Abstract
HIV-1 gp41 cytoplasmic tail (CT) is highly conserved among HIV-1 isolates, particularly the region designated lentivirus lytic peptide (LLP1–2), which includes two α-helical domains LLP1 and LLP2. Although the gp41 CT is recognized as a modulator of viral fusogenicity, little is known about the regulatory mechanism of this region in the viral fusion process. Here we report that anti-LLP1–2 and anti-LLP2 antibodies (IgG) inhibited HIV-1 Env-mediated cell fusion and bound to the interface between effector and target cells at a suboptimal temperature (31.5 °C), which slows down the fusion process and prolongs the fusion intermediate state. This suggests that LLP1–2, especially the LLP2 region located inside the viral membrane, is transiently exposed on the membrane surface during the fusion process. Synthetic LLP2 peptide could bind to the gp41 six-helix bundle core with high binding affinity. These results suggest that the gp41 CT may interact with the gp41 core, via the surface-exposed LLP2 domain, to regulate Env-mediated membrane fusion.
It is generally agreed that the human immunodeficiency virus (HIV)4 envelope glycoprotein (Env) transmembrane subunit gp41 plays an essential role during viral infection by mediating membrane fusion. Based on the crystal structure of the gp41 core, Chan et al. (1, 2) have proposed a membrane fusion model for HIV-1 entry. In this model, the Env surface subunit gp120 binds to both CD4 and the chemokine receptor (CCR5 or CXCR4) on the target cells to trigger a conformational change of gp41, i.e. association between its N- and C-terminal heptad repeats (NHR and CHR, respectively) to form a six-helix bundle (6-HB), which is thought to bring the membranes of both virus and target cells into close proximity for facilitating membrane fusion (1, 3–6). However, the events following 6-HB formation prior to completion of the fusion process are unclear, and the regulation of the fusion process remains to be elucidated. By using suboptimal temperature (31.5 °C) to slow down the fusion process, Golding et al. (7) have shown that the anti-bundle specific antibodies could bind to the 6-HB located between effector and target cells and block virus-cell and cell-cell fusion. These results suggest that 6-HBs are formed prior to membrane fusion, and after drawing the viral and target cell membranes together, they may perform additional functions in the late stage of membrane fusion. Meanwhile, apart from the important documented roles of the NHR, CHR, and the 6-HB in fusion reactivity, completion of the entire fusion process may also require the involvement of other regions in gp41, such as the membrane-spanning domain (aa 666–682 of the HXB2 strain of HIV-1) and the gp41 cytoplasmic tail (CT) (8–10).
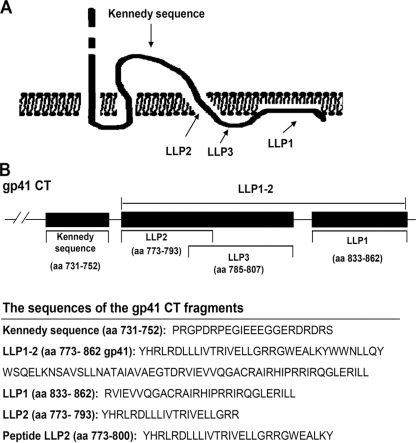
As members of the lentiviruses, HIV-1, HIV-2, and simian immunodeficiency virus have a relatively long cytoplasmic tail (CT) (∼150 amino acids), compared with other retroviruses with cytoplasmic domains consisting of only 20–30 amino acids (9). The HIV-1 gp41 CT consists of the “Kennedy” polypeptide sequence (aa 731–752) and the lentivirus lytic peptide (LLP1–2, aa 773–862), which contains three highly conserved α-helix domains as follows: LLP1 (aa 833–862), LLP2 (aa 773–793), and LLP3 (aa785–807) (11). The HIV-1 gp41 CT has many functions, including interaction with the plasma membrane, decrease of bilayer stability, alteration of membrane ionic permeability, and mediation of cell killing (10, 12–16). Recent studies have shown that the gp41 CTs of HIV and simian immunodeficiency virus correlate with their infectivity and Env-mediated cell-cell fusion (10, 15). The mutations or truncations of gp41 CT can affect HIV-1 Env-mediated cell-cell fusion, presumably because of modulation of the fusogenicity of Env (17, 18). Kalia et al. (15) discovered that one Env having a mutation in the gp41 CT LLP2 region (MX2) exhibited a decrease of about 90% in fusogenicity compared with the wild-type Env and that the mutation did not correspond to apparent defects in the levels of cell surface Env expression. However, in some other studies, certain point mutations and particularly truncations of gp41 CT were shown to induce faster fusion kinetics (19–22). Recently, Wyss et al. (10) have also demonstrated that the gp41 CT LLP2 domain restricted Env fusogenicity during Env processing, as did the murine leukemia viruses, where cleavage of a membrane-interactive R peptide at the C terminus is required for Env to become fusogenic. Although the combined data imply that gp41 CT could affect cell-cell fusion, this mechanism also remains vague and controversial. Therefore, further study of the characteristics of gp41 CT is warranted, especially that of its added effect on Env-mediated fusion reactivity. It is generally believed that the entire gp41 CT is located inside the cell. However, existing experimental and computational evidence suggest that the Kennedy sequence might lie outside the cell, because the antibodies against the Kennedy sequence can bind to virus-infected cells and neutralize HIV-1 infectivity (23, 24).
Our study focuses on the gp41 CT LLP1–2 region, especially the LLP1 and LLP2 regions that have been directly proven to be associated with the membrane fusion process (10, 15), and aims to determine whether LLP1–2 directly or indirectly participates in the fusion reaction and, if so, to understand the underlying mechanism(s) of this event. Here we report, for the first time, that during the HIV-1 fusion process, the LLP1–2 region of gp41 CT, in particular the LLP2 domain, may be transiently exposed on the surface of the effector cell in the presence of the target cell. Synthetic peptide derived from the LLP2 sequence could interact with the gp41 6-HB core. These results suggest that gp41 CT may regulate the fusogenicity of viral Env through the interaction of the transiently exposed LLP2 region with the gp41 core on virions or HIV-1-infected cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Animals—3T3 cells stably expressing CD4 and CXCR4 (3T3-T4-CXCR4) were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 100 IU/ml of penicillin and streptomycin. CHO cells stably transfected with HIV-1 (HXB2) Env expressing vector pEE 14 (designated CHO-WT) or control pEE 14 vector (designated CHO-EE) were cultured in glutamine-deficient minimal essential medium (GMEM-S) containing 400 μm methionine sulfoximine (Sigma).
Expression and Purification of gp41 CT Fragments—The gp41 CT fragments LLP1–2, LLP1, and LLP2 (Fig. 1) were expressed in Escherichia coli using the plasmids pGEX-6p-1-LLP1–2, pGEX-6p-1-LLP1, and pGEX-6p-1-LLP2, respectively, as described previously (25). The polypeptide N36-L8-C34, its mutants, and 3NC were constructed and expressed as described previously (26–28), and further purified by anion-exchange Q column (GE Healthcare). Peptide LLP2 was synthesized by a standard solid phase Fmoc (N-(9-fluorenyl)methoxycarbonyl) method and purified to homogeneity (>95% purity) by high pressure liquid chromatography.
FIGURE 1.
The locations and sequences of the gp41 CT fragments. A, putative model of the HIV-1 gp41 CT as proposed by Cleveland et al. (23). The Kennedy sequence may be exposed on the exterior of the virus membrane. B, sequences of the gp41 CT fragments.
Production and Detection of Anti-gp41 CT Polyclonal Antibodies—New Zealand White rabbits (1.7–1.9 kg) were immunized subcutaneously with a dose of 100 μg of purified recombinant protein LLP1–2 or peptide LLP2 in complete Freund's adjuvant and boosted three times at 2-week intervals with an identical dose of LLP1–2 or LLP2 in incomplete Freund's adjuvant. Rabbit antisera were collected, and the anti-LLP1–2 and anti-LLP2 IgG were purified with protein G affinity column (GE Healthcare), from which LLP1–2-, LLP1-, and LLP2-specific IgG were further isolated using N-hydroxysuccinimide-activated Sepharose 4 Fast Flow beads (GE Healthcare) conjugated with the corresponding proteins or peptides, respectively. The flow-through fractions were also collected as controls. Antibody concentrations were established by spectrophotometry.
HIV-1 Env-mediated Cell-Cell Fusion Assay—Wells of a 96-well plate were coated with 1 × 106 3T3 cells stably expressing CD4 and CXCR4 (3T3-T4-CXCR4) and incubated overnight at 37 °C. Simultaneously, CHO-WT cells were pre-stimulated with 7 mm sodium butyrate for about 20 h and then 2×104 cells per well were added in the absence or presence of anti-gp41 CT-specific IgG or normal rabbit IgG. After coculturing for 6 h at 5% CO2, 31.5 °C, the plate was gently transferred to a 5% CO2, 37 °C environment. After a further 9–16 h, syncytia were counted under a microscope. The percentage of inhibition of syncytium formation was calculated using the following formula: % inhibition = (1 - (number of syncytia in a well containing an inhibitor)/number of syncytia in a well containing no inhibitor) × 100.
Flow Cytometry Assay—CHO-WT cells or a mixture of CHO-WT cells and 3T3-T4-CXCR4 cells (1 × 105) were incubated with anti-gp41 CT-specific IgG or normal rabbit IgG in a 50-μl final volume at 31.5 °C for 2 h in GMEM containing 2% goat serum. Then the mixtures were transferred onto ice for 1 h. After thorough washing with TBS and 2% goat serum (pH 7.4), cells were incubated with FITC-conjugated goat anti-rabbit IgG (Dako, 1:40 dilution in TBS with 2% goat serum) for 1 h on ice. Finally, the mixtures were washed three times using 200 μl of TBS with 2% goat serum and subsequently analyzed in a FACSCalibur (BD Biosciences) with CellQuest™ software (BD Biosciences).
Immunofluorescence—To observe binding of LLP1–2- and LLP2-specific IgG to fusion intermediates, Env-expressing CHO-WT and 3T3-T4-CXCR4 cells, respectively, were blocked with blocking buffer (2% normal goat serum in TBS) for 30 min at 31.5 °C, followed by incubation with rabbit LLP1–2-specific IgG, LLP2-specific IgG, or normal rabbit IgG (50 μg/ml) for 2 h at 31.5 °C in GMEM containing 2% goat serum. Then the cells were immediately placed on ice for 1 h. After the cells were harvested, they were washed twice with ice-cold blocking buffer and incubated with FITC-conjugated goat anti-rabbit IgG (Dako, 1:40 dilution in TBS with 2% goat serum). Finally, the cells were washed again, fixed, applied to poly-l-lysine-coated glass slides, mounted, and observed under the confocal microscope.
Enzyme-linked Immunosorbent Assay (ELISA)—To determine the specific antibody binding to the gp41 CT fragments, ELISA was carried out as described previously (29). Briefly, the respective recombinant proteins and peptides (50 μg/ml) dissolved in 0.1 m Tris (pH 8.8) were used to coat 96-well polystyrene plates, which were then blocked with Tris-buffered solution (pH 8.5) containing 0.25% gelatin and 0.1% Tween 20. After three washes with TBS (pH 7.4) containing 0.1% Tween 20 (TBS-T), the respective IgGs were added to the wells at the indicated concentrations. Then swine-anti-rabbit IgG and the substrate o-phenylenediamine were added sequentially. The absorbance at 450 nm (A450) was recorded with an ELISA reader. Each sample was tested in triplicate. Binding of the mAb NC-1 and the recombinant LLP1–2 and LLP2, respectively, to the peptides N36, C34, or the 6-HB modeled by N36/C34, N36-L8-C34, 3NC, and N36-L8-C34 mutants, was determined as described previously (30). Briefly, 96-well polystyrene plates were coated with 100 μl of LLP1–2 (20 μg/ml), LLP2 (20 μg/ml), NC-1 (2 μg/ml), or 0.25% gelatin (as a negative control), respectively, and blocked with TBS (pH 8.5) containing 0.25% gelatin and 0.1% Tween 20. After three washes, the N36/C34 mixture, the isolated N36 or C34 peptide (5 μg/ml for each peptide) (Fig. 5A, panel a), recombinant N36-L8-C34 and 3NC at indicated concentrations (Fig. 5A, panel b), and N36-L8-C34 mutants (5 μg/ml) (Fig. 7A) were added, respectively. After incubation at 37 °C for 45 min, rabbit anti-N36/C34 IgG, which can recognize both the individual peptides N36 and C34 and 6-HBs, peroxidase-conjugated anti-rabbit IgG and substrate o-phenylenediamine were added sequentially. Absorbance (A450) values were recorded. The relative binding activity (%) of the N36-L8-C34 (NC) mutants to LLP2 peptide was calculated on the basis that the binding activity of the wild-type N36-L8-C34 (NC WT) to LLP2 peptide equals 100% (Fig. 7A).
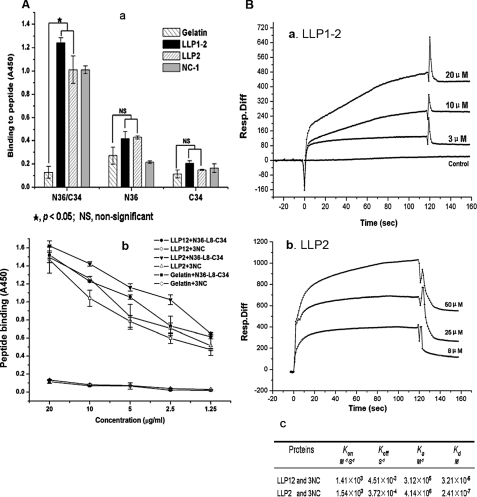
FIGURE 5.
Interaction of LLP1–2 or LLP2 with HIV-1 gp41 6-HB core. A, ELISA was performed to determine the binding activity of the recombinant protein LLP1–2, synthetic peptide LLP2, and mAb NC-1 to the peptides N36 and C34 and the 6-HB formed by the N36/C34 mixture, respectively, at 1 μm concentration (panel a) or the binding activity of the recombinant LLP1–2 and peptide LLP2 to the 6-HBs modeled N36-L8-C34 and 3NC, respectively, at graded concentrations (panel b). The data represent triplicate determinations that were performed at least three times. All results are presented as mean ± S.D. WINKS 4.651 evaluation program was used to assess the statistical significance of differences between groups. B, detection by SPR of the binding activity of the recombinant protein LLP1–2 (panel a) or peptide LLP2 (panel b) to the 6-HB formed by 3NC at graded concentrations. The association (Kon) and dissociation (Koff) rate constants as well as the dissociation equilibrium constant (Kd), calculated as described under “Experimental Procedures,” are shown in B, panel c.
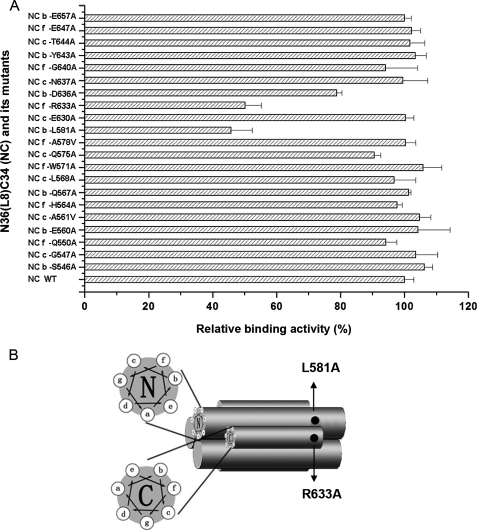
FIGURE 7.
A, relative binding activity of 6-HBs formed by N36-L8-C34 (NC) and its mutants to the synthetic peptide LLP2. Each mutant of 6-HB bears a single Ala or Val substitution at the b, c, and f positions of the helix wheels of the N- and C-helices in 6-HB. The binding activity was detected by ELISA. The data represent triplicate determinations that were performed at least three times. All results are presented as mean ± S.D. B, schematic shows the potential binding sites of LLP2 in gp41 6-HB.
Surface Plasmon Resonance (SPR) Assay—The kinetics of the binding affinity of the gp41 CT LLP1–2 and LLP2 to gp41 core modeled by recombinant protein 3NC was determined by SPR using the Biacore system (GE Healthcare) according to the biomolecular interaction analysis (BIA) technology manual. In brief, the CM5 sensor chip was immobilized with 3NC (100 μg/ml) by amine coupling, and the unreacted sites were blocked with ethanolamine. The association reaction was initiated by injecting 40 μl of polypeptide LLP1–2 or LLP2 in HEPES buffer (pH 7.4) with 0.05% Triton X-100 at a flow rate of 20 μl/min. The dissociation reaction was done by washing with running buffer (10 mm HEPES (pH 7.4) containing 0.15 m NaCl, 3.4 mm EDTA and 0.005% v/v surfactant P20). At the end of the cycle, the sensor chip surface was regenerated with 10 mm HCl for 30 s. The association (Kon) and dissociation (Koff) rate constants were estimated from the response curve, and the dissociation equilibrium constant (Kd) was calculated using the following equation: Kd = Koff/Kon, as described previously (31).
RESULTS
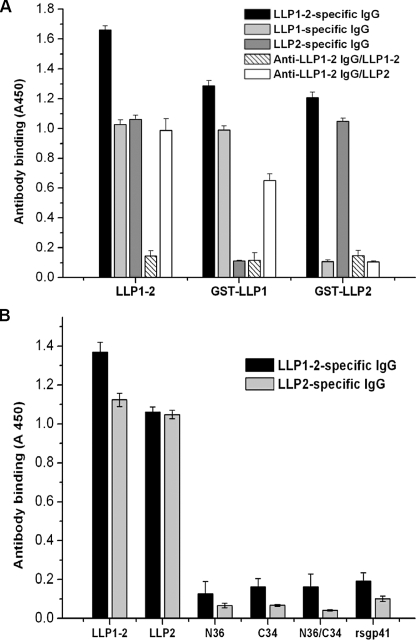
Isolation and Characterization of Antibodies Specific for the gp41 CT Fragments LLP1–2, LLP1, and LLP2—The gp41 CT fragment LLP1–2, expressed in E. coli using the plasmids pGEX-6p-1, and the synthetic peptide LLP2 were used for immunization of rabbits. Anti-LLP1–2 and anti-LLP2 IgG, respectively, were purified from the rabbit antisera with protein G affinity column (GE Healthcare). Then LLP1–2-, LLP1-, and LLP2-specific IgG were isolated from anti-LLP1–2 and anti-LLP2 IgG using the GST-LLP1–2-, GST-LLP1-, and GST-LLP2-bound N-hydroxysuccinimide-activated Sepharose (4 Fast Flow, GE Healthcare), respectively. The flow-through fractions from LLP1–2- and LLP2-bound Sepharose 4 were collected as LLP1–2-specific IgG-depleted anti-LLP1–2 IgG (designated anti-LLP1–2 IgG/LLP1–2) and LLP2-specific IgG-depleted anti-LLP1–2 IgG (designated anti-LLP1–2 IgG/LLP2), respectively. The binding specificity of the IgG obtained was determined by ELISA. As shown in Fig. 2A, LLP1–2-specific IgG bound with LLP1–2, GST-LLP1, and GST-LLP2. LLP1-specific IgG reacted with both LLP1–2 and GST-LLP1 but did not interact with GST-LLP2, whereas LLP2-specific IgG bound to both LLP1–2 and GST-LLP2 but did not recognize GST-LLP1. These results verify the specificity of the antibodies directed against the corresponding gp41 CT fragments. No LLP1–2- and LLP2-specific IgGs were detected in the flow-through fractions from the Sepharose 4B columns conjugated with LLP1–2 (anti-LLP1–2 IgG/LLP1–2) and LLP2 (anti-LLP1–2 IgG/LLP2), respectively, suggesting that the corresponding IgGs were depleted from the anti-LLP1–2 IgG fraction. Interestingly, LLP2-specific IgG predominated over the LLP1-specific IgG in the anti-LLP1–2 IgG (data not shown). In the next step, the LLP1–2- and LLP2-specific antibodies were used for detecting the potential interaction of the gp41 CT fragments with the peptides derived from the gp41 NHR or CHR or 6-HB. To exclude the possibility of the anti-gp41 CT antibodies binding nonspecifically to the extracellular domain of gp41, an ELISA was performed. As shown in Fig. 2B, anti-LLP1–2 and anti-LLP2 IgGs only recognize LLP1–2 and LLP2 but did not react with the rsgp41, nor with the peptides N36 and C34 as well as the 6-HB formed by N36/C34, confirming that anti-gp41 CT antibodies do not cross-react with the gp41 ectodomain.
FIGURE 2.
Binding specificity of antibodies directed against various anti-gp41 CT fragments. A, binding of LLP1–2- and LLP2-specific IgG to recombinant proteins LLP1–2, GST-LLP1, and GST-LLP2 in ELISA. B, binding of LLP1–2- and LLP2-specific IgG to recombinant protein LLP1–2, peptides LLP2, N36, C34, 6-HB modeled by N36/C34, and rsg41 in ELISA. The data represent triplicate determinations that were performed at least three times. All results are presented as mean ± S.D.
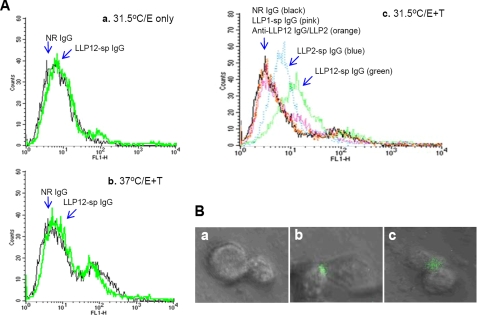
Binding of Anti-gp41 CT Antibodies to the Effector Cells in the Presence of Target Cells at Suboptimal Temperature—It was reported previously that the main part of the gp41 CT, i.e. the LLP1–2 region, is hidden within the HIV-1 virion (32) and cannot be recognized by the antibodies specific for the gp41 CT sequences (aa 799–817 and 844–863) (23). However, we believe that part of the gp41 LLP1–2 region may be exposed on the cell surface during the membrane fusion process, interacting with the extracellular domain of gp41, thus regulating viral fusogenicity via the “insideout” signaling pathway. To determine the potential exposure of gp41 CT fragments, flow cytometry assay was performed to detect the binding of LLP1–2-, LLP1-, and LLP2-specific IgG to the effector (E) cells that express HIV-1 Env (CHO-WT cells) in the absence or presence of the target (T) cells, which express CD4 and the coreceptor CXCR4 (3T3-T4-CXCR4 cells), at a physiological temperature (37 °C) or a suboptimal temperature (31.5 °C). The results show that LLP1–2-specific IgG did not bind to effector cells in the absence of target cells at 37 °C (data not shown) nor at 31.5 °C (Fig. 3A, panel a). This antibody was also unable to bind to effector cells in the presence of target cells at 37 °C (Fig. 3A, panel b). However, both LLP1–2- and LLP2-specific IgG bound significantly with the effector cells in the presence of target cells at 31.5 °C (Fig. 3A, panel c). In contrast, the LLP1-specific IgG and the LLP2-specific IgG-depleted anti-LLP1–2 IgG (anti-LLP-12 IgG/LLP2) were incapable of binding to effector cells under the same conditions (Fig. 3A, panel c). These results suggest that the LLP2, and not the LLP1 domain in the gp41 CT LLP1–2 region, may be exposed on the surface of the effector cell when it interacts with the target cell for cell-to-cell fusion, resulting in binding of the anti-LLP2 antibody to the effector cell. However, this binding is only detectable when the effector cell interacts with the target cell at suboptimal temperature, which can prolong the fusion intermediate state. Otherwise, the antibody (IgG) may not have access to the narrow space of the interface between the effector and target cell during the conventional fusion process at physiological temperatures. This assumption is further supported by results from confocal microscopy analysis. At suboptimal temperature, normal rabbit IgG could not bind to effector cells in the presence of target cells (Fig. 3B, panel a). However, both LLP1–2-specific IgG (Fig. 3B, panel b) and LLP2-specific IgG (Fig. 3B, panel c) bound significantly to the interfaces of the effector/target cells. None of these antibodies bound to effector cells in the absence of target cells at either 37 or 31.5 °C, nor in the presence of target cells at 37 °C (data not shown).
FIGURE 3.
Binding of anti-gp41 CT antibodies to effector (E) cells (CHO-WT cells) in the presence or absence of target (T) cells (3T3-T4-CXCR4 cells) at physiological temperature (37 °C) or suboptimal temperature (31. 5 °C). A, flow cytometric analysis. Purified normal rabbit (NR) IgG or anti-gp41 CT antibodies were incubated for 2 h with effector cells in the absence of target cells at 31.5 °C (panel a), or in the presence of target cells at 37 °C (panel b), or in the presence of target cells at 31.5 °C (panel c). The mixtures were transferred onto ice and incubated for 1 h, followed by washes to remove unbound antibodies. The cells were then incubated with FITC-conjugated goat anti-rabbit IgG for 1 h on ice, before analysis in a FACSCalibur. B, confocal microscopy analysis. Purified normal rabbit (NR) IgG (panel a) or LLP1–2-specific IgG (panel b), or LLP2-specific IgG (panel c) was incubated with effector cells in the presence of target cells at 31.5 °C for 2 h. The cells were stained with FITC-conjugated goat anti-rabbit IgG as described above and analyzed under a confocal microscope. The presence of green color in the E/T cell junction areas indicates the binding sites of anti-LLP1–2 or anti-LLP2 antibodies.
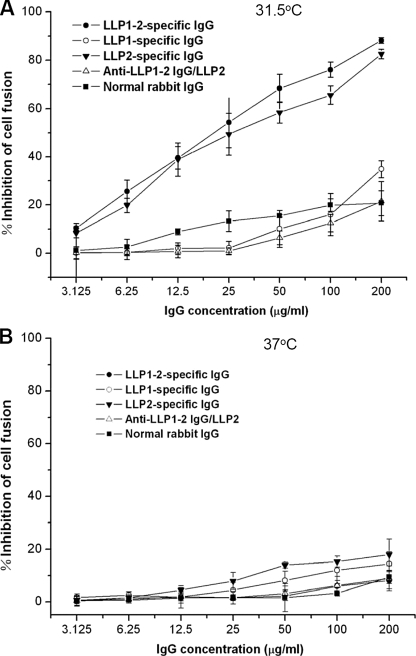
Inhibition of Env-mediated Cell Fusion by Anti-gp41 CT Antibodies at Suboptimal Temperature—We further hypothesized that during the membrane fusion process, the gp41 CT LLP2 domain may be transiently exposed on the surface of effector cells at a suboptimal temperature to interact with the extracellular domain for regulating viral fusogenicity. Binding of the anti-LLP2 antibodies to the exposed LLP2 domain may affect the membrane fusion process. Therefore, the inhibitory activity of antibodies against gp41 CT fragments on Env-mediated cell-to-cell fusion at physiological and suboptimal temperatures was determined by using a syncytium formation assay. As shown in Fig. 4A, both LLP1–2- and LLP2-specific IgG showed potent inhibitory activity against Env-mediated syncytium formation when effector and target cells were cocultured at a suboptimal temperature (31.5 °C), whereas normal rabbit IgG, LLP1-specific IgG, and the LLP2-specific IgG-depleted anti-LLP1–2 IgG (anti-LLP-1–2 IgG/LLP2) exhibited no significant inhibition of cell-cell fusion. However, none of the anti-gp41 CT antibodies could inhibit Env-mediated syncytium formation when effector and target cells were cocultured at 37 °C (Fig. 4B). This result suggests that the LLP1–2- and LLP2-specific IgG may bind to the LLP2 domain, which is transiently exposed on the effector cell surface during the interaction between effector and target cells, thereby interfering with gp41 CT-mediated cell-cell fusion. Similarly, the antibodies may not be able to access the E/T interface during interaction of effector cell with target cell at physiological temperatures, resulting in lack of cell-cell fusion inhibition.
FIGURE 4.
Inhibition of anti-gp41 CT antibodies on HIV-1 Env-mediated cell-cell fusion at suboptimal temperature. CHO-WT cells and 3T3-T4-CXCR4 cells were cocultured for 6 h in the presence of LLP1–2-, LLP1-, LLP2-specific IgG, anti-LLP1–2 IgG/LLP2, and normal rabbit IgG (control), respectively, at graded concentrations at 31.5 °C (A) or 37 °C (B) and then cocultured for additional 16 h at 37 °C. The syncytia were then counted under a microscope. The data represent triplicate determinations that were performed at least three times. All results are presented as mean ± S.D.
Interaction of LLP1–2 and LLP2 with gp41 6-HB Core—The next logical question is which molecule LLP1–2 or LLP2 will interact with after its surface exposure. We hypothesized that LLP1–2 or LLP2 may affect the HIV-1 Env-mediated membrane fusion by interacting with either the gp41 NHR or CHR region at an earlier stage or with the gp41 6-HB core at the later stage of cell-cell fusion. We thus tested whether LLP1–2 could bind to NHR-peptide N36, or CHR-peptide C34, or 6-HB core modeled by N36/C34. Surprisingly, recombinant protein LLP1–2 and synthetic peptide LLP2 strongly bound to the 6-HB core formed by the N36/C34 mixture, but exhibited no significant binding to the individual N36 and C34 peptides (Fig. 5A). To confirm binding between LLP2 (or LLP1–2) and the 6-HB core, we expressed and purified the polypeptides N36-L8-C34 (26) and 3NC, which can automatically form 6-HBs in physiological solution (27, 28). As shown in Fig. 5A, panel b, both the recombinant protein LLP1–2 and peptide LLP2 strongly bound to N36-L8-C34 and 3NC in a dose-dependent manner. In the real time SPR spectrum assay, similar results were obtained, i.e. both LLP1–2 (Fig. 5B, panel a) and LLP2 (Fig. 5B, panel b) bound to 3NC in a dose-dependent manner with high binding affinity. However, the data did not fit the 1:1 Langmuir binding and other existing models, which may be attributed to the fact that N36-L8-C34 and 3NC form typical 6-HBs in physiological solution (26–28, 31), whereas the recombinant LLP1–2 and peptide LLP2 were present as monomer, dimer, trimer, tetramer, and high ordered polymers in phosphate-buffered saline (data not shown), consistent with findings reported by Lee et al. (33). Therefore, one molecule of N36-L8-C34 or 3NC may interact with varying numbers of LLP1–2 or LLP2 molecules in different multimeric forms.
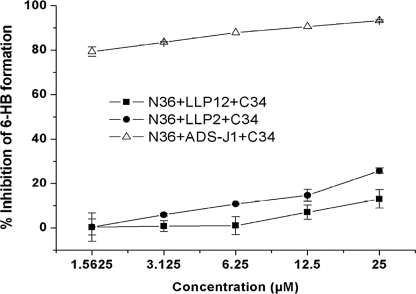
Next, we investigated whether the gp41 CT fragments LLP1–2 and LLP2 could inhibit 6-HB formation between gp41 NHR and CHR peptides N36 and C34 as detected by ELISA, using mAb NC-1 that binds specifically to the N36-C34 complex (i.e. 6-HB) but not to the individual peptides (34). Unlike the fusion inhibitors ADS-J1 (29), neither LLP1–2 nor LLP2 could inhibit 6-HB formation (Fig. 6). The above results indicate that the gp41 CT LLP2 domain may not affect the membrane fusion at an earlier stage but rather at the late stage of fusion process by interacting with the gp41 6-HB core.
FIGURE 6.
Inhibition of the recombinant protein LLP1–2 and synthetic peptide LLP2, respectively, on gp41 6-HB formation as measured by ELISA. ADS-J1, a potent inhibitor of gp41 6-HB formation (31), was used as a control. The data represent triplicate determinations that were performed at least three times. All results are presented as mean ± S.D.
Binding Site Mapping with 6-HB Mutants—To examine the binding sites of the LLP2 domain in gp41 6-HB, we expressed and purified 23 different N36-L8-C34 mutants, in which the amino acid residues at the b, c, and f positions of the helix wheels of N- and C-helices in the 6-HB were replaced by Ala or Val. These sites are located on the surface of 6-HB, and mutations at these sites are not expected to disrupt 6-HB conformation. The binding of the synthetic peptide LLP2 to the 23 respective mutants was determined as described under “Experimental Procedures.” To verify the feasibility of the binding assay for detecting 6-HB mutants, we first established the association of the anti-N36/C34 rabbit polyclonal antibody with each mutant, and we found that this polyclonal antibody bound to most of the mutants with similar affinity (data not shown). Then the interaction between LLP2 and each mutant was detected using the anti-N36/C34 rabbit antibody. The results indicate that peptide LLP2 bound to the 6-HB mutants L581A and R633A to a much lesser extent than to the wild-type 6-HB and other 6-HB mutants (Fig. 7A), suggesting that Leu-581 and Arg-633 are possible binding sites of LLP2. Interestingly, these two sites are close to each other in the 6-HB, near the loop, and located separately on the C-terminal region of NHR, and N-terminal region of CHR (Fig. 7B).
DISCUSSION
Recent studies on gp41 CT, especially the LLP regions, have increasingly focused on the potential relationship between gp41 CT and the fusion process (10, 15), which can be attributed mainly to several factors as follows. (i) The HIV entry process is considered to be an attractive target for anti-HIV drug design, as blocking HIV entry into target cells leads to prevention of viral infection (6, 35). Hence, it is very important to further determine the mechanisms underlying the fusion process to identify new and effective drug targets. (ii) The domains in gp41 involved in interaction and destabilization of the viral and host membranes may play an essential role in the fusion process. It has been reported that the tryptophan-rich region, directly adjacent to the membrane-proximal external region, and other segments in gp41 may bind to the surfaces of phospholipid membranes, resulting in conformational change and destabilization. Thus, these domains may possibly interact with biological membranes, taking part in fusion reactivity (6, 16, 36–40). (iii) Accumulated data indicate that gp41 CT LLP regions, which can interact with phospholipid membranes, may be involved in modulating cell-cell fusion by certain as-yet-undefined pathways (10, 15, 17–21). It has been widely recognized that Gag matrix proteins mediate a mechanism of inside-out regulation of HIV-1 Env fusogenicity (18, 41, 42). In immature virions, Gag precursor protein interacts with the gp41 CT to limit Env-mediated fusion. During virion maturation, proteolytic cleavage of the p55 Gag precursor protein is required to generate maximal Env fusion activity. It has been proposed that the LLP domain may regulate Env fusogenicity by associating with plasma membranes and/or interacting with cellular proteins to alter conformation of the gp41 ectodomain (10). However, the mechanism for this insideout signaling effect has not yet been clearly defined.
In this study, we attempted to investigate whether the important functional domains in the gp41 CT region, such as LLP1–2, LLP1, and LLP2, interact directly with the extracellular domain of gp41 to regulate the viral fusogenicity. We first generated and purified a series of antibodies specific for the gp41 CT fragments LLP1–2, LLP1, and LLP2, respectively. These antibodies were then used to probe potential gp41 CT region(s) exposed on the surface of HIV-1 Env expressing effector cells serving as models of virus-infected cells or cell-free virions. Flow cytometric analysis showed that both LLP1–2- and LLP2-specific IgG bound significantly with effector cells (CHO-WT cells) in the presence of target cells (3T3-T4-CXCR4 cells) at a suboptimal temperature (31.5 °C), whereas LLP1-specific IgG and the LLP2-specific IgG-depleted anti-LLP1–2 IgG could not bind to effector cells under the same conditions (Fig. 3A, panel c). These results suggest that the LLP2 domain located in the gp41 CT LLP1–2 region may be exposed on the surface of the effector cell after its interaction with the target cell. This finding was confirmed by confocal microscopy analysis, which showed that LLP2-specific IgG bound significantly to the interfaces between effector and target cells when both E/T cells were cocultured at 31.5 °C (Fig. 3B, panel c). This study has benefited from a report by Golding et al. (7) who demonstrated that some antibodies specific for gp41 may not have access to the corresponding gp41 region during the Env-mediated virus-cell or cell-cell fusion process at a physiological temperature (37 °C), but it can bind to their epitopes when effector and target cells are cocultured at a suboptimal temperature (31.5 °C), which can slow down fusion process and prolong the intermediate state. Indeed, none of the antibodies against the gp41 CT fragments could bind to the effector cells in the presence of target cells at 37 °C, even if some domain(s) in the LLP1–2 region may be exposed.
To investigate the potential role of the gp41 CT LLP2 domain in the Env-mediated cell-cell fusion, we determined the inhibitory activity of the anti-LLP1–2 and anti-LLP2 antibodies on syncytium formation. Remarkably, both LLP1–2- and LLP2-specific IgG exhibited potent inhibitory activity against Env-mediated cell-cell fusion when effector and target cells were cocultured at 31.5 °C, whereas normal rabbit IgG, LLP1-specific IgG, and the LLP2-specific IgG-depleted anti-LLP1–2 IgG showed no significant inhibition of cell-cell fusion under the same conditions (Fig. 4A). Similarly, none of the anti-gp41 CT antibodies could inhibit Env-mediated syncytium formation when effector and target cells were cocultured at 37 °C (Fig. 4B). These results further support our assumption that the anti-gp41 CT antibodies may bind to the transiently exposed LLP2 domain during membrane fusion and interfere with the fusion process, resulting in inhibition of syncytium formation. However, caution should be taken in interpreting the results obtained from experiments when cells expressing HIV-1 Env rather than cell-free virions are used, because cell-associated Env may not act identically as virus-associated Env to mediate membrane fusion.
We then proposed that LLP2 may interact with some functional region(s) in the extracellular domain of gp41 to carry out its function of regulating viral fusogenicity. Our study as well as other previous ones have shown that the gp41 NHR and CHR regions and the gp41 core formed by the association of these two regions play critical roles in Env-mediated membrane fusion and are important targets for HIV fusion inhibitors. We thus investigated first by ELISA and SPR whether the LLP2 peptide could bind to any of these functional domains. We found that the LLP2 peptide bound neither to the NHR peptide nor the CHR peptide, but bound with high affinity to the gp41 6-HB formed by the NHR and CHR peptides (Fig. 5). This result was confirmed by using different gp41 peptides and recombinant proteins containing the NHR and/or CHR sequences, including N36 peptide/C34 peptide, N36-L8-C34, and 3NC, all of which have been previously proven to form stable 6-HBs (26–28, 31). We further demonstrated that the binding site of LLP2 may be located at the Leu-581 and Arg-633 positions, overlapping both N- and C-helices in the 6-HB (Fig. 7B). Because the LLP2 peptide could not bind to NHR or CHR, nor block the 6-HB formation (Fig. 6), we thus proposed that the gp41 CT LLP2 domain may regulate viral fusogenicity by affecting the late stage of membrane fusion, i.e. after association of NHR and CHR to form 6-HB core, but prior to completion of membrane fusion. However, one should be cautious when interpreting results obtained from experiments using cells expressing HIV-1 Env rather than cell-free virions, because cell-associated Env and virus-associated Env may not function identically.
Because the LLP2-specific antibody inhibits HIV-1 Env-mediated syncytium formation at suboptimal temperatures (Fig. 4A), one may assume that the interaction between LLP2 and the gp41 6-HB core may promote HIV-1 fusion, and removal of this peptide from the CT may result in inhibition of viral fusion. Indeed, Sodroski and co-workers (20) demonstrated that deletion of the fragment (aa 754–797) corresponding to the LLP2 peptide of the gp41 CT resulted in 67–80% reduction of HIV-1-mediated syncytium formation, whereas truncation of the fragment (aa 846–856) overlapping the LLP1 peptide or the entire CT (aa 726–856) had no significant effect on cell-cell fusion. Kalia et al. (15) showed that mutations in gp41 CT LLP2 region caused about 90% decrease in fusogenicity. However, others reported that truncation of gp41 CT could induce faster fusion kinetics or increased fusion efficiency (10, 18, 19). These findings suggest that truncations or mutations of the sequences in different regions of the gp41 CT may result in distinct changes in the viral fusion activity. The inhibitory activity of the LLP2-specific antibody on the HIV-1 Env-mediated syncytium formation may not be attributed to its blockage of association between the LLP2 and the gp41 core, but rather ascribed to its steric hindrance effect by binding to the exposed LLP2 at the interface between the effector and target cells, in a way like the gp41 6-HB-specific mAbs that bind to the gp41 core and inhibit the HIV-1 Env-mediated cell-cell fusion at suboptimal temperatures (7). We believe that the gp41 core may participate in the inside-out regulation mediated by the interactions between gp41 CT and Gag matrix proteins as described above.
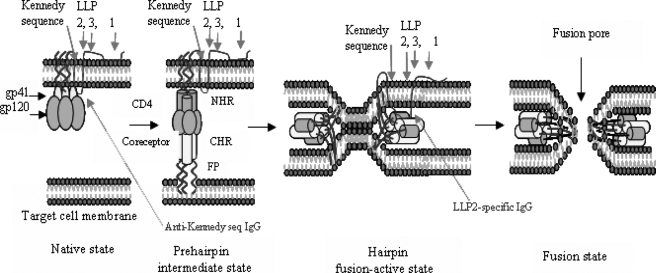
Based on the results presented here, we further developed the Env-mediated fusion model proposed by Chan and Kim (2). After gp120 binds to CD4 and a coreceptor, gp41 changes conformation by inserting its N-terminal fusion peptide into the target cell membrane exposing the NHR and CHR regions. Then NHR and CHR associate to form a 6-HB, which may bring the viral and cell membranes into close proximity for fusion. At this stage, the gp41 CT LLP2 domain may be exposed, triggered possibly by the Gag protein, to interact with the 6-HB to regulate the viral fusion process (Fig. 8). For clarity, only one copy of the gp41 CT is shown in the figure. In reality, it is expected that the gp41 CT forms a trimer or another oligomeric form during the membrane fusion process because the gp41 extracellular domain forms a fusogenic core in the trimer-of-heterodimer (or 6-HB) form at this stage. Although there is no conclusive evidence to prove this assumption, Lee et al. (33) have shown that folding of the gp41 CT as a multimeric structure is a required step for gp41-mediated viral entry. Further investigation of the effect of gp41 CT oligomerization on its interaction with the gp41 core and on the virus fusion process is warranted.
FIGURE 8.
A schematic diagram of interaction of gp41 CT with 6-HB during Env-mediated membrane fusion. In the native state, the Kennedy sequence in HIV-1 gp41 CT may be exposed on the exterior of the virion (the target for antibodies against Kennedy sequence) (23). After gp120 binds to CD4 and a coreceptor, gp41 changes conformation by inserting its N-terminal fusion peptide into the target cell membrane exposing the NHR and CHR. Then NHR and CHR associate to form a 6-HB, which brings the viral and cell membrane into close proximity, resulting in formation of the fusion pore. In the fusion-active state, the exposed gp41 CT, in particular the LLP2 domain, may interact with the gp41 6-HB to regulate the viral fusion process.
All components of gp41 involved in virus fusion and entry may serve as targets for HIV fusion inhibitors. The best example is the gp41 NHR region, the target of the CHR peptides, e.g. SJ-2176 (43), C34 (44), and T-20 (brand name Fuzeon), which is the first peptidic HIV fusion inhibitor approved by the United States Food and Drug Administration for clinical use. CHR is the target for NHR peptides, e.g. N36 (44) and 5-helix (45), whereas the fusion peptide is the target for a novel peptidic fusion inhibitor identified from human body liquid, VIRIP (46). We believe that the gp41 CT region, particularly the LLP2 domain, may also serve as an attractive target for development of HIV-1 fusion and entry inhibitors.
Acknowledgments
We thank Veronica L. Kuhlemann for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI46221. This work was also supported by the Emphases Project of NSFC-30530680 and 973-2006CB504203 (to Y. H. C.) and the Outstanding Overseas Chinese Scholars Fund of Chinese Academy of Sciences Grant 2005-2-6 (to S. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HIV, human immunodeficiency virus; HIV-1, HIV type 1; CT, cytoplasmic tail; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; ELISA, enzyme-linked immunosorbent assay; FITC, fluorescein isothiocyanate; SPR, surface plasmon resonance; TBS, Tris buffered solution; GMEM, glutamine-deficient minimal essential medium; 6-HB, six-helix bundle; mAb, monoclonal antibody; aa, amino acid.
References
- 1.Chan, D. C., Fass, D., Berger, J. M., and Kim, P. S. (1997) Cell 89 263-273 [DOI] [PubMed] [Google Scholar]
- 2.Chan, D. C., and Kim, P. S. (1998) Cell 93681 -684 [DOI] [PubMed] [Google Scholar]
- 3.Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J., and Wiley, D. C. (1997) Nature 387426 -430 [DOI] [PubMed] [Google Scholar]
- 4.Tan, K., Liu, J., Wang, J., Shen, S., and Lu, M. (1997) Proc. Natl. Acad. Sci. U. S. A. 9412303 -12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caffrey, M., Cai, M., Kaufman, J., Stahl, S. J., Wingfield, P. T., Covell, D. G., Gronenborn, A. M., and Clore, G. M. (1998) EMBO J. 174572 -4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascual, R., Moreno, M. R., and Villalain, J. (2005) J. Virol. 795142 -5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golding, H., Zaitseva, M., de Rosny, E., King, L. R., Manischewitz, J., Sidorov, I., Gorny, M. K., Zolla-Pazner, S., Dimitrov, D. S., and Weiss, C. D. (2002) J. Virol. 766780 -6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrov, A. S., Jacobs, A., Finnegan, C. M., Stiegler, G., Katinger, H., and Blumenthal, R. (2007) Biochemistry 461398 -1401 [DOI] [PubMed] [Google Scholar]
- 9.Hunter, E., and Swanstrom, R. (1990) Curr. Top. Microbiol. Immunol. 157187 -253 [DOI] [PubMed] [Google Scholar]
- 10.Wyss, S., Dimitrov, A. S., Baribaud, F., Edwards, T. G., Blumenthal, R., and Hoxie, J. A. (2005) J. Virol. 7912231 -12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, M. A., Garry, R. F., Jaynes, J. M., and Montelaro, R. C. (1991) AIDS Res. Hum. Retroviruses 7 511-519 [DOI] [PubMed] [Google Scholar]
- 12.Chen, S. S., Lee, S. F., and Wang, C. T. (2001) J. Virol. 759925 -9938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chernomordik, L., Chanturiya, A. N., Suss-Toby, E., Nora, E., and Zimmerberg, J. (1994) J. Virol. 687115 -7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comardelle, A. M., Norris, C. H., Plymale, D. R., Gatti, P. J., Choi, B., Fermin, C. D., Haislip, A. M., Tencza, S. B., Mietzner, T. A., Montelaro, R. C., and Garry, R. F. (1997) AIDS Res. Hum. Retroviruses 131525 -1532 [DOI] [PubMed] [Google Scholar]
- 15.Kalia, V., Sarkar, S., Gupta, P., and Montelaro, R. C. (2003) J. Virol. 773634 -3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kliger, Y., and Shai, Y. (1997) Biochemistry 365157 -5169 [DOI] [PubMed] [Google Scholar]
- 17.Edwards, T. G., Wyss, S., Reeves, J. D., Zolla-Pazner, S., Hoxie, J. A., Doms, R. W., and Baribaud, F. (2002) J. Virol. 762683 -2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zingler, K., and Littman, D. R. (1993) J. Virol. 672824 -2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahamyan, L. G., Mkrtchyan, S. R., Binley, J., Lu, M., Melikyan, G. B., and Cohen, F. S. (2005) J. Virol. 79 106-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabuzda, D. H., Lever, A., Terwilliger, E., and Sodroski, J. (1992) J. Virol. 663306 -3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spies, C. P., and Compans, R. W. (1994) Virology 2038 -19 [DOI] [PubMed] [Google Scholar]
- 22.Wilk, T., Pfeiffer, T., and Bosch, V. (1992) Virology 189167 -177 [DOI] [PubMed] [Google Scholar]
- 23.Cleveland, S. M., McLain, L., Cheung, L., Jones, T. D., Hollier, M., and Dimmock, N. J. (2003) J. Gen. Virol. 84591 -602 [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, R. C., Henkel, R. D., Pauletti, D., Allan, J. S., Lee, T. H., Essex, M., and Dreesman, G. R (1986) Science 2311556 -1559 [DOI] [PubMed] [Google Scholar]
- 25.Zhu, Y., Lu, L., Chao, L., and Chen, Y. H. (2007) Chem. Biol. Drug Des. 70 311-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, J. H., Liu, Z. Q., Liu, S., Jiang, S., and Chen, Y. H. (2006) FEBS Lett. 5804807 -4814 [DOI] [PubMed] [Google Scholar]
- 27.Qiao, Z. S., Kim, M., Reinhold, B., Montefiori, D., Wang, J. H., and Reinherz, E. L. (2005) J. Biol. Chem. 28023138 -23146 [DOI] [PubMed] [Google Scholar]
- 28.Root, M. J., Kay, M. S., and Kim, P. S. (2001) Science 291884 -888 [DOI] [PubMed] [Google Scholar]
- 29.Jiang, S., Lin, K., Zhang, L., and Debnath, A. K. (1999) J. Virol. Methods 80 85-96 [DOI] [PubMed] [Google Scholar]
- 30.Huang, J. H., Yang, H. W., Liu, S., Li, J., Jiang, S., and Chen, Y. H. (2007) Biochem. J. 403565 -571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, J. H., Lu, L., Lu, H., Chen, X., Jiang, S., and Chen, Y. H. (2007) J. Biol. Chem. 2826143 -6152 [DOI] [PubMed] [Google Scholar]
- 32.Wyma, D. J., Kotov, A., and Aiken, C. (2000) J. Virol. 749381 -9387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S. F., Wang, C. T., Liang, J. Y., Hong, S. L., Huang, C. C., and Chen, S. S. (2000) J. Biol. Chem. 27515809 -15819 [DOI] [PubMed] [Google Scholar]
- 34.Jiang, S., Lin, K., and Lu, M. (1998) J. Virol. 7210213 -10217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckert, D. M., and Kim, P. S. (2001) Annu. Rev. Biochem. 70777 -810 [DOI] [PubMed] [Google Scholar]
- 36.Dimitrov, A. S., Rawat, S. S., Jiang, S., and Blumenthal, R. (2003) Biochemistry 4214150 -14158 [DOI] [PubMed] [Google Scholar]
- 37.Contreras, L. M., Aranda, F. J., Gavilanes, F., Gonzalez-Ros, J. M., and Villalain, J. (2001) Biochemistry 403196 -3207 [DOI] [PubMed] [Google Scholar]
- 38.Peisajovich, S. G., and Shai, Y. (2001) J. Mol. Biol. 311249 -254 [DOI] [PubMed] [Google Scholar]
- 39.Peisajovich, S. G., Epand, R. F., Pritsker, M., Shai, Y., and Epand, R. M. (2000) Biochemistry 391826 -1833 [DOI] [PubMed] [Google Scholar]
- 40.Sackett, K., and Shai, Y. (2002) Biochemistry 414678 -4685 [DOI] [PubMed] [Google Scholar]
- 41.Wyma, D. J., Jiang, J., Shi, J., Zhou, J., Lineberger, J. E., Miller, M. D., and Aiken, C. (2004) J. Virol. 783429 -3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celma, C. C., Manrique, J. M., Affranchino, J. L., Hunter, E., and Gonzalez, S. A. (2001) Virology 283253 -261 [DOI] [PubMed] [Google Scholar]
- 43.Jiang, S., Lin, K., Strick, N., and Neurath, A. R. (1993) Nature 365 113. [DOI] [PubMed] [Google Scholar]
- 44.Lu, M., and Kim, P. S. (1997) J. Biomol. Struct. Dyn. 15465 -471 [DOI] [PubMed] [Google Scholar]
- 45.Liu, S., Jing, W., Cheung, B., Lu, H., Sun, J., Yan, X., Niu, J., Farmar, J., Wu, S., and Jiang, S. (2007) J. Biol. Chem. 2829612 -9620 [DOI] [PubMed] [Google Scholar]
- 46.Munch, J., Standker, L., Adermann, K., Schuz, A., Schindler, M., Chinnadurai, R., Pohlmann, S., Chaipan, C., Biet, T., Peters, T., Meyer, B., Wilhelm, D., Lu, H., Jing, W. G., Jiang, S. B., Forssmann, W. G., and Kirchhoff, F. (2007) Cell 129263 -275 [DOI] [PubMed] [Google Scholar]