Abstract
Tripeptidyl peptidase I (TPP I) is the first mammalian representative of a family of pepstatin-insensitive serine-carboxyl proteases, or sedolisins. The enzyme acts in lysosomes, where it sequentially removes tripeptides from the unmodified N terminus of small, unstructured polypeptides. Naturally occurring mutations in TPP I underlie a neurodegenerative disorder of childhood, classic late infantile neuronal ceroid lipofuscinosis (CLN2). Generation of mature TPP I is associated with removal of a long prosegment of 176 amino acid residues from the zymogen. Here we investigated the inhibitory properties of TPP I prosegment expressed and isolated from Escherichia coli toward its cognate protease. We show that the TPP I prosegment is a potent, slow-binding inhibitor of its parent enzyme, with an overall inhibition constant in the low nanomolar range. We also demonstrate the protective effect of the prosegment on alkaline pH-induced inactivation of the enzyme. Interestingly, the inhibitory properties of TPP I prosegment with the introduced classic late infantile neuronal ceroid lipofuscinosis disease-associated mutation, G77R, significantly differed from those revealed by wild-type prosegment in both the mechanism of interaction and the inhibitory rate. This is the first characterization of the inhibitory action of the sedolisin prosegment.
Tripeptidyl peptidase I (TPP I)2 is an acidic lysosomal hydrolase sequentially removing tripeptides en bloc from the N terminus of small, unstructured polypeptides (for a review, see Ref. 1). The natural substrate(s) of the enzyme still remains elusive. TPP I is widely distributed in the human body, with a high expression level in brain tissue (2, 3). Naturally occurring mutations in TPP I underlie a devastating neurodegenerative disorder of early childhood, classic late infantile neuronal ceroid lipofuscinosis (CLN2) (4).
On the basis of significant sequence homology and inhibitory studies, TPP I was assigned to a family of pepstatin-insensitive serine-carboxyl proteases recently renamed sedolisins (for a review, see Ref. 5). The best characterized members of this family are bacterial proteases: kumamolisin, a thermostable endopeptidase isolated from Bacillus novosp. MN-32, and a proteinase from Pseudomonas sp. 101 (PSCP, sedolisin). High resolution crystal structures resolved for kumamolisin and PSCP revealed a subtilisin-like fold and the Ser-Glu-Asp catalytic triad distinct from that of classical serine proteases equipped with Ser-His-Asp (for a review, see Ref. 5). Although the crystal structure of TPP I has not yet been established, inihibitory and mutational analyses identified Ser475 as an active site nucleophile (6) and the involvement of Glu272 and Asp276 (7) in the catalytic reaction. TPP I is the sole mammalian representative of sedolisins identified to date.
Like most proteolytic enzymes, TPP I is synthesized as an inactive precursor, zymogen (8, 9). The prepro-TPP I consists of a 19-amino acid (aa) signal peptide cleaved off when the newly forming polypeptide chain is inserted into the endoplasmic reticulum lumen, the 176-aa prosegment (or prodomain), and a 368-aa catalytic domain. As we demonstrated earlier, the removal of the prosegment from a purified TPP I proenzyme (pro-TPP I) occurs via an autocatalytic, intramolecular mechanism, whereby the mature enzyme does not significantly participate in its own generation (10). Efficient in vitro processing and autoactivation of the proenzyme is triggered by lowering the pH of the proenzyme solution to below 4.5. When pro-TPP I is activated at pH 4.5 and above, it is processed slowly and generates additional 6- and 14-aa N-terminal extensions in the newly formed polypeptide, rendering it enzymatically inactive (10). However, as we showed earlier, the presence of polyanionic compounds, such as glycosaminoglycans (GAGs), allows for an efficient activation of TPP I proenzyme at higher pH, up to pH 5.5 (11). Given that endogenous TPP I with active site nucleophile Ser475 mutated is able to generate mature enzyme in cultured fibroblasts from patients (9), the heteroprocessing of pro-TPP I also may occur in vivo, at least in some cell types.
Prosegments of proteases are separate functional modules, and up-to-date multiple functions have been ascribed to them (for a review, see Ref. 12). Isolated prosegments often show the ability to assist in fast refolding of denatured enzymes. Apart from these chaperone-like activities, prosegments may assist in targeting of the protease to specific organelles or in membrane association. But what appears to be a primary function of most prosegments is regulation of enzymatic activities of cognate proteases by means of selective inhibition either in cis or in trans.
The length of the prosegment of various proteases varies distinctly, i.e. only two aa residues are found in granzymes (13), and as many as several hundred residues are found in enterokinase (14). The length of TPP I prosegment (176 aa) suggests that it could easily fold into an independent domain, and thus, in addition to active site shielding, it might also have other functions (i.e. facilitate the folding or stabilize the structure of the enzyme). The association of naturally occurring missense mutations in the TPP I prosegment region with the CLN2 disease process supports this supposition (see NCL Resource on the World Wide Web).
In the work presented below, we investigated the inhibitory properties of TPP I prosegment. We show that wild-type (WT) TPP I prosegment acts as a potent, slow-binding inhibitor of its parent enzyme. We also demonstrate that the inhibitory properties of TPP I prosegment with the introduced CLN2 disease-associated mutation G77R significantly differed from those revealed by WT prosegment in both the mechanism of interaction and the inhibitory rate. This is the first characterization of the inhibitory properties of the prosegment of members of the sedolisin family.
EXPERIMENTAL PROCEDURES
Materials—Antiserum R413 was raised in rabbits against the prosegment of human TPP I purified from Escherichia coli, by using the standard procedure described (9). Anti-His antibody was obtained from GeneTex (San Antonio, TX). ECL kit reagents were from GE Healthcare. The bicinchoninic acid (BCA) kit was from Pierce. TPP I substrate (AAF-AMC) was obtained from Bachem Bioscience (King of Prussia, PA). All other chemicals were of the highest purity available and were purchased either from Fisher or Sigma.
Expression and Purification of WT and G77R TPP I Prosegment—A plasmid encoding the entire TPP I cDNA inserted in pcDNA3.1 Hygro (9) was used as a PCR template to amplify the prosegment of TPP I encompassing aa 20–195. The following primers were used: Kpnprocln2 (forward), 5′-tgggtaccagttacagcccggagcccgaccag; Salcln2 (reverse), 5′-gtgtcgacttagcctacagtccctgtcacctgcgg. PCR product was digested with KpnI and SalI (restriction sites are underlined) and ligated with plasmid pET45b (Novagen, Madison, WI) digested with the same enzymes. The resulting construct (pET45b-PST) encodes the prosegment polypeptide with the following additional amino acid sequence at the N terminus: MAHHHHHHVGT. The G77R mutation was introduced by using the overlap PCR technique, as described previously (7), and the following primers: forward, 5′-ccagctctcctcaatacagaaaatacctgaccc; reverse, 5′-gggtcaggtattttctgtattgaggagagctgg. Construct integrity was verified by dideoxy-mediated PCR sequencing of the entire insert. pET45b-PST was expressed in E. coli strain BL21 (DE3). ∼200 ml of LB medium was inoculated and cultured overnight in the presence of 75 μg/ml carbenicillin. The overnight culture was centrifuged at 5000 × g for 5 min, and the bacterial pellet was resuspended in 2 liters of the fresh medium and shaken at 37 °C for 1 h, at which time the expression of the prosegment was induced by the addition of 0.5 mm isopropyl-β-d-galactoside. After a 4-h induction, cells were pelleted by centrifugation at 5000 × g for 5 min and resuspended in 50 mm Tris, 50 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 0.5 mm EDTA, 5% glycerol, pH 7.4 (resuspension buffer), at a ratio of 10 ml per 1 g of original cell paste, and lysozyme was added, to reach a final concentration of 100 μg/ml. The suspension was stirred at room temperature for 10 min and then sonicated. Afterward, Triton X-100 was added to a final concentration of 1%, and the lysate was stirred for an additional 15 min at room temperature. Inclusion bodies were pelleted at 8000 × g for 15 min and washed twice with resuspension buffer containing 1% Triton X-100 and twice with resuspension buffer without Triton X-100. Washed inclusion bodies were suspended in resuspension buffer (10 ml/g of original paste), tris(2-carboxyethyl)phosphine was adjusted to 5 mm, and the sample was briefly sonicated. Afterward, sarcosyl was added to achieve a final concentration of 5% (denaturation buffer), and the solution was stirred at room temperature for 1 h and then centrifuged at 20,000 × g for 15 min. The protein concentration in the supernatant was measured by using a BCA method. The denatured prosegment was refolded by a rapid dilution into the buffer containing 500 mm l-Arg, 100 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 50 mm Tris, pH 8.5, to a final concentration of 100 μg/ml and continuous stirring for ∼18 h. Afterward, the prosegment was concentrated by salting out with the help of ammonium sulfate. Ammonium sulfate was added, with continuous stirring, to the refolded prosegment on an ice bath in a stepwise manner to a final saturation of 50%, followed by centrifugation at 4000 × g for 10 min. The resulting pellet was dissolved in 10 mm Tris, pH 8.0, 100 mm NaCl, adjusted to 10 mm immidazole, and passed through a column containing 10 ml of Ni2+ resin (Qiagen, Valencia, CA). The column was washed in the same buffer and eluted with 300 mm immidazole. The eluate was dialyzed against 6–8 kDa cut-off membrane in two changes of 1 liter each of 10 mm Tris, pH 8.0, 0.05 mm EDTA, 0.1 mm tris(2-carboxyethyl)phosphine. Purified prosegment was concentrated on Millipore Centriplus-20 centrifugation filters (cut-off 10 kDa), clarified by high speed centrifugation, and stored at -20 °C.
Proenzyme Purification—Pro-TPP I was purified, as described earlier (10), with the following modifications: the Bio-Rad Bio-Logic system was used instead of the Amersham Biosciences Akta Purifier, and, as the last step, heparin affinity chromatography was introduced as follows. Pro-TPP I in 10 mm Tris, pH 7.4, was diluted 10-fold to 50 mm sodium acetate, pH 5.7 (buffer A), and injected onto 1 ml of heparin-agarose (Bio-Rad) cartridge, washed, and eluted with a gradient of buffer A and 1 m NaCl in 20 mm Tris, pH 8.0 (buffer B), over 20 min at 1 ml/min. Fractions containing pro-TPP I (centered at ∼280 mm NaCl) were pooled, concentrated, and buffer-exchanged back to 10 mm Tris, pH 8.0, on Millipore Centriplus-20 centrifugation filters (50 kDa cut-off) and stored at -20 °C.
Proenzyme Activation and Mature TPP I Purification—Mature TPP I was obtained by activating the proenzyme at ∼100 μg/ml for 2 h in 50 mm sodium acetate, pH 3.5, 0.05% Triton X-100 at 37 °C, followed by purifying the resulting enzyme on a heparin-agarose cartridge as above, with the exception that both A and B solutions were buffered by 50 mm sodium acetate, pH 5.0. Purified enzyme was concentrated and stored at -20 °C.
SDS-PAGE and Western Blotting—SDS-PAGE and Western blotting were performed, as described (9).
The Effect of Prosegment on TPP I Activity—To assess the effect of the added prosegment on the activity of the mature enzyme, we used time progression assays of TPP I activity toward a reporter substrate (AAF-AMC). Each assay was performed in either three or four replicates in a 100-μl reaction mixture of 50 mm sodium acetate, 0.1% Triton X-100, containing enzyme (0.1 nm) and various concentrations of substrate and inhibitor. The reaction was initiated by adding enzyme to the premixed solution of prosegment and substrate. The release of the AMC was monitored continuously at room temperature with intermittent shaking by using a GENios fluorescent plate reader (Tecan USA, Research Triangle Park, NC) with excitation and emission wavelengths of 355 and 460 nm, respectively.
Our initial experiments showed that WT TPP I prosegment, similar to prosegments of many other proteolytic enzymes (16), acts as a slow-binding inhibitor of the mature enzymes. Because of the time-dependent formation of the enzyme-inhibitor (EI) complex, it is possible to determine pre-steady state kinetic parameters using the progress curves of product formation. Formation of the product as a function of time for an enzyme inhibited by a slow-binding inhibitor is described by the following integrated equation (17),
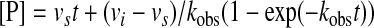 |
(Eq. 1) |
where [P] is the concentration of the product at any time t, vi and vs are the initial and steady state (i.e. final) velocities of the reaction in the presence of inhibitor, and kobs is the apparent first-order rate constant characterizing the establishment of the steady-state equilibrium. Progress curves were submitted to a nonlinear regression curve analysis using the software GraphPad Prism version 4.0 (GraphPad Software), which allows determination of the individual parameters vi, vs, and kobs for each curve. For time-dependent inhibitors, which are binding the enzymes in an apparent two-step fashion as the enzyme undergoes an isomerization, the reaction can be written as follows,
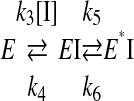 |
REACTION 1 |
The relationship of apparent dissociation constants for EI and E*I complex and the calculated kobs is expressed in the following equation.
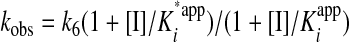 |
(Eq. 2) |
The overall inhibition constant Ki* is given by the following relationship (18).
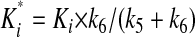 |
(Eq. 3) |
For uncompetitive inhibitors, the Kiapp and Ki are related by the following formula (19).
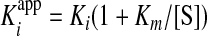 |
(Eq. 4) |
Proteolytic Susceptibility of the Prosegment—∼450 ng of TPP I was mixed with ∼1 μg of WT prosegment in 100 mm sodium acetate, pH 3.0–6.0, 0.1% Triton X-100 in a total volume of 20 μl and was incubated at 37 °C for 2 h and then analyzed on Coomassie Blue-stained SDS-polyacrylamide gels.
RESULTS
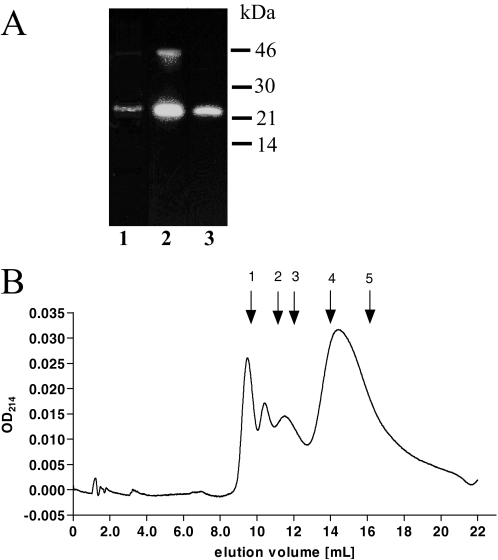
Prosegment Expression and Purification—His-tagged TPP I prosegment construct was prepared by using PCR and T7 polymerase promoter-based pET45b expression vector. The construct was used to transform E. coli strain BL21, and expression was induced with isopropyl-β-d-galactoside, as described under “Experimental Procedures.” The recombinant prosegment is predicted to comprise Ser20–Gly195 of TPP I proenzyme fused to N-terminal extension MAHHHHHHVGT containing a polyhistidine stretch facilitating the purification procedure. Because the majority of the expressed TPP I prosegment was found to be insoluble, inclusion bodies were isolated, and the prosegment was extracted under denaturing conditions, refolded, and further purified by using Ni2+ affinity resin. The identity and the purity of the isolated WT protein was assessed on Coomassie Brilliant Blue R-stained SDS-PAGE (Fig. 1A, lane 1) and Western blotting with R413 (Fig. 1A, lane 2) and anti-His monoclonal antibodies (Fig. 1A, lane 3). The vast majority of TPP I prosegment appeared as a single band migrating at ∼22 kDa. A small amount of the prosegment dimer visible on Coomassie-stained gel was easily recognizable by R413, anti-TPP I prosegment antibodies, and anti-His monoclonal antibodies after longer exposure of the blots. The amount of the dimer was a few-fold higher when the gel was run under nonreducing conditions (not shown). Gel permeation chromatography revealed that, assuming the globular structure of the prosegment, its majority migrated as ∼22.6 kDa, which is in close agreement with the predicted absolute mass of ∼20.8 kDa. Although some of the protein eluted in the void volume (Fig. 1B), the monomeric peptide under nondenaturing conditions constituted ∼70% of the entire preparation. Because the purified prosegment easily aggregated at the concentration above 1 mg/ml (not shown), the stock solutions were kept at the prosegment concentration below 1 mg/ml. The purity, migration pattern, and immunoreactivity to R413 and anti-His monoclonal antibodies of G77R prosegment were similar to those of WT protein (not shown).
FIGURE 1.
SDS-PAGE and immunoblotting (A) and gel filtration (B) analysis of purified WT TPP I prosegment. A, ∼500 ng of purified prosegment was loaded on reducing SDS-PAGE and analyzed after Coomassie Brilliant Blue R staining of the gel (lane 1), and 50 ng of purified prosegment was analyzed by Western blotting with R413 (lane 2) and monoclonal antibody anti-His (lane 3). B, ∼100 μg of purified prosegment was loaded onto a 1 × 25-cm Superdex75 gel permeation column and run at 0.5 ml/min in 200 mm NaCl, 20 mm Tris, pH 7.0. The eluate was monitored at 214 nm. Arrows 1–5 denote the elution position of molecular weight standards run individually on the same column, which correspond to the following proteins: blue dextran 2000 (exclusion limit) (1); bovine serum albumin, 66 kDa (2); ovalbumin, 45 kDa (3); chymotrypsinogen, 25 kDa (4); RNase A, 13.7 kDa (5).
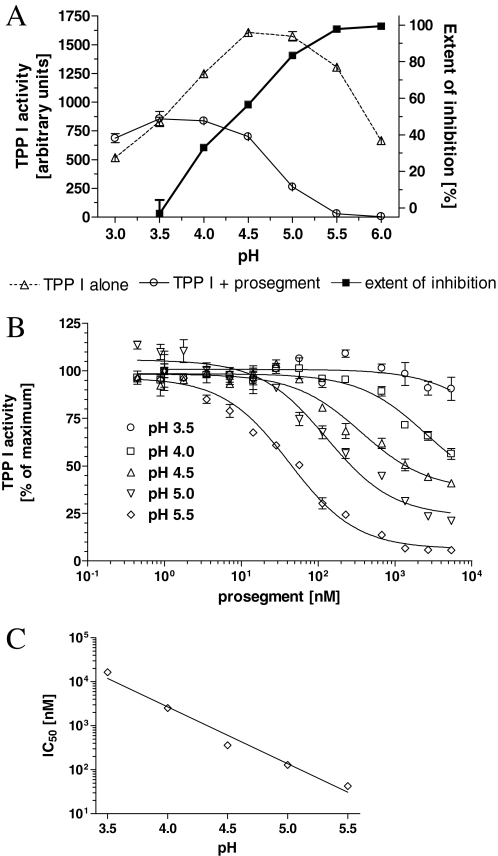
Inhibition of TPP I by the WT Prosegment—To investigate the ability of the WT prosegment to inhibit TPP I, the enzyme was incubated with the prosegment for 2 h at pH varying from 3.0 to 6.0, and residual TPP I activity toward the reporter substrate, AAF-AMC, was measured, as described under “Experimental Procedures.” As demonstrated in Fig. 2A, TPP I alone displayed a broad peak of maximum activity between pH 4.5 and 5.0, as previously shown (10, 20). The presence of the prosegment (at 1 μm) led from a significant to complete inhibition of TPP I activity at pH between 3.5 and 6.0. At pH 3.0, the prosegment did not demonstrate any inhibitory activity, and the increase of the inhibitory activity between pH 5.0 and 6.0 was small. The plot of the extent of inhibition revealed a linear relationship between pH and the inhibitory activity of prosegment in pH range 3.5–5.5.
FIGURE 2.
pH profile of TPP I inhibition by its prosegment. A, 1 nm TPP I was preincubated for 2 h at room temperature with prosegment at ∼1 μm in 50 mm sodium acetate, 0.05% Triton X-100, at the indicated pH, and then the residual enzyme activity was measured upon 2-fold dilution into the substrate solution AAF-AMC (final concentration 100 μm) dissolved in the same buffer at corresponding pH. B, TPPI at 1 nm was preincubated for 2 h at room temperature with the indicated concentrations of the prosegment, in 50 mm sodium acetate, 0.05% Triton X-100, pH 3.5–5.5, and residual activity was measured upon 2-fold dilution with substrate solution (final concentration 100 μm). The IC50 values were then calculated with the help of curve-fitting software. C, the IC50 values calculated from curve fitting of data presented in Fig. 3B replotted as a function of pH. Note the semilogarithmic scale in C.
To quantify the inhibitory activity of the prosegment at different pH values, TPP I was preincubated with the prosegment at various concentrations, and the concentration at which the inhibition reached 50% (IC50) was calculated and plotted versus pH (Fig. 2, B and C). The inhibition of TPP I by its prosegment was again dependent on pH, with IC50 ranging from >16 μm to ∼43 nm at pH from 3.5 to 5.5. These data demonstrate that the prosegment binds to and strongly inhibits its cognate enzyme in a pH-dependent fashion.
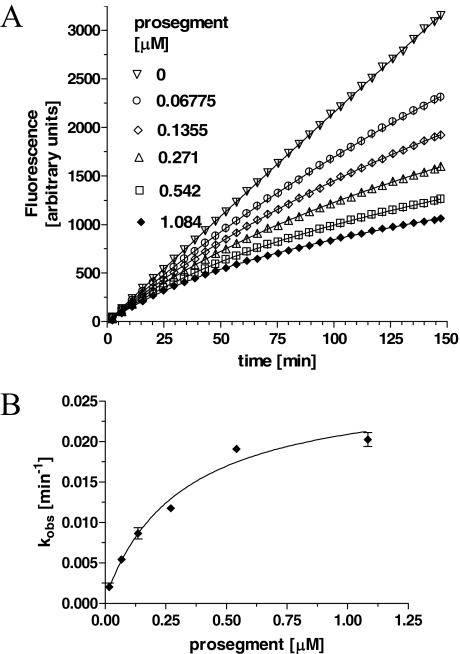
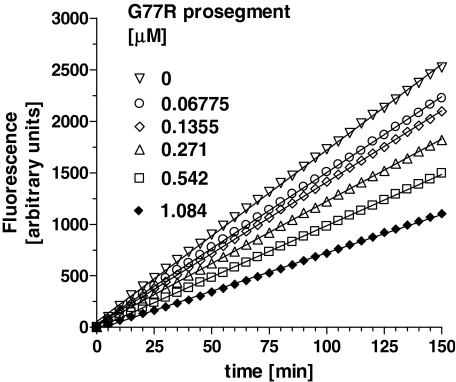
It is well documented that prosegments of many proteases are slow binding and, often times, tight binding inhibitors of their cognate proteases. Our preliminary studies also indicated that the interaction between the prosegment and TPP I obeyed the slow-binding inhibition kinetics. To assess the time-dependent inhibitory capacity of the prosegment, the real time progress curves were generated for TPP I activity toward AAF-AMC in the presence of increasing concentrations of prosegment at pH 5.0. As demonstrated in Fig. 3A, in the absence of the prosegment, TPP I displayed maximal steady-state activity during the entire time range of the assay, suggesting that under the experimental conditions employed, there was no significant depletion of the substrate. In the presence of the prosegment, the enzymatic reaction at first displayed a linear character, followed by a hyperbolic curve, hence displaying a time-dependent characteristic. By using an equation (Equation 1) describing the development of enzymatic reaction in the presence of a slow-binding inhibitor, the experimental data were curve-fitted to calculate the apparent rate constant (kobs) for establishment of the final steady-state equilibrium. When the calculated kobs values were plotted as function of prosegment concentration, the biphasic hyperbolic curve was obtained (Fig. 3B), suggesting that a faster equilibrium between the prosegment and the enzyme was followed by a slower dissociating enzyme-inhibitor complex (E*I). Nonlinear regression analysis of the data from Fig. 3B according to Equation 2 revealed an overall inhibition constant Ki* in the low nanomolar range (∼3.6 nm) (Table 1). These results indicate a two-step, slow, tight inhibition mechanism of TPP I by its prosegment.
FIGURE 3.
Time-dependent inhibition of TPP I by the prosegment as a function of prosegment concentration. A, TPP I at 0.5 nm was incubated with substrate at 100 μm and at the indicated varying concentrations of prosegment in 100 μl of total volume of 50 mm sodium acetate, pH 5.0, on a 96-well microtiter plate, and AMC fluorescence was acquired continuously with intermittent shaking. All data points are the average of quadruplicate measurements. Only every 10th data point collected is shown for clarity. B, the calculated kobs rates from A were replotted as a function of prosegment concentration.
TABLE 1.
Kinetic parameters of TPP I interaction with its prosegment (at pH 5.0) Values of rate constants for TPP I inhibition by its prosegment were calculated from real time progression curves of enzymatic reactions run at room temperature in sodium acetate buffer (50 mm, pH 5.0), 0.05% Triton X-100 and analyzed by curve-fitting software and Equations 1, 2, 3, 4. IC50 was calculated from a dose-dependent inhibition profile.
| Parameter | Value (μm) |
|---|---|
| IC50 | 0.1292 |
| Ki | 0.15815 |
| Ki* | 0.00355 |
| k5 | 0.0261106 |
| k6 | 0.0005994 |
| k5/k6 | 43.56 |
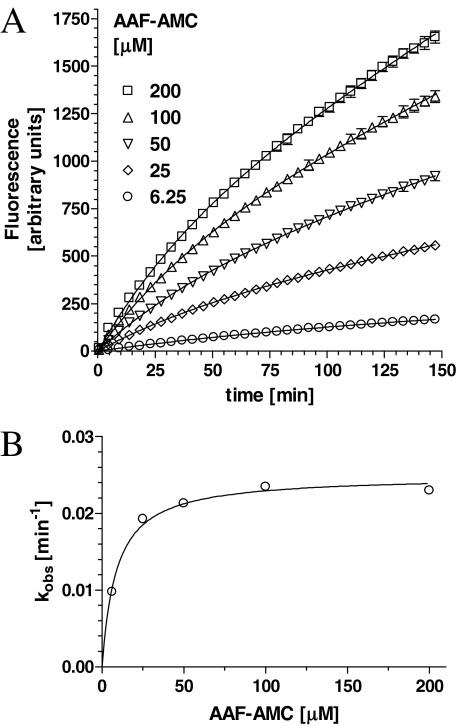
To assess the mode of TPP I inhibition by its prosegment, the time-dependent curves were generated in the presence of constant inhibitor concentrations and increasing concentration of the substrate (Fig. 4A). Plotting of the rate of inhibition versus substrate concentration (Fig. 4B) revealed again the saturable inhibition, suggestive of the uncompetitive type of interaction.
FIGURE 4.
Inhibition of TPP I by prosegment as a function of substrate concentration. A, TPP I at 0.5 nm was incubated with prosegment at 0.5 μm and at varying concentrations of AAF-AMC, as indicated, in 50 mm sodium acetate, pH 5.0. Only every 10th data point is shown for clarity. B, the kobs rates calculated from curve-fitting analysis in A were replotted as a function of substrate concentration.
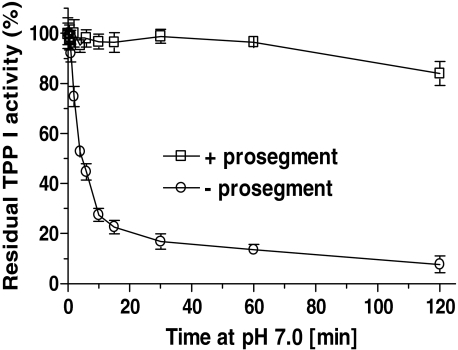
Protective Effect of the Propeptide on the Inactivation of TPP I by Alkaline pH—Prosegments of some proteases are believed to play the role of intramolecular chaperone, facilitating folding and refolding of their cognate protease. TPP I is an acidic protease that is quickly inactivated under alkaline pH conditions (21). To evaluate whether the interaction in trans of the prosegment with the protease might stabilize the enzyme, we studied the alkaline pH-mediated inactivation of the enzyme in the presence and absence of the prosegment. TPP I was incubated in 20 mm Tris, pH 7.0, and the residual enzymatic activity was assessed at pH 3.5. As shown in Fig. 5, in the absence of prosegment, the enzyme was quickly denatured with a rate constant of ∼0.107/min, amounting to a half-life of 6.472 min, whereas in the presence of the prosegment, the inactivation rate was reduced to ∼0.035/min, and ∼84% of the activity was preserved even after over 2 h of incubation.
FIGURE 5.
Prosegment protects its cognate enzyme against alkaline inactivation. TPPI at 10 nm was incubated either in the presence of prosegment at 660 nm or with bovine serum albumin at 10 μg/ml at 25 °C in 20 mm Tris, pH 7.0, 0.05% Triton X-100 for the indicated periods, and then the residual enzyme activity was measured at pH 3.5 and 25 °C upon 5-fold dilution.
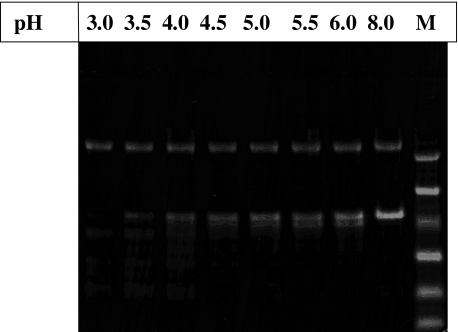
Proteolytic Susceptibility of TPP I Prosegment—To investigate the ability of TPP I to degrade an exogenously provided prosegment, they were mixed together at varying pH values (pH 3.0–6.0), incubated at 37 °C for 2 h, and analyzed by SDS-PAGE. As demonstrated in Fig. 6, TPP I was able to partially degrade its prosegment, producing several smaller peptides. However, although the degradation under the conditions employed was very efficient only at low pH conditions (up to pH 3.5), there were only limited degradation and small changes in the amount of the prosegment remaining in the pH range 4.0–6.0.
FIGURE 6.
Proteolytic susceptibility of the prosegment. ∼1 μg of the purified prosegment was incubated in the presence of ∼450 ng of TPP I in 0.1% Triton X-100, 50 mm sodium acetate, pH as indicated, for 2 h at 37 °C and analyzed on 10% Coomassie Blue-stained SDS-PAGE (M, molecular weight markers used: 45, 30, 21, 14.3, 6.5, and 3.5 kDa).
The Effect of G77R Mutation on Inhibitory Properties of the Prosegment—To evaluate whether the pathogenic mutation, G77R, affects the inhibitory properties of the prosegment toward its cognate protease, a real time kinetics assay was performed at varying concentrations of the inhibitor (prosegment). Unlike the WT prosegment, the G77R mutant exhibited a dose-dependent rapid equilibrium inhibition of TPP I, as demonstrated by the linearity of the progress curves (Fig. 7). The Ki of 0.285 μm for competitive inhibition for G77R prosegment was calculated by using the Cheng and Prusoff relationship (22).
FIGURE 7.
Inhibition of TPP I by the G77R mutant prosegment as a function of prosegment concentration. A, TPP I at 0.5 nm was incubated with substrate at 100 μm and at the indicated varying concentrations of G77R prosegment in a 100-μl total volume on a 96-well microtiter plate, and AMC fluorescence was acquired continuously with intermittent shaking. All data points are the average of quadruplicate measurements. Only every fifth data point collected is shown for clarity.
DISCUSSION
Our studies revealed that the prosegment of TPP I is a potent inhibitor of its cognate enzyme with an overall inhibition constant (Ki*) of ∼3.6 nm at pH 5.0. This inhibition constant is comparable with low nanomolar values obtained for inhibition of subtilases and proprotein convertases (PCs) by their corresponding prosegments (23–26). Also, similarly to most subtilases and subtilase-like proteases, TPP I prosegment is a slow-binding inhibitor of its cognate enzyme. To document the nature of the inhibition mechanism, we performed on-line analysis using a fixed concentration of enzyme and inhibitor and varying concentrations of substrate. An exponential function of the kobs versus substrate concentration we obtained is indicative of a two-step mechanism of inhibition. Thus, once bound to the enzymatically active TPP I, the prosegment yields a bimolecular complex, which undergoes further modification in the presence of excess inhibitor.
Prosegments appear to adopt stably folded structures when bound to the protease domain in their cognate proenzymes and have regions of multiple interfacial contacts with catalytic domains (12), which govern their affinities and specificities (27). Although the majority of isolated prosegments of proteases are either unstructured (23) or possess a loosely packed structure (28), the strongest inhibitory properties demonstrate those isolated prosegments, which reveal a folded tertiary structure (25). For some subtilases, the interaction of the isolated prosegment with the active site of the enzyme may be important for optimal inhibition, although it is not essential for complex formation (29). In this regard, the initial binding of TPP I prosegment to its cognate protease might slowly induce (additional) more rigid structure of the prosegment, which in turn would clamp the enzyme much more efficiently, explaining the apparent two-step mechanism of inhibition.
Our studies demonstrated saturable inhibition of TPP I by its prosegment as a function of substrate concentration, thus suggesting an uncompetitive model of interaction. Although the inhibitory effect of many prosegments analyzed in the past was of a competitive type, the prosegments of several subtilases and PCs behaved as mixed type inhibitors of their cognate enzymes (24, 30, 31). Given that mixed inhibition can lead to an uncompetitive kinetics (32), it may be difficult to unequivocally distinguish these two modes of inhibition (33). The high resolution structure of the intact prokumamolisin, as the first prosedolisin, has been resolved recently (38), and it has been shown that the prosegment exhibits a half-β sandwich core docking to the catalytic domain, similarly to the equivalent subtilases and PCs. Given the overall structural similarity of sedolisins (5), a similar arrangement of the TPP I prosegment and catalytic domain of TPP I might be expected. The small size of the substrate cavity of TPP I predicted by computer modeling (34) rather precludes the retrofitting of the in trans-supplied prosegment, which favors the idea that TPP I prosegment cannot act as a purely competitive inhibitor. Accordingly, our data suggest that when applied in trans, the inhibitor strongly interacts with region(s) distinct from the active site, most likely with several sites on the enzyme.
We have shown that the ability of the prosegment to inhibit TPP I activity strongly depends on pH. These results suggest that multiple ionic interactions are involved in the establishment of the enzyme-prosegment complex. This robust pH dependence of the activity of in trans-provided TPP I prosegment toward its cognate enzyme correlates well with the pH-dependent activation of the proenzyme in vitro. TPP I proenzyme activates quickly at low pH, whereas at higher pH (4.5–5.5) the proenzyme is activated slowly and is processed to a polypeptide containing N-terminal extensions (10). However, most of the activity present in a preparation of the proenzyme incubated at pH 4.5 could be recovered by further lowering the pH, suggesting that the already cleaved-off prosegment remains tightly bound to the catalytic domain at pH 4.5. Limited (5-fold) dilution of the enzyme preinhibited at pH 5.0 and immediate measurement of the remaining activity at pH 3.5 and 5.0 demonstrated that the amount of released enzyme is ∼5 times higher at pH 3.5 than at pH 5.0 (not shown). These data indicate that the tight complex between the prosegment and TPP I is reversible, and its stability is a consequence of a slow dissociation rate of the prosegment held to the enzyme by strong ionic interactions.
Our studies also demonstrated that the alkaline pH-induced inactivation of TPP I was almost completely averted in the presence of the prosegment. Although the half-life of the enzyme alone under the experimental conditions employed was only ∼6.5 min, the presence of the prosegment led to the preservation of ∼84% activity after 2 h of incubation. This protective effect of the prosegment is significantly stronger than that afforded by the GAGs we demonstrated earlier (11). Although the binding site of GAGs and the prosegment on the TPP I molecule appear to partially overlap, the binding of GAGs was not saturable, and the interaction of the prosegment with its cognate enzyme was tight and saturable. It was documented that alkaline pH-induced inactivation of chicken cathepsin L was accompanied by the loss of α-helical content (35). Thus, it is reasonable to propose that the TPP I prosegment, by tightly binding its parent protease, might stabilize the activity of TPP I at alkaline pH by preventing pH-induced unfolding of the protein.
We also showed that upon extended incubation of the prosegment with TPP I (at a ratio of ∼1:5, enzyme/prosegment), the prosegment was partially degraded. Also, the prosegments of some subtilases (25, 36) and PCs (37) were found to be degraded by their cognate enzymes, although distinctly more efficiently than the TPP I prosegment. Obviously, degradation of the prosegment by TPP I might potentially compound our kinetic measurement results. However, our kinetic assays were performed under pseudo-first order conditions; thus, degradation of the prosegment was negligible under the experimental conditions employed. Accordingly, despite significant differences between the IC50 values we obtained in the pH range of 4.5–5.5, the degradation of the prosegment was similar in this pH range. Thus, the measured kinetic constants reflect the true inhibitory property of the prosegment and not the ability of the enzyme to degrade the prosegment under the tested conditions.
The majority of ∼56 mutations identified to date in CLN2 patients are either nonsense or intron/exon junction site mutations (see NCL Resource on the World Wide Web). Only G77R and S153P are pure missense mutations within the prosegment region of the proenzyme. On the molecular level, G77R mutation leads to a significant endoplasmic reticulum retention of the newly synthesized proenzyme (2-fold elongation of its half-life in comparison with WT proenzyme), its arrested secretion, and a dramatic decrease in the specific activity of the mature enzyme in the lysosomes (to ∼10% of the WT values),3 suggesting an altered endoplasmic reticulum folding pathway of mutated TPP I. When we analyzed presently the inhibitory properties of the heterologously expressed G77R prosegment against its cognate protease, we found not only that the inhibitory potency of the mutant prosegment was greatly diminished but also that its inhibition mechanism differed significantly from that of its WT counterpart. Although the WT prosegment acted as a slow-binding inhibitor, the G77R prosegment demonstrated a rapid equilibrium behavior, binding instantly and mostly, if not exclusively, to the substrate binding site of the enzyme. One possible explanation of this phenomenon is that the replacement of a small uncharged glycine residue by a large, positively charged arginine could lead to a disruption of important eletrostatic/hydrophobic interactions between the prosegment and its cognate enzyme. This, in turn, might interfere either with the precise fitting of the prosegment with the catalytic domain and/or the conformational, time-dependent fine tuning of the binding, resulting in a weaker binding of mutated than WT prosegment to the catalytic pocket.
The role of the prosegment in subtilases and PCs is 2-fold. First, it inhibits its cognate enzyme, which allows for the prevention of undue activation in terms of location and time. Second, it plays the role of intramolecular chaperone, assisting in folding the catalytic domain by lowering the transition state energy, allowing the conversion of a collapsed metastable intermediate to a native enzyme (39). Given our earlier observation that the prosegment of TPP I is indispensable for the expression of active TPP I,4 similar to other sedolisins (15, 40), and the results of our present studies demonstrating that the prosegment of TPP I is a potent, slow-binding inhibitor of its cognate protease, we can conclude that the prosegment of TPP I also plays a dual role acting as an intramolecular chaperone and as an inhibitor of its cognate enzyme. Future resolution of the crystal structure of TPP I proenzyme will provide more insight into these phenomena.
Acknowledgments
We thank Maureen Stoddard Marlow for copy editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant NS047355. This work was also supported by the New York State Office for Mental Retardation and Developmental Disabilities. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TPP I, tripeptidyl peptidase I; AMC, aminomethylcoumarin; GAG, glycosaminoglycan; CLN2, classic late infantile neuronal ceroid lipofuscinosis; aa, amino acid(s); WT, wild type.
M. Walus et al., unpublished results.
A. A. Golabek, N. Dolzhanskaya, M. Walus, K. E. Wisniewski, and E. Kida, unpublished results.
References
- 1.Golabek, A. A., and Kida, E. (2006) Biol. Chem. 387 1091-1099 [DOI] [PubMed] [Google Scholar]
- 2.Kida, E., Golabek, A. A., Walus, M., Wujek, P., Kaczmarski, W., and Wisniewski, K. E. (2001) J. Neuropathol. Exp. Neurol. 60 280-292 [DOI] [PubMed] [Google Scholar]
- 3.Kurachi, Y., Oka, A., Itoh, M., Mizuguchi, M., Hayashi, M., and Takashima, S. (2001) Acta Neuropathol. (Berl.) 102 20-26 [DOI] [PubMed] [Google Scholar]
- 4.Sleat, D. E., Donnelly, R. J., Lackland, H., Liu, C. G., Sohar, I., Pullarkat, R. K., and Lobel, P. (1997) Science 277 1802-1805 [DOI] [PubMed] [Google Scholar]
- 5.Wlodawer, A., Li, M., Gustchina, A., Oyama, H., Dunn, B. M., and Oda, K. (2003) Acta Biochim. Pol. 50 81-102 [PubMed] [Google Scholar]
- 6.Lin, L., Sohar, I., Lackland, H., and Lobel, P. (2001) J. Biol. Chem. 276 2249-2255 [DOI] [PubMed] [Google Scholar]
- 7.Walus, M., Kida, E., Wisniewski, K. E., and Golabek, A. A. (2005) FEBS Lett. 579 1383-1388 [DOI] [PubMed] [Google Scholar]
- 8.Lin, L., and Lobel, P. (2001) Biochem. J. 357 49-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golabek, A. A., Kida, E., Walus, M., Wujek, P., Mehta, P., and Wisniewski, K. E. (2003) J. Biol. Chem. 278 7135-7145 [DOI] [PubMed] [Google Scholar]
- 10.Golabek, A. A., Wujek, P., Walus, M., Bieler, S., Soto, C., Wisniewski, K. E., and Kida, E. (2004) J. Biol. Chem. 279 31058-31067 [DOI] [PubMed] [Google Scholar]
- 11.Golabek, A. A., Walus, M., Wisniewski, K. E., and Kida, E. (2005) J. Biol. Chem. 280 7550-7561 [DOI] [PubMed] [Google Scholar]
- 12.Khan, A. R., and James M. N. (1998) Protein Sci. 7 815-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smyth, M. J., O'Connor, M. D., and Trapani, J. A. (1996) J. Leukocyte Biol. 60 555-562 [DOI] [PubMed] [Google Scholar]
- 14.Kitamoto, Y., Yuan, X., Wu, Q., McCourt, D. W., and Sadler, J. E. (1994) Proc. Natl. Acad. Sci. U. S. A. 16 7588-7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oyama, H., Hamada, T., Ogasawara, S., Uchida, K., Murao, S., Beyer, B. B., Dunn, B. M., and Oda, K. (2002) J. Biochem. (Tokyo) 131 757-765 [DOI] [PubMed] [Google Scholar]
- 16.Fox, T., de Miguel, E., Mort, J. S., and Storer, A. C. (1992) Biochemistry 31 12571-12576 [DOI] [PubMed] [Google Scholar]
- 17.Morrison, J. F., and Walsh, C. T. (1988) Adv. Enzymol. Relat. Areas Mol. Biol. 61 201-301 [DOI] [PubMed] [Google Scholar]
- 18.Williams, J. W., Morrison, J. F., and Duggleby, R. G. (1979) Biochemistry 18 2567-2573 [DOI] [PubMed] [Google Scholar]
- 19.Copeland, R A. (2000) Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd Ed., John Wiley & Sons, Inc., New York
- 20.Ezaki, J., Takeda-Ezaki, M., Oda, K., and Kominami, E. (2000) Biochem. Biophys. Res. Commun. 268 904-908 [DOI] [PubMed] [Google Scholar]
- 21.Junaid, M. A., Wu, G., and Pullarkat, R. K. (2000) J. Neurochem. 74 287-294 [DOI] [PubMed] [Google Scholar]
- 22.Cheng, Y.-C., and Prusoff, W. H. (1973) Biochem. Pharmacol. 22 3099-3108 [DOI] [PubMed] [Google Scholar]
- 23.Shinde, U., Li, Y., and Inouye, M. (1995) Intramolecular Chaperones and Protein Folding (Shinde, U., and Inouye, M., eds) pp. 1-34, R. G. Landes Co., Austin, TX
- 24.Zhong, M., Munzer, J. S., Basak, A., Benjannet, S., Mowla, S. J., Decroly, E., Chretien, M., and Seidah, N. G. (1999) J. Biol. Chem. 274 33913-33920 [DOI] [PubMed] [Google Scholar]
- 25.Huang, H. W., Chen, W. C., Wu, C. Y., Yu, H. C., Lin, W. Y., Chen, S. T., and Wang, K. T. (1997) Protein Eng. 10 1227-1233 [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., Hu, Z., Jordan, F., and Inouye, M. (1995) J. Biol. Chem. 270 25127-25232 [DOI] [PubMed] [Google Scholar]
- 27.Groves, M. R., Coulombe, R., Jenkins, J., and Cygler, M. (1998) Proteins 32 504-514 [PubMed] [Google Scholar]
- 28.Gutiérrez-González, L. H., Rojo-Domínguez, A., Cabrera-González, N. E., Pérez-Montfort, R., and Padilla-Zúñiga, A. J. (2006) Arch. Biochem. Biophys. 446 151-160 [DOI] [PubMed] [Google Scholar]
- 29.Jean, L., Hackett, F., Martin, S. R., and Blackman, M. J. (2003) J. Biol. Chem. 278 28572-28579 [DOI] [PubMed] [Google Scholar]
- 30.Lesage, G., Tremblay, M., Guimond, J., and Boileau, G. (2001) FEBS Lett. 508 332-336 [DOI] [PubMed] [Google Scholar]
- 31.Basak, A., Ernst, B., Brewer, D., Seidah, N. G., Munzer, J. S., Lazure, C., and Lajoie, G. A. (1997) J. Pept. Res. 49 596-603 [DOI] [PubMed] [Google Scholar]
- 32.Frieden, C. (1964) J. Biol. Chem. 239 3522-3531 [PubMed] [Google Scholar]
- 33.Evin, G., Devin, J., Castro, B., Menard, J., and Corvol, P. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 48-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wlodawer, A., Durell, S. R., Li, M., Oyama, H., Oda, K., and Dunn, B. M. (2003) BMC Struct. Biol. 3 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dufour, E., Dive, V., and Toma, F. (1998) Biochim. Biophys. Acta. 955 58-64 [DOI] [PubMed] [Google Scholar]
- 36.Siezen, R. J., and Leunissen, J. A. (1997) Protein Sci. 6 501-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudreault, A., Gauthier, D., and Lazure, C. (1998) J. Biol. Chem. 273 31574-31580 [DOI] [PubMed] [Google Scholar]
- 38.Comellas-Bigler, M., Maskos, K., Huber, R., Oyama, H., Oda, K., and Bode, W. (2004) Structure 12 1313-1323 [DOI] [PubMed] [Google Scholar]
- 39.Eder, J., Rheinnecker, M., and Fersht, A. R. (1993) J. Mol. Biol. 233 293-304 [DOI] [PubMed] [Google Scholar]
- 40.Oda, K., Takahashi, T., Tokuda, Y., Shibano, Y., and Takahashi, S. (1994) J. Biol. Chem. 269 26518-26524 [PubMed] [Google Scholar]