activates the endothelial cell nitric-oxide synthase (eNOS)2 enzyme to produce nitric oxide (6–8), and over several hours shear increases expression of this enzyme (9–11). We have shown previously that this is mediated by a transient activation of NFκB and binding of the NFκB subunits to a shear stress response element (SSRE) located 984 base pairs 5′ to the transcription start site in the human eNOS promoter and that mutation of this site completely abrogates the increase in eNOS promoter activity caused by shear (12).
NFκB encompasses a family of transcription factors including RelA/p65, RelB, c-Rel, p50, and p52 (13–15). In most unstimulated cells, NFκB is sequestered in the cytoplasm by its physical association with IκB. Upon activation by a variety of stimuli, the IκB subunit is phosphorylated and degraded, allowing the p50/p65 dimer to translocate to the nucleus to transactivate a variety of genes. These events are regulated by upstream signals, including Iκ kinase and alternate pathways (16). Generally, NFκB-dependent gene transcription occurs in response to diverse pro-inflammatory stimuli such as the cytokine tumor necrosis factor α, double-stranded RNA, lipopolysaccharide, reactive oxygen species, and high glucose. A departure from this pro-inflammatory role of NFκB seems to be its activation in the endothelium by shear stress, which in turn promotes induction of cytoprotective proteins. In addition to eNOS, protective genes activated by NFκB in response to shear include manganese superoxide dismutase, Bcl2, and GADD45β (17). It is unclear how shear modulates a beneficial effect of NFκB activation that contrasts to deleterious effects caused by its activation in response to inflammatory cytokines.
Another level of complexity in understanding how shear affects eNOS expression relates to epigenetic phenomenon. Eukaryotic genes are assembled into units referred to as nucleosomes containing eight histone proteins clustered on 146 bp of DNA (18). Histones must be unraveled from DNA to permit access of transcription factors. This process is promoted by histone acetylation, a process that is mediated by the transcription coactivator histone acetyltransferases such as p300 and its homolog, the cAMP response element-binding protein (CREB)-binding protein (CBP) and is antagonized by histone deacetylases (18). In addition to histone acetylation, p300/CBP also acetylates transcription factors, a process that enhances their DNA binding. In the case of NFκB, acetylation of p65 reduces its association with IκB and therefore promotes its activation (19). Moreover, p65 enhances histone acetylation and chromatin remodeling via an interaction with specific histone deacetylases (19). There is also an interplay between p300/CBP, NFκB subunits, and poly(ADP-ribose) polymerase-1 that promotes histone acetylation, also via histone deacetylase association (20). An important step for NFκB activation is phosphorylation of p65 by upstream kinases such as casein kinase-2 and protein kinase A. This enhances p65 DNA binding and promotes its association with p300/CBP, which in turn acetylates p65 and further increases its DNA binding efficiency (21).
Although these steps in NFκB activation and the importance of p300/CBP have been documented to occur in response to inflammatory stimuli, the manner in which shear affects these events has not been investigated, particularly in the context of eNOS transcription. In the present studies, we sought to determine how these upstream events might be modulated by shear and if they could affect eNOS mRNA levels in human endothelial cells. In preliminary studies we found that both p50 and p65 were present in nuclear extracts of human endothelial cells not exposed to shear and that the levels of these were only modestly increased by shear. These findings suggested that shear likely has additional effects, beyond simply promoting p50 and p65 nuclear translocation, to promote binding of these subunits to the eNOS promoter and transcriptional initiation. We hypothesized that acetylation of NFκB subunits and acetylation of histones might play a critical role in modulating chromatin remodeling, activation of p65 and p50, and initiation of eNOS transcription in response to shear.
EXPERIMENTAL PROCEDURES
Cell Culture and Shear Apparatus—Primary cultures of human umbilical vein endothelial cells (HUVECs) were purchased from the Emory Skin Diseases Research Center. HUVECs were grown on 0.05% gelatin-coated tissue culture plates in M199 supplemented with 20% fetal calf serum, 13.3 units/ml heparin, 40 μg/ml endothelial mitogen (Biomedical Technologies), 1% l -glutamine and penicillin-streptomycin. Cells were used between the 3rd and 6th passages. Shear (15 dynes/cm2) was applied using a cone-in-plate viscometer with a 1° angle in an incubator at 37 °C in 5% CO2.
Nuclear Extraction and Western Blotting—Nuclear fractions were extracted using a commercially available kit (Pierce Biotechnology). 15 μg of nuclear proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted with p50 or p65 antibodies (Santa Cruz Biotechnology). Immunoreactive bands were visualized by enhanced chemiluminescence (Amersham Biosciences).
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assays were performed using kits from Upstate or Imgenex according to the manufacturer's instructions. Briefly, cells were fixed by adding formaldehyde directly to the medium (final concentration 1%). 10 min later, the formaldehyde-mediated cross-linking reaction was stopped by adding glycine (90 mm). The cells were washed with ice-cold phosphate-buffered saline, pH 7.2, and harvested in 500 μl of phosphate-buffered saline supplemented with protease inhibitors (Roche Applied Science). The cells were pelleted by low speed centrifugation (1000 × g) and lysed in 200 μl of lysis buffer containing protease inhibitors. Chromatin was sheared to an average DNA fragment size of 300–400 bp using sonication (six 10-s bursts at one-fifth maximum potency). Antibodies used for immunoprecipitation included rabbit anti-p50 and anti-p65 (Santa Cruz Biotechnology) and anti-acetylated histone 3 and 4 (Cell Signaling). Fourteen PCR primer pairs were used to amplify 150-bp sequential regions of the eNOS promoter from –1604 to +50, each overlapping by ∼30 bp (Table 1).
TABLE 1.
Primers used for ChIP assay
| eNOS primer 1 Forward | -1604 to -1469 | AGCATCTGATGCTGCCTG |
| eNOS primer 1 Reverse | ATCCCAAGTGTGGCTTCC | |
| eNOS primer 2 Forward | -1490 to -1346 | ATGGAAGCCACACTTGGG |
| eNOS primer 2 Reverse | TTGTCCCATGACCCCAAG | |
| eNOS primer 3 Forward | -1365 to -1222 | GCTTGGGGTCATGGGACAA |
| eNOS primer 3 Reverse | GGGTCTGCCAGTTTTGGA | |
| eNOS primer 4 Forward | -1250 to -1106 | CCTCCGATCCTCCAAAAC |
| eNOS primer 4 Reverse | GGCCAGACTCCTCTGAAA | |
| eNOS primer 5 Forward | -1126 to -984 | CTTTTCAGAGGAGTCTGGCC |
| eNOS primer 5 Reverse | CGTGATTTCGAGACCTCA | |
| eNOS primer 6 Forward | -1010 to -863 | TCAGTCTCTGAGGTCTCG |
| eNOS primer 6 Reverse | TACACCAGCACTCTCCAG | |
| eNOS primer 7 Forward | -890 to -748 | CTGAAGTGCCTGGAGAGT |
| eNOS primer 7 Reverse | CAGGTCAGCAGAGAGACT | |
| eNOS primer 8 Forward | -770 to -627 | CCCTAGTCTCTCTGCTGACC |
| eNOS primer 8 Reverse | CCAATTTCCTGGAACCCC | |
| eNOS primer 9 Forward | -649 to -508 | TGTGGGGGTTCCAGGAAA |
| eNOS primer 9 Reverse | AGAACTCCTGGATCCCCA | |
| eNOS primer 10 Forward | -530 to -389 | AGGGTGGGGATCCAGGAGTT |
| eNOS primer 10 Reverse | GGGTTGGGCAGAAGGTGA | |
| eNOS primer 11 Forward | -448 to -307 | CTGAGAGTGTGGGCTGCCAT |
| eNOS primer 11 Reverse | CCACCAGGGGGTCATAAA | |
| eNOS primer 12 Forward | -328 to -188 | ACCTTTATGACCCCCTGGTGGCTCT |
| eNOS primer 12 Reverse | GGGCCGGACGCCTGGGTT | |
| eNOS primer 13 Forward | -218 to -60 | GGAACCCAGGCGTCCGGC |
| eNOS primer 13 Reverse | GGCAGTGGGAGGGGGCTC | |
| eNOS primer 14 Forward | -90 to +50 | GCCAGCACTGGAGAGCCC |
| eNOS primer 14 Reverse | CCTGGGCCACGCTCTTCA |
Immunoprecipitation and Immunoblotting—200 μg of nuclear proteins were incubated overnight at 4 °C with anti-p65 antibody and then exposed to 50% slurry of protein A/G Plus-agarose beads (Santa Cruz Biotechnology) for 2 h. After two washes in 0.1% Tween 20 phosphate-buffered saline buffer, the immunoprecipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-p300 (Santa-Cruz Biotechnology) or anti-acetylated lysine (Cell Signaling) antibodies.
Histone Acetyltransferase Activity Assay—Histone acetyltransferase activity was determined using a commercially available kit (Upstate Biotechnology) according to the manufacturer's instruction. Briefly, 500 μg of nuclear protein in 500 μl of lysis buffer was precleared with 50 μl of protein G-agarose bead slurry at 4 °C for 30 min and then immunoprecipitated with 5 μg of anti-p300 antibody at 4 °C for 2 h. Histone acetyltransferase activity was determined by monitoring the transfer of [3H]acetyl-CoA to a histone H4 peptide. Normal mouse IgG was used rather than the histone acetyltransferase antibody as a negative control.
Chromatin DNA Accessibility Assay—Accessibility of DNA at the SSRE site was analyzed using chromatin accessibility by real-time PCR as described previously (22). Basically, intact nuclei from the HUVECs were isolated and purified on a sucrose cushion using the Nuclei Isolation Kit (Sigma NUC-201) according to the vendor's instruction. 2 × 105 nuclei/treatment were digested with 10 units of BsaI (New England Biolabs) for 2 h at 50°C. Genomic DNA was purified by phenol/chloroform extraction and phenol precipitation, and 50 ng were analyzed by quantitative PCR (Light Cycler-SYBR Green real-time PCR; Roche Diagnostics) by amplification of a fragment flanking the SSRE (BsaI recognition site). DNA from undigested nuclei was used as a positive amplification control, while digested naked DNA was used as a restriction digestion control and negative amplification control. Under these conditions, chromatin openness or SSRE accessibility in this case is inversely proportional to the level of PCR amplification.
Small Interfering RNA (siRNA) Transfection and Quantitative PCR—Control siRNA and siRNA against p300 were obtained from Santa Cruz Biotechnology, and HUVECs were transfected according to the manufacturer's instruction. Briefly, 2 × 106 cells were seeded on 100-mm plates the day before transfection. The medium was switched to Opti-MEM and either control siRNA or anti-p300 siRNA in Oligofectamine was added to the culture medium for 4 h (final concentration 50 nmol/liter), after which the medium was replaced with normal HUVEC medium. eNOS mRNA expression was quantified by quantitative real-time reverse transcription PCR using a LightCycler (Roche Applied Science) thermocycler. Total RNA was extracted using RNeasy kit (Qiagen) and reverse-transcribed to cDNA using Superscript III reverse transcriptase (Invitrogen) and random primers. Real-time PCR cycling conditions included an initial denaturation (2 min, 95 °C), followed by repeated cycles of denaturation (15 s, 95 °C), annealing (10 s, 65 °C), and extension (12 s, 72 °C), with acquisition of fluorescence at the end of each extension. The primers used were sense: 5′-AATCCTGTATGGCTCCGAGA-3′ and antisense: 5′-ATGCTGTTGAAGCGGATCTT-3′ for eNOS; sense: 5′-CCAAGATCCAACTACGAGCT-3′ and anti-sense: 5′-GAACGTCTGCCCTATCAACT-3′ for 18S. eNOS RNA expression was normalized to 18S.
Statistical Analysis—Data are presented as means ± S.E. of the mean. Analysis of variance was used to compare results of experiments; when differences were observed, a Student-Neuman-Keuls post-hoc test was employed.
RESULTS
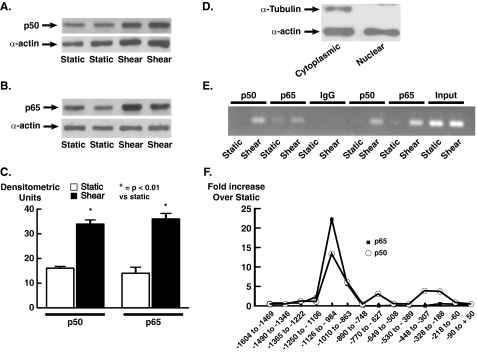
Translocation of NFκB Subunits p50 and p65 into the Nuclei and Binding to the eNOS Promoter in Response to Shear Stress—We have shown previously that the acute (<6 h) increase in eNOS mRNA is entirely dependent on NFκB and that inhibition of NFκB activation completely prevents this response (12). To determine whether shear promotes nuclear translocation of either p50 or p65 in human endothelial cells, we exposed HUVECs to either static conditions or shear for 30 min. At base line, the nuclei of HUVECs contained both p50 and p65, and 30 min of laminar shear increased nuclear levels of both of these by ∼2-fold (Fig. 1, A–C). Given that the increase in eNOS mRNA caused by shear is >5-fold, this suggested that the DNA binding and transcriptional activity of these NFκB subunits is regulated by shear in a manner that is independent of nuclear translocation. To determine whether DNA binding of either p50 or p65 to the endogenous human eNOS gene is affected by shear, we performed ChIP assays using overlapping primers allowing amplification of eNOS promoter regions from –1600 to +50 (Table 1). The shear stress response element in the human eNOS promoter is –976 bp upstream from the transcription start site (12). In cells exposed to static conditions, there was minimal specific association of either p50 or p65 with any aspect of the eNOS promoter (Fig. 1, E and F), but 30 min of shear increased binding of p50 and p65 to the SSRE. The apparent increase in DNA binding of p50 and p65 to this region was striking, averaging >20-fold for p65 and >10-fold for p50 (Fig. 1, D and F). Binding of these factors to other portions of the eNOS promoter was not affected by shear.
FIGURE 1.
Effect of unidirectional laminar shear on nuclear levels of the NFkB subunits p50 and p65 and binding of these to the eNOS promoter. Western blots are shown of nuclear extracts for p50 (A) and p65 (B). C shows mean densitometry analysis of three to four separate experiments for each. Data are mean ± S.E. of the mean. D shows a Western blot for the cytoplasmic protein α-tubulin and α-actin (present in both the nucleus and cytoplasm) from a nuclear preparation (lane 1) and a cytoplasmic preparation (lane 2). E shows ChIP analysis of binding of p50 and p65 to the eNOS promoter. P50 and p65 were immunoprecipitated from nuclear extracts and segments of eNOS DNA amplified by PCR. Input refers to nuclear extracts before immunoprecipitation. IgG was used as an immunoprecipitation control. F shows mean values for densitometric analysis of three similar experiments. As evident, shear only increased nuclear levels of p50 and p65 by 2-fold but dramatically increased binding of these subunits to the endogenous eNOS promoter.
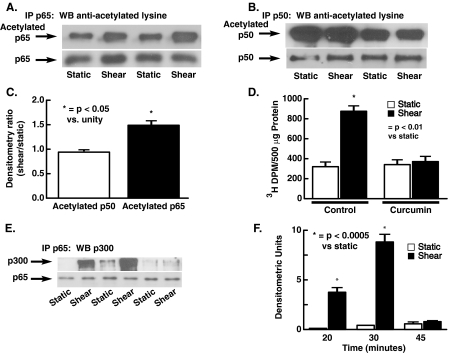
Acetylation of p65 by p300 during Translocation into the Nuclei in Response to Shear—Previous studies have indicated that p300/CBP plays a key role in NFκB subunit acetylation and that p50 and p65 are essential in modulating eNOS transcription in response to shear (23, 24). To determine whether p50 and p65 were acetylated in response to shear, these proteins were immunoprecipitated from HUVEC nuclear extracts and Western blots performed with anti-acetylated lysine antibodies. These studies indicated that both p65 and p50 were acetylated at base line but that 30 min of shear consistently increased p65 acetylation while having no effect on p50 acetylation (Fig. 2, A–C). In additional experiments, we demonstrated that shear increased p300 acetyltransferase activity, as evidenced by the ability of immunoprecipitated p300 to acetylate histone substrate (Fig. 2D). This increase in acetyltransferase activity was blocked by curcumin (80 mm), a known inhibitor of HAT activity (Fig. 2D). Co-immunoprecipitation experiments indicated that within 20 min shear also markedly increased association of p300 with p65 (Fig. 2E). This binding of p65 and p300 persisted for 30 min but was no longer evident following 45 min of shear. Taken together, these data indicate that shear stress activates p300 acetyltransferase activity and that p300 associates with and acetylates p65.
FIGURE 2.
Acetylation of p50 and p65 at base line and in response to laminar shear and association of p65 with p300. The NFκB subunits p50 and p65 were immunoprecipitated and Western blots using anti-lysine antibody performed. As shown in A and B, both p50 and p65 are acetylated at base line; however, 30 min of shear increases acetylation of p65 (A and C). C shows densitometric analysis of A and B, with the density of acetylated lysines in response to shear expressed as a ratio to the average static sample on the same blot. Shear (15 dynes/cm2 × 20 min) also increases acetyltransferase activity of p300, and this increase is prevented by the histone acetyltransferase inhibitor curcumin (80 mm in D, n = 3 for each). p300 was immunoprecipitated and acetyltransferase activity measured using histone substrate as described under “Experimental Procedures.” E shows the effect of shear on the association between p65 and p300. Mean data for densitometric analysis of three separate experiments are shown in F. Data are mean ± S.E. of the mean.
Chromatin Acetylation and Remodeling in Response to Shear—To determine whether shear could promote acetylation of histones associated with the eNOS promoter, antibodies against acetylated histones 3 and 4 and primers 5, 7, and 10 (Table 1) were used in ChIP assays. These experiments demonstrated that 15 min of shear stress stimulates acetylation of histones H3 and H4 at the site of SSRE within the eNOS promoter (Fig. 3, A–C) and this acetylation extended 3′ toward the eNOS coding region. This was associated with chromatin opening, as demonstrated by accessibility of native chromatin at the SSRE site. Native DNA was digested with the restriction enzyme Bsa1, the recognition site of which corresponds to the SSRE. Therefore, BsaI would cut the SSRE not protected by nucleosomes in the chromatin. The digested SSRE could be quantified by a reduction in a PCR product spanning the SSRE. In absence of BsaI digestion, quantitative PCR yielded high levels of product that were not different between shear and static samples (Fig. 3D), indicating a good efficiency of PCR amplification in sheared and unsheared samples. This product was reduced by about half in DNA from unsheared cells treated with Bsa1 but was completely absent in DNA from sheared cells treated with Bsa1 (Fig. 3C). Phenol denaturation of nuclear DNA, to remove chromatin, also eliminated the ability to amplify the SSRE following Bsa1 exposure. These data indicate that shear stimulates chromatin opening in the eNOS gene at the SSRE site and that this was associated with histone acetylation at this site.
FIGURE 3.
Effect of shear on histones associated with the eNOS promoter. A–C show mean data of ChIP assays examining acetylation of histones 3 and 4 surrounding the eNOS promoter at base line and following 30 min of shear (15 dynes/cm2). Data are from three to four experiments. D shows opening of the eNOS promoter in response to shear. Nuclei were purified from sheared and unsheared cells, and 2 × 105 nuclei/treatment were digested with 10 units of BsaI. DNA was purified, and a fragment flanking the SSRE (BsaI recognition site) was amplified by quantitative PCR. DNA from undigested nuclei was used as a positive amplification control, while digested purified genomic (naked) DNA was used as a restriction digestion control and negative amplification control. In the case of closed chromatin (histone-protected promoter), BSA1 should fail to cut this site, allowing ultimate amplification of this region using PCR. As evident, shear leads to almost complete opening of the histones surrounding the shear response element. Data are mean ± S.E. of the mean. E shows localization of primers used in experiments in A–C.
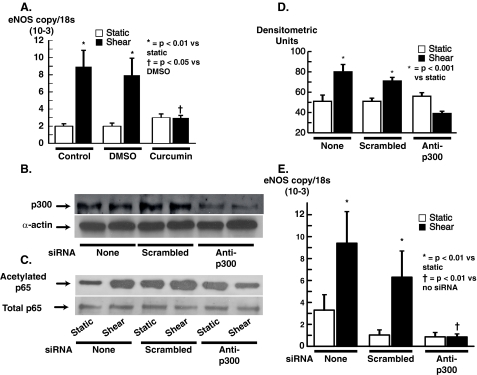
Role of p300 Acetylation Activity in Modulating eNOS Transcription in Response to Shear Stress—The above experiments demonstrated that shear stimulates not only p65 and histone acetylation but also chromatin opening at the site of the eNOS SSRE and that these events were mediated by p300. We performed additional experiments to determine the role of p300 in modulating the increase in eNOS mRNA that occurs in response to shear. Pretreatment of HUVECs with curcumin (80 μm) completely blocked the increase in eNOS mRNA caused by 6 h of shear, whereas the vehicle DMSO had no effect on this response (Fig. 4A). To specifically examine the role of p300, HUVECs were transfected with either a non-silencing mRNA or an siRNA against p300 and subjected to shear. Although non-silencing siRNA had no effect on levels of p300 in HUVECs, siRNA against p300 markedly reduced levels of this protein (Fig. 4B). Following 30 min of shear, nuclear fractions were extracted and acetylation of p65 was examined using immunoprecipitation with an antibody against p65 and Western blotting with an anti-acetylated lysine antibody. These experiments showed that the increase in acetylation of p65 caused by shear was prevented in cells transfected with siRNA against p300 but was unaffected by transfection with non-silencing siRNA (Fig. 4, C and D). Importantly, the increase in eNOS mRNA caused by shear stress was totally eliminated by transfection with p300 siRNA but was not altered by non-silencing siRNA (Fig. 4E).
FIGURE 4.
Role of p300 in eNOS transcription in response to laminar shear. A shows effect of the p300 inhibitor curcumin on eNOS transcription in response to 6 h of shear (n = 4–5/group). B shows effect of p300 siRNA on p300 protein levels as determined by Western blot. As evident, p300 siRNA effectively reduced p300 levels whereas control (Scrambled) siRNA did not. Down-regulation of p300 with siRNA prevented acetylation of p65 as shown in the example in C and in the mean data (n = 4) presented in D. Inhibition of p300 expression with siRNA completely prevented the increase in eNOS mRNA as determined by real-time quantitative PCR (D, n = 4–5). Data are mean ± S.E. of the mean.
DISCUSSION
In the present study we have shown that unidirectional laminar shear not only causes a modest nuclear translocation of the NFκB subunits p50 and p65 but also markedly stimulates binding of both of these to the eNOS promoter near the SSRE region. Our findings demonstrated that this is dependent on activation of p300, which in turn acetylates p65 and histones in proximity to the human eNOS SSRE. This is associated with opening of chromatin structure surrounding the SSRE. Activation of p300 is critical for the increase in eNOS mRNA in response to shear, as its pharmacological inhibition using curcumin or down-regulation by siRNA completely abrogated the increase in eNOS caused by this mechanical force. These findings provide new insight into how shear stress modulates eNOS expression via both transcriptional and epigenetic mechanisms.
Given that NFκB-mediated transcriptional events are suppressed in resting cells, the finding that human endothelial cell nuclei contain substantial amounts of p50 and p65 under basal conditions suggested that events other than nuclear translocation were responsible for shear induction of eNOS transcription. Indeed, the absence of p50 and p65 binding to the endogenous eNOS promoter at base line and the striking increase in binding of these with shear strongly support this conclusion. Our data indicate that activation of p300 histone acetyltransferase activity is an early event in shear modulation of gene expression. Of note, Wang et al. (25) have recently shown that resting HUVEC contain both p50 and p65 and that the eosinophil-derived oxidant hypothiocyanate (HOSCN) causes further translocation of these. In contrast to the modest changes (2- to 3-fold) in nuclear levels of p50 and p65, HOSCN markedly stimulated binding of these subunits to a consensus NFκB sequence in a fashion almost entirely analogous to our current findings.
Of note, oxidative stress has been shown to stimulate p300 association with p65 and histone acetylation in a fashion similar to the effect of shear observed in the present study (26). Laminar shear can transiently stimulate production of reactive oxygen species in endothelial cells (27), and hydrogen peroxide and lipid peroxides stimulate eNOS expression (28, 29). It is interesting to speculate that our current findings could be accounted for by an acute burst of reactive oxygen species production in response to shear. Although this scenario is possible, in previous studies we have been unable to block the increase in eNOS expression caused by shear using antioxidants, suggesting that redox-independent pathways are likely responsible for p300 activation in response to this mechanical stimulus (28).
There is substantial interest in how extracellular forces such as shear signal intracellular events. The platelet endothelial cell adhesion molecule (PECAM) has recently been implicated in this process. Inclusion of PECAM in a heterologous expression system yields a mechanosensory complex sufficient to confer responsiveness to flow (30). Relevant to our present studies, shear activation of Akt is absent in cells lacking PECAM (31). Huang and Chen (32) have recently shown that Akt phosphorylation of p300 on serine 1834 is essential for its histone acetyltransferase activity. It is therefore possible that PECAM modulation of p300 phosphorylation underlies the early activation of this enzyme in response to shear. These extracellular events ultimately lead to activation of the small G protein rac1, rearrangement of the endothelial cell actin cytoskeleton, and NFκB activation (33).
In summary, these studies identify a previously unidentified role for p300 in modulation of eNOS transcription in human endothelial cells. Laminar shear increases p300 acetyltransferase activity, which in turn coordinates opening of chromatin near the shear stress response element in the eNOS promoter and activation of p65 and thus promotes p65 binding to the eNOS promoter. Inhibition of p300 either pharmacologically or by small interfering RNA completely prevents the increase in eNOS caused by shear. p300 also modulates activity of other transcription factors, including KLF2 (34), and it is probable that its influence extends beyond regulation of NFκB activity.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grants HL39006, HL38206, HL59248, and DK51257 and by National Institutes of Health Program Project Grants HL58000 and HL075209. This work was also supported by a Department of Veterans Affairs merit grant. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: eNOS, endothelial nitric-oxide synthase; NFκB, nuclear factor κ B; ChIP, chromatin immunoprecipitation assay; HUVEC, human umbilical vein endothelial cell; siRNA, small interfering RNA; SSRE, shear stress response element; CBP, cAMP response element-binding protein (CREB)-binding protein.
References
- 1.Davies, P. F. (1995) Physiol. Rev. 75 519–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James, N. L., Harrison, D. G., and Nerem, R. M. (1995) FASEB J. 9 968–973 [DOI] [PubMed] [Google Scholar]
- 3.Berk, B. C., Corson, M. A., Peterson, T. E., and Tseng, H. (1995) J. Biomech. 28 1439–1450 [DOI] [PubMed] [Google Scholar]
- 4.Malek, A. M., and Izumo, S. (1995) J. Biomech. 28 1515–1528 [DOI] [PubMed] [Google Scholar]
- 5.Nerem, R. M., Harrison, D. G., Taylor, W. R., and Alexander, R. W. (1993) J. Cardiovasc. Pharmacol. 21 Suppl. 1, S6–S10 [DOI] [PubMed] [Google Scholar]
- 6.Dimmeler, S., Fleming, I., Fisslthaler, B., Hermann, C., Busse, R., and Zeiher, A. M. (1999) Nature 399 601–605 [DOI] [PubMed] [Google Scholar]
- 7.Boo, Y. C., Hwang, J., Sykes, M., Michell, B. J., Kemp, B. E., Lum, H., and Jo, H. (2002) Am. J. Physiol. 283 H1819–H1828 [DOI] [PubMed] [Google Scholar]
- 8.Fleming, I., and Busse, R. (1999) Cardiovasc. Res. 43 532–541 [DOI] [PubMed] [Google Scholar]
- 9.Nishida, K., Harrison, D. G., Navas, J. P., Fisher, A. A., Dockery, S. P., Uematsu, M., Nerem, R. M., Alexander, R. W., and Murphy, T. J. (1992) J. Clin. Investig. 90 2092–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Searles, C. D. (2006) Am. J. Physiol. 291 C803–C816 [DOI] [PubMed] [Google Scholar]
- 11.Mattart, M., Mazzolai, L., Chambaz, C., Hayoz, D., Brunner, H. R., and Silacci, P. (2003) Biorheology 40 289–297 [PubMed] [Google Scholar]
- 12.Davis, M. E., Grumbach, I. M., Fukai, T., Cutchins, A., and Harrison, D. G. (2004) J. Biol. Chem. 279 163–168 [DOI] [PubMed] [Google Scholar]
- 13.Li, Q., and Verma, I. M. (2002) Nat. Rev. Immunol. 2 725–734 [DOI] [PubMed] [Google Scholar]
- 14.De Martin, R., Hoeth, M., Hofer-Warbinek, R., and Schmid, J. A. (2000) Arterioscler. Thromb. Vasc. Biol. 20 E83–E88 [DOI] [PubMed] [Google Scholar]
- 15.Rothwarf, D. M., and Karin, M. (1999) Sci. STKE (1999) RE1. [DOI] [PubMed]
- 16.Viatour, P., Merville, M. P., Bours, V., and Chariot, A. (2005) Trends Biochem. Sci. 30 43–52 [DOI] [PubMed] [Google Scholar]
- 17.Partridge, J., Carlsen, H., Enesa, K., Chaudhury, H., Zakkar, M., Luong, L., Kinderlerer, A., Johns, M., Blomhoff, R., Mason, J. C., Haskard, D. O., and Evans, P. C. (2007) FASEB J. 21 3553–3561 [DOI] [PubMed] [Google Scholar]
- 18.Khorasanizadeh, S. (2004) Cell 116 259–272 [DOI] [PubMed] [Google Scholar]
- 19.Blasko, E., Glaser, C. B., Devlin, J. J., Xia, W., Feldman, R. I., Polokoff, M. A., Phillips, G. B., Whitlow, M., Auld, D. S., McMillan, K., Ghosh, S., Stuehr, D. J., and Parkinson, J. F. (2002) J. Biol. Chem. 277 295–302 [DOI] [PubMed] [Google Scholar]
- 20.Hassa, P. O., and Hottiger, M. O. (2002) Cell Mol. Life Sci. 59 1534–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh, S., and Karin, M. (2002) Cell 109 (Suppl.) S81–S96 [DOI] [PubMed] [Google Scholar]
- 22.Goriely, S., Van Lint, C., Dadkhah, R., Libin, M., De Wit, D., Demonte, D., Willems, F., and Goldman, M. (2004) J. Exp. Med. 199 1011–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanden Berghe, W., Vermeulen, L., Delerive, P., De Bosscher, K., Staels, B., and Haegeman, G. (2003) Adv. Exp. Med. Biol. 544 181–196 [DOI] [PubMed] [Google Scholar]
- 24.Grumbach, I. M., Chen, W., Mertens, S. A., and Harrison, D. G. (2005) J. Mol. Cell. Cardiol. 39 595–603 [DOI] [PubMed] [Google Scholar]
- 25.Wang, J. G., Mahmud, S. A., Thompson, J. A., Geng, J. G., Key, N. S., and Slungaard, A. (2006) Blood 107 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman, I., Marwick, J., and Kirkham, P. (2004) Biochem. Pharmacol. 68 1255–1267 [DOI] [PubMed] [Google Scholar]
- 27.Laurindo, F. R., Pedro Mde, A., Barbeiro, H. V., Pileggi, F., Carvalho, M. H., Augusto, O., and da Luz, P. L. (1994) Circ. Res. 74 700–709 [DOI] [PubMed] [Google Scholar]
- 28.Drummond, G. R., Cai, H., Davis, M. E., Ramasamy, S., and Harrison, D. G. (2000) Circ. Res. 86 347–354 [DOI] [PubMed] [Google Scholar]
- 29.Ramasamy, S., Parthasarathy, S., and Harrison, D. G. (1998) J. Lipid Res. 39 268–276 [PubMed] [Google Scholar]
- 30.Tzima, E., Irani-Tehrani, M., Kiosses, W. B., Dejana, E., Schultz, D. A., Engelhardt, B., Cao, G., DeLisser, H., and Schwartz, M. A. (2005) Nature 437 426–431 [DOI] [PubMed] [Google Scholar]
- 31.Fleming, I., Fisslthaler, B., Dixit, M., and Busse, R. (2005) J. Cell Sci. 118 Pt. 18, 4103–4111 [DOI] [PubMed] [Google Scholar]
- 32.Huang, W. C., and Chen, C. C. (2005) Mol. Cell. Biol. 25 6592–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzima, E., Del Pozo, M. A., Kiosses, W. B., Mohamed, S. A., Li, S., Chien, S., and Schwartz, M. A. (2002) EMBO J. 21 6791–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huddleson, J. P., Ahmad, N., Srinivasan, S., and Lingrel, J. B. (2005) J. Biol. Chem. 280 23371–23379 [DOI] [PubMed] [Google Scholar]