Abstract
Expression of the human T-cell leukemia virus type 1 (HTLV-1) oncoprotein Tax is correlated with cellular transformation, contributing to the development of adult T-cell leukemia. In this study, we investigated the role of Tax in the regulation of the ZNF268 gene, which plays a role in the differentiation of blood cells and the pathogenesis of leukemia. We demonstrated that ZNF268 mRNA was repressed in HTLV-1-infected cells. We also showed that stable and transient expression of HTLV-1 Tax led to repression of ZNF268. In addition, by using reporter constructs that bear the human ZNF268 promoter and its mutants, we showed that Tax repressed ZNF268 promoter in a process dependent on a functional cAMP-responsive element. By using Tax, cAMP-responsive element-binding protein (CREB)-1, CREB-2, and their mutants, we further showed that Tax repressed ZNF268 through the CREB/activating transcription factor pathway. Electrophoretic mobility shift assays and chromatin immunoprecipitation demonstrated the formation of the complex of Tax·CREB-1 directly at the cAMP-responsive element both in vitro and in vivo. These findings suggest a role for ZNF268 in aberrant T-cell proliferation observed in HTLV-1-associated diseases.
Human T-cell leukemia virus type I (HTLV-1)3 is the first discovered human retroviral pathogen. It has been firmly implicated with the etiology of an aggressive malignancy known as adult T-cell leukemia and of a neurological progressive inflammatory syndrome called tropical spastic paraparesis or HTLV-1-associated myelopathy (1, 2). Tax was originally discovered as a trans-activator modulating the synthesis or function of a wide variety of cellular regulatory factors that control gene expression, cell replication and differentiation, cell cycle, apoptosis, and genome stability. Thus, Tax is widely regarded as a key factor in the HTLV-1 pathogenic mechanism (3, 4). Tax mediates the transition from latency to virion production by interacting with specific host proteins associated with cellular transcription pathways, such as nuclear factor κB (NF-κB) (5–7), cAMP-responsive element-binding protein/activating transcription factor (CREB/ATF) (8–10), serum response factor (11–13), stimulatory protein 1 (14), and activating protein 1 (15, 16). Through interactions with cellular transcription factors, Tax potently activates transcription from the viral promoter and enhancer elements of many cellular genes involved in host cell proliferation (17–19). In comparison, transcriptional repression by Tax on β-polymerase, lck, c-myb, and p53 promoters (20), reported recently from several studies, is less well understood.
Many studies have suggested that regulation through the CREB/ATF pathway by Tax plays an important cellular role (21). A model for Tax-mediated transcription through the CREB/ATF pathway is that a CREB dimer binds to the Tax-responsive elements (14), which have a high similarity with cAMP-responsive element (CRE) and interact with a Tax homodimer. This CREB·Tax·TRE ternary complex can then influence TATA-binding protein to regulate the initiation by RNA polymerase II (22).
The ZNF268 gene is one of the typical KRAB-containing zinc finger genes, cloned and characterized from an early human embryonic cDNA library (23). KRAB-containing zinc finger genes represent a subfamily within a large family of zinc finger genes, and they typically act as transcriptional repressors (24). Several different alternatively spliced transcripts have been isolated for the ZNF268 gene, and developmental expression studies have suggested that ZNF268 plays a role in the differentiation of blood cells and the development of human fetal liver (25, 26). Analysis of the ZNF268 gene promoter shows that the ZNF268 gene utilizes an intragenic promoter element to control its transcription, similar to some other genes involved in the diseases development and progression, such as WT-1. In addition, in HeLa cells, CREB-2 has been shown to play an important role during the regulation of ZNF268 expression (27).
By using a recombinant expression cloning (SEREX) approach to identify tumor-associated antigens in chronic lymphocytic leukemia, Krackhardt et al. (28) identified 14 antigens, KW-1 to -14. Among them, KW-4 was found to be one of the several known alternatively spliced transcripts of the ZNF268 gene. These results suggest that the ZNF268 gene plays a role in the differentiation of blood cells and the pathogenesis of leukemia. Thus, considering the ability of Tax to transcriptionally regulate cellular gene expression as a likely mechanism for Tax-mediated transformation and leukemogenesis (22), the aim of this study was to investigate if Tax plays a role in the regulation of ZNF268 expression and to determine the underlying molecular mechanisms. Our results showed that HTLV-1 Tax was able to repress ZNF268 gene expression and that CREB-1 was involved in this repression of ZNF268 by Tax.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Cell Culture—The sequence from –37 to +938 containing the intragenic promoter element and a series of truncation mutants of the human ZNF268 promoter were inserted into the promoterless luciferase expression vector pGL3 (Promega) (27). pGL3(–37/+938)-p53-mut (+596 to +621), pGL3(–37/+938)-Ets-mut (+606 to +631), pGL3(–37/+938)-CREB-mut (+724 to +749), pGL3(–37/+938)-AP1-mut (+722 to +746), and pGL3(–37/+938)-C/EBP-mut (+728 to +752) were constructed by using the overlapping extension PCR method with pGL3(–37/+938) plasmid as described previously (27). pcDNA-Tax expresses the wild-type Tax, M22 expresses a Tax mutant that can activate CREB/ATF but not NF-κB, and M47 expresses a Tax mutant that can activate NF-κB but not CREB/ATF (29). The Tax ORF was amplified by PCR from pcDNA-Tax using primers of Tax1 and Tax2. The PCR products were cloned into EcoRI and XhoI sites of pCMV-Tag2B to generate plasmid pCMV-Tag2B-Tax. pEGFP-C1 expresses GFP protein. pcDNA-CREB-1 and pcDNA-CREB-2 express CREB-1 and CREB-2, respectively. pcDNA-CREB-1-dominant negative (DN) expresses CREB-1 dominant negative mutant (S133A), as described (30, 31). The corresponding primers used for cloning are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Oligonucleotide sequence (5′ to 3′) | Locationa |
|---|---|---|
| PDT8 | CGAGGTACCCTCTGTGAATGTCACCTC | -37 to -20 |
| PD2T1 | GTTAAGCTTCTCCTCCAAACCCTGAAG | +938 to +921 |
| PD2T2 | GTTAAGCTTCTACGTATGTCGCACAGG | +760 to +743 |
| PD2T3 | GTTAAGCTTGACACCAATGGCTCAACG | +540 to +523 |
| p53-wt-F | CTGGCCAGGAAGGCCTGAGCTTCCGGb | +596 to +621 |
| p53-mut-F | CTGGCCAGGAAGTAAGGAGCTTCCGG | +596 to +621 |
| Ets-wt-F | AGGCCTGAGCTTCCGGGTCATCTTAG | +606 to +631 |
| Ets-mut-F | AGGCCTGAGCTGAAGGGTCATCTTAG | +606 to +631 |
| CRE-wt-F | GCCTCTCCATGACGCAATTCCTGTGC | +724 to +749 |
| CRE-mut-F | GCCTCTCCATGCATCAATTCCTGTGC | +724 to +749 |
| AP1-wt-F | TTGCCTCTCCATGACGCAATTCCTG | +722 to +746 |
| AP1-mut-F | TTGCCTCTCCAGTCCGCAATTCCTG | +722 to +746 |
| C/EBP-wt-F | CTCCATGACGCAATTCCTGTGCGAC | +728 to +752 |
| C/EBP-mut-F | CTCCATGACGCCCTTCCTGTGCGAC | +728 to +752 |
| PECS3 | GCAGATATGAGAATCCAGCT | +287 to +306 |
| PE41 | GCTACGTATGTCGCACAGGAATTG | +761 to +738 |
| PECS11 | ACCTGGCCAGGAAGGCCTGAG | +594 to +614 |
| PECA | TGAAGGGGCAGCAGAATAGA | +925 to +906 |
| PU | CGAGGTACCAGAAGACATACAAATGGCCAAC | -1790 to -1769 |
| PDT1A | CTACTGTGACTGATGTAAGA | -1381 to -1400 |
| Tax1 | CTAATTGAATTCGGAATTCGATCCACCATGGC | |
| Tax2 | GCCGCGGTCTCGAGTTTTCAGACTTCTGTTTCG | |
| CREBU | GGAGAAGCTTGTACCACCGGTAACTAAATGAC | |
| CREBD | GAGAGCGGCCGCTTATTAATCTGATTTGTGGCAGTAAA | |
| CREB-DN-F | CTTTCAAGGAGGCCTGCCTACAGGAAAATTT | |
| CREB-DN-R | AAATTTTCCTGTAGGCAGGCCTCCTTGAAAG | |
| EgrU | ACCGAATTCATGGCCGCGGCCAAGGCCGAGATGC | |
| EgrD | ATCGTCGACCTTAGCAAATTTCAATTGTCCTGGG |
Shown are the oligonucleotide positions, where +1 is the transcription start site of the ZNF268 gene.
Nucleotides in bold are the mutated binding sites of the corresponding base.
HEK293 and HeLa cells (CCTCC, Wuhan, China) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Invitrogen), penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a 5% CO2 incubator. Jurkat and Hut-102 cells (CCTCC) were maintained in RPMI medium (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen), penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a 5% CO2 incubator.
Transfection and Luciferase Assays—HeLa cells were chosen for stably expressing Tax protein due to the ease with which stably transfected cell lines were established. The cells were transfected with the Tax expression vector pCMV-Tag2B-Tax or empty vector pCMV-Tag2B using Sofast™ transfection reagent (Sunma, China) according to the manufacturer's instructions. Stable transfectants were obtained after 2–3 weeks of selection with 600 μg/ml Geneticin (G-418) and screened for FLAG-tagged Tax protein expression by Western blotting as described below. HEK293 and HeLa cells were seeded in 24-well plates at 75% confluence and co-transfected with luciferase reporter vectors and the indicated recombinant plasmids by mixing 0.2 μg of firefly luciferase reporter vectors and the internal control Renilla luciferase reporter construct, pRL-TK (Promega) (firefly luciferase reporter construct and pRL-TK in a ratio of 20:1), which contains the Renilla luciferase gene driven by the herpes simplex virus thymidine kinase promoter and 0.4 μg of plasmids with 1.5 μl of Sofast™ transfection reagent (Sunma) according to the manufacturer's instructions. Jurkat cells were seeded in 24-well plates and transfected at about 1 × 105 cells/well using DMRIE-C transfection reagent (Invitrogen), following the manufacturer's instructions. Phyto-hemagglutinin (final concentration 1 μg/ml; Sigma) and phorbol 12-myristate 13-acetate (final concentration 50 ng/ml; Sigma) were added to each well 4 h after transfection. After incubation for 48 h, the cells were harvested for luciferase assays.
Semiquantitative RT-PCR—Total RNA was extracted by using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription was performed with total RNA as the template. Specific mRNA was amplified by RT-PCR using primers PE41 and PECS3 for the ZNF268 gene and EgrU and EgrD for the egr-1 gene (Table 1). The PCRs were individually optimized so that each reaction fell within the linear range of product amplification. The PCR analysis of β-actin was used as the internal control.
Quantitative RT-PCR—Quantitative RT-PCR was performed using the Rotor-Gene 2000 real time PCR system (Rotor-Gene, Sydney, Australia) with 20× SYBR Green I PCR mix reagent in a 25-μl volume in triplicate according to the manufacturer's instructions. As a control, the mRNA level of β-actin was determined for each RNA sample and was used to correct for experimental variations. ZNF268 mRNA was amplified by RT-PCR using primers PE41 and PECS3.
Western Blot Analysis—Lysates of cells were prepared in the lysis buffer, containing 100 mm NaCl, 10 mm Tris-HCl (pH 8.0), 1 mm EDTA (pH 8.0), 1% Triton X-100, and 1 mm phenylmethylsulfonyl fluoride, followed by centrifugation at 12,000 × g for 10 min. Supernatants were collected, mixed with an equal volume of SDS-PAGE 2× sample buffer, aliquoted, and stored at –80 °C until used.
Aliquots (50 μg) were analyzed on a 10% polyacrylamide gel and transferred to nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk for 1 h at 37°C. Subsequently, it was incubated with anti-Tax monoclonal antibody (Tab172) for 2 h at 37°C. Membranes were then incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit antibodies (Zhongshan Biotechnology, China) at 37 °C. Protein bands were detected using SuperSignal chemiluminescence (Pierce).
Electrophoretic Mobility Shift Assay (EMSA)—HEK293 cells incubated in serum-free medium for 24 h were washed with cold phosphate-buffered saline twice and scraped into 1 ml of cold phosphate-buffered saline. Cells were harvested by centrifugation for 15 s and incubated in 2 packed cell volumes of buffer A (10 mm, HEPES, pH 8.0, 0.5% Nonidet P-40, 1.5 mm MgCl2, 10 mm KCl, 200 mm sucrose, and 0.5 mm dithiothreitol) for 5 min at 4 °C with flicking. The crude nuclei were collected by centrifugation for 30 s, and pellets were rinsed with buffer A, resuspended in 1 packed cell volume of buffer B (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, and 1.0 mm dithiothreitol), and incubated on a rocking platform for 30 min at 4 °C. Nuclei were clarified by centrifugation for 5 min, and the supernatants were diluted 1:1 with buffer C (20 mm HEPES, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 20% glycerol, and 1.0 mm dithiothreitol). Protease inhibitor mixture tablets were added to all buffers. Nuclear extracts were frozen in liquid N2 and stored at –70 °C until use. Probes were generated by annealing single-stranded oligonucleotides (sequences are listed in Table 1) containing the cognate promoter regions of the ZNF268 gene and labeled at the ends with [γ-32P]ATP using T4 polynucleotide kinase (TaKaRa).
EMSAs were performed with 4 μg of nuclear extract in binding buffer (20 mm Hepes, pH 7.9, 0.1 mm EDTA, pH 8.0, 75 mm KCl, 2.5 mm MgCl2, and 1 mm dithiothreitol) containing 1 μg of poly(dI-dC). To ensure the specific binding of transcription factors to the probe, unlabeled wild-type and mutated double-stranded oligonucleotide competitors were preincubated at a 100-fold molar excess for 10 min prior to probe addition. For supershift experiments, 2 μg of anti-Tax monoclonal antibody (Tab172) or anti-CREB-1 monoclonal antibody (Epitomics) were incubated with nuclear extracts on ice for 30 min before adding to the binding buffer. Samples were then electrophoresed on 5% nondenaturing polyacrylamide, 0.5× Tris/glycine/EDTA gels, and the gels were dried and subjected to autoradiography.
Chromatin Immunoprecipitation (ChIP)—The assay was done as previously described (27). Monolayer of HEK293 cells (80% confluent) were incubated for 24 h after transfection and then were serumstarved for 24 h. Formaldehyde was added to the culture medium to a final concentration of 1%. The cells were then washed twice in phosphate-buffered saline, scraped, and lysed in lysis buffer (1% SDS, 10 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, 50 mg/ml both aprotinin and leupeptin) for 10 min on ice. The lysates were sonicated on ice, and the debris was removed by centrifugation at 12,000 rpm for 15 min at 4 °C. One-fourth of the supernatant was used as DNA input control. The remaining supernatant was diluted 10-fold with dilution buffer (0.01% SDS, 1% Triton X-100, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0, and 150 mm NaCl) and incubated with anti-Tax monoclonal antibody or anti-phospho-CREB-1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) overnight at 4 °C. Immunoprecipitated complexes were collected using protein A/G-agarose beads. The pellets were washed with dialysis buffer (2 mm EDTA, 50 mm Tris-HCl, pH 8.0). Samples were incubated at 67 °C for 5 h to reverse formaldehyde cross-link. DNA was precipitated with ethanol and extracted three times with phenol/chloroform. Finally, pellets were resuspended in TE buffer and subjected to PCR amplification using primers PECS11/PECA (+594 to +925) (Table 1) for the promoter region containing the CREB/ATF binding site and PU/PDT1A for an upstream region as a negative control (Table 1). The PCR products were resolved by agarose gel electrophoresis.
RESULTS
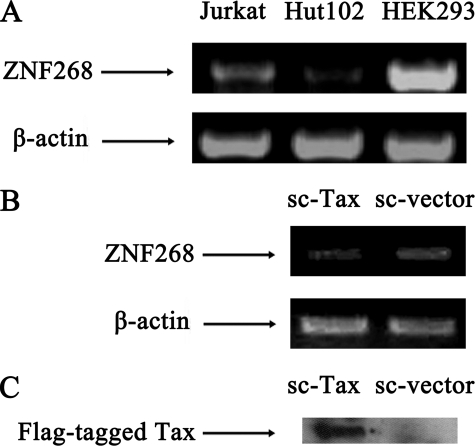
Analysis of ZNF268 Expression in HTLV-1-infected Cell Lines and Stably Transfected Tax-expressing Cell Line—To determine whether HTLV-1 infection affects the expression of ZNF268, we compared ZNF268 mRNA levels in HTLV-1-infected T-cell line Hut-102 with uninfected T-cell line Jurkat and HEK293 cell line. The result showed that Hut-102 expressed a much lower level of ZNF268 mRNA compared with Jurkat and HEK293 cell lines. This finding suggested that ZNF268 expression was repressed in HTLV-1-infected cells (Fig. 1A).
FIGURE 1.
ZNF268 mRNA was repressed in HTLV-1-infected cells and in cells stably expressing Tax. A, specific mRNA was amplified by RT-PCR using primers PE41 and PECS3 for the ZNF268 gene. As an internal control, the mRNA level of β-actin was also analyzed. ZNF268 mRNA level in the Hut-102 T-cell line infected by HTLV-1 was compared with the noninfected Jurkat T-cell line and HEK293 cell line. B, stable cell lines were generated from transfection of HeLa cells with pCMV-Tag2B-Tax for expressing Tax protein (sc-Tax) and the empty vector pCMV-Tag2B (sc-vector) as a control. Total RNA was isolated from the transfected cells and was amplified by RT-PCR using primers PE41 and PECS3 for the ZNF268 gene. Again, the mRNA level of β-actin was used as an internal control. C, Western blot analysis showing the expression of FLAG-tagged Tax in sc-Tax but not in sc-vector-transfected cell line.
To investigate whether Tax from HTLV-1 was responsible for the repression of the expression of ZNF268, we generated stably transfected cells overexpressing the Tax protein in HeLa cells, since it is easy to generate stable HeLa cells, and ZNF268 is expressed in HeLa cells (32). Transfections were done with pCMV-Tag2B-Tax for stably expressing Tax protein and the empty vector pCMV-Tag2B as a control. The transfected cells were cultured in the medium containing 600 μg/ml Geneticin (G-418), and the G418-resistant clones were obtained after 2–3 weeks. Tax expression was confirmed by Western blotting of the FLAG-tagged Tax protein expression (Fig. 1C). Analysis of ZNF268 expression in the transfected cells showed that the sc-Tax cell line has a reduced level of ZNF268 mRNA compared with the sc-vector cell line (Fig. 1B), suggesting that Tax was responsible for the repression of ZNF268 in HTLV-1-infected cells.
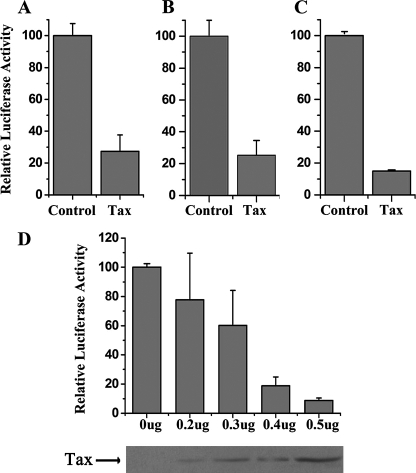
Tax Represses the ZNF268 Promoter—To investigate the regulation of ZNF268 by Tax, we transfected the Tax expression plasmid pcDNA-Tax with a reporter plasmid carrying the luciferase gene under the control of ZNF268 promoter into HEK293 cells, Jurkat cells, and HeLa cells, respectively. Cells were cultured for 24 h and then serum-starved for an additional 24 h and harvested. Cell lysates were analyzed for luciferase activity. The luciferase assay demonstrated that Tax dramatically repressed the ZNF268 promoter in all three different cell lines when compared with transfection with the empty vector pcDNA-3.1 (Fig. 2, A–C). Because of the higher levels of endogenous ZNF268 gene expression in HEK293 cells (Fig. 1) and the ease with which HEK293 cells were transfected, most of our transfection studies below were done in HEK293 cells.
FIGURE 2.
HTLV-1 Tax represses the ZNF268 promoter. HEK293 (A), Jurkat (B), and HeLa (C) cells were co-transfected with plasmid expressing Tax (0.4 μg in 24-well plates) and the reporter plasmid in which the luciferase gene is under the control of the ZNF268 promoter. The empty vector pcDNA-3.1 was used as a control. Relative luciferase activity was determined with standard procedures. Values correspond to an average of at least three independent experiments done in duplicate. D, dose-dependent repression by Tax. HEK293 cells were co-transfected with different amounts of the plasmid expressing Tax along with the reporter plasmid, and relative luciferase activity was determined. Values correspond to an average of at least three independent experiments done in duplicate. At the bottom, the 40-kDa Tax was detected by Western blot from the same transfected cells.
To determine whether this repression of the ZNF268 promoter by Tax was dependent on the amount of Tax, different concentrations of pcDNA-Tax plasmid along with plasmid carrying the reporter gene were co-transfected into HEK293 cells. Luciferase activity assays showed that ZNF268 promoter activity decreased as the concentration of plasmid DNA increased (Fig. 2D), indicating that the repression of ZNF268 promoter by Tax was dose-dependent. To confirm the expression of Tax in transfected cells, transfected cells were harvested as described above. Western blot analysis was carried out using monoclonal antibody against Tax (Tab172). The amount of Tax expressed increased with increasing amounts of the plasmid DNA (Fig. 2D).
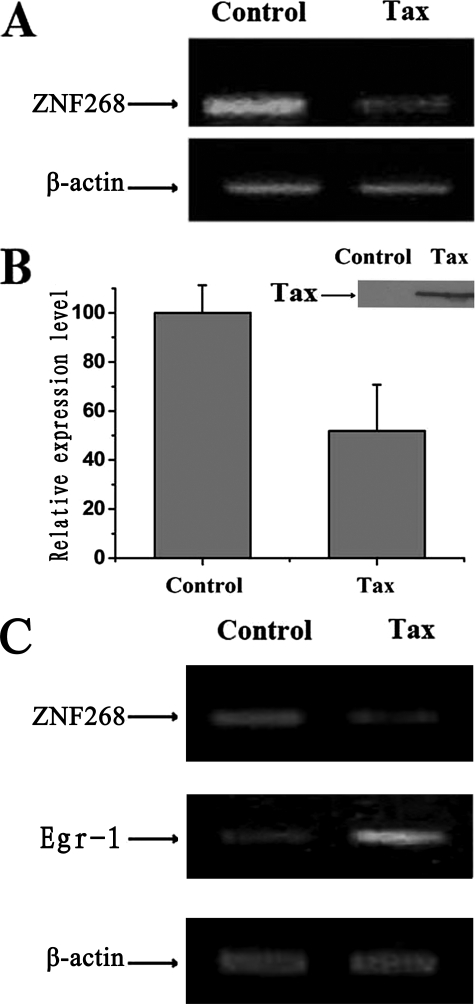
Tax Represses Endogenous ZNF268 mRNA Level—To determine whether transient expression of Tax also affected the endogenous ZNF268 gene in HEK293 cells as observed above with the stably transfected HeLa cells, plasmid (pcDNA-Tax) expressing Tax or control plasmid (pcDNA-3.1) was transfected into HEK293 cells. Total RNA of the transfected cells was isolated and used for semiquantitative RT-PCR and quantitative real time PCR (Fig. 3, A and B) as described above. The results showed that Tax repressed the amount of endogenous ZNF268 mRNA in the HEK293 cells (Fig. 3, A and B). To determine the specificity of the repression of ZNF268 by Tax, we carried out a similar analysis in Jurkat cells. It has been shown that in Jurkat cells, Tax activates egr-1 (33). Consistently, we found that transient transfection of Tax expression plasmid led to increased expression of egr-1 (Fig. 3C). In these same cells, Tax also repressed ZNF268, just like in HEK293 cells (Fig. 3C). These results demonstrate gene-specific repression of ZNF268 by Tax.
FIGURE 3.
HTLV-1 Tax represses the endogenous ZNF268 gene in HEK293 and Jurkat cells. A, semiquantitative RT-PCR analysis of the expression of ZNF268 mRNA. Cells were transfected with Tax expression plasmid or the empty vector pcDNA-3.1 as a control. Specific mRNA was amplified by RT-PCR using primers PE41 and PECS3 for ZNF268. As an internal control, the mRNA level of β-actin was also analyzed. Note the reduced level of ZNF268 mRNA in the Tax-transfected cells. B, quantitative real time PCR analysis of the expression of ZNF268 mRNA in cells transfected with Tax expression plasmid or control pcDNA-3.1. The mRNA level of β-actin was determined by real time PCR for each RNA sample and was used to normalize the ZNF268 expression. ZNF268 mRNA was amplified by using primers PE41 and PECS3. Inset, the 40-kDa Tax was detected in the cells transfected with pcDNA-Tax but not pcDNA-3.1 (Control). C, semiquantitative RT-PCR analysis shows that endogenous ZNF268 is also repressed by Tax in Jurkat cells, whereas a known Tax-inducible gene, egr-1, is up-regulated as expected. Cells were transfected with the empty vector pcDNA-3.1 or Tax expression plasmid. Specific mRNA was amplified by RT-PCR using primers PE41 and PECS3 for ZNF268 and EgrU and EgrD for egr-1. As a control, the mRNA level of β-actin was also analyzed.
Again, to confirm the expression of Tax in the transfected cells, transfected cells were treated and harvested as described above. Western blot analysis was carried out using monoclonal antibody against Tax (Tab172). Tax was detected in cells transfected with the plasmid pcDNA-Tax but not present in cells transfected with control plasmid pcDNA-3.1 (Fig. 3B, inset).
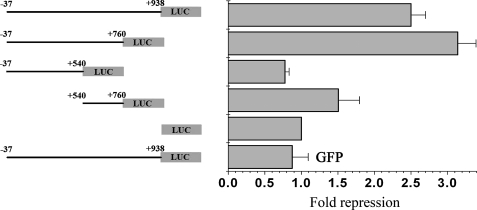
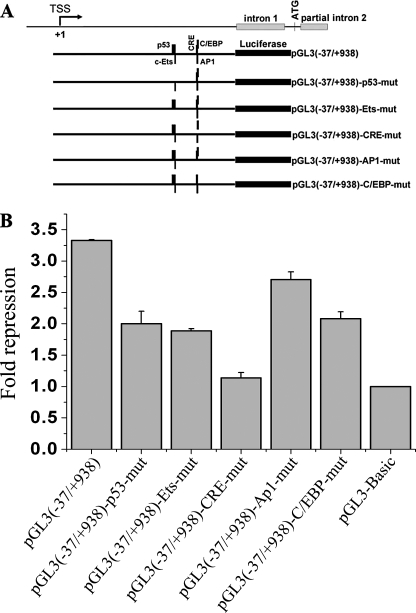
The CRE Is Required for the Repression of ZNF268 Expression by Tax—To define the cis-regulatory element that was responsive to Tax, a series of mutants with truncation or site-specific mutations in the ZNF268 promoter were generated (Figs. 4 and 5). HEK293 cells were co-transfected with a plasmid carrying the Tax gene (pcDNA-Tax) and plasmids containing the luciferase reporter gene driven by mutated ZNF268 promoters. The results of luciferase assays indicated that +540 to +760 of the ZNF268 promoter was necessary and sufficient for the repression by Tax (Fig. 4). This repression by Tax was specific, since transfection with a GFP expression plasmid did not affect the promoter activity (Fig. 4, bottom row). Furthermore, site-specific mutational analysis revealed that mutations in the CRE significantly diminished the repression of ZNF268 promoter activity by Tax (Fig. 5).
FIGURE 4.
Deletion analysis of cis-regulatory elements of the ZNF268 promoter important for repression by Tax. Diagrams of deletion mutants of ZNF268 promoter are shown on the left, and the transfection results are shown on the right. Tax expression plasmid or the empty vector pcDNA-3.1 (Control) and plasmids containing the luciferase reporter gene driven by individual ZNF268 promoter mutants were co-transfected into HEK293 cells. GFP expression plasmid was used as an unrelated protein control to confirm the specificity of the repression of ZNF268 by Tax. The pGL3-Basic plasmid was transfected as the promoterless negative control. Promoter activities were determined by measuring the relative luciferase activity in transfected cell lysates from three independent experiments. To determine the relative repression, the promoter activity from cells transfected with the empty vector pcDNA-3.1 was divided by that from Tax-transfected cells. Note that repression was observed for all promoter constructs containing the sequences from +540 to +760.
FIGURE 5.
Site-specific mutation analysis of cis-regulatory elements in the ZNF268 promoter important for repression by Tax. A, schematic diagrams of the promoter constructs with various transcription factor binding sites mutated as indicated. B, Tax expression plasmid and plasmids containing the luciferase reporter gene driven by individual ZNF268 promoter mutants were co-transfected into HEK293 cells. The pGL3-Basic plasmid was transfected as the promoterless negative control. Promoter activities were determined as before. Note that only mutations in the CRE significantly reduced the repression by Tax.
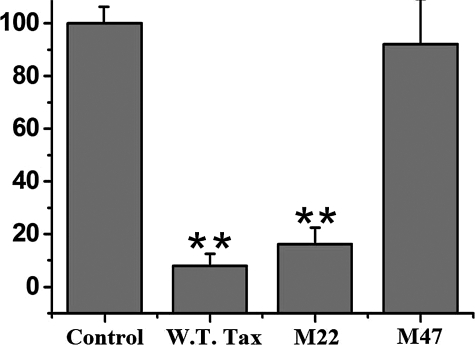
To further elucidate the mechanism of this repression, we employed two Tax mutants that have the missense mutations in Tax that functionally segregated two pathways: Tax M22, which is capable of activating CREB/ATF but not the NF-κB transcription factors, and Tax M47, which can activate NF-κB but not CREB/ATF (29). Transient transfection studies demonstrated that Tax M22 strongly repressed the ZNF268 promoter, whereas Tax M47 was ineffective in this respect (Fig. 6). These results, together with the findings from the promoter mutants above, indicate that Tax exerts its effect on the ZNF268 promoter through the CREB/ATF pathway.
FIGURE 6.
Effects of Tax mutants on the ZNF268 promoter. The pcDNA-3.1 (empty vector control) and plasmids expressing Tax (wild type; W.T. Tax) or its mutant M22, which can affect transcriptional activation by CREB/ATF but not NF-κB, or M47, which can affect transcriptional activation by NF-κB but not CREB/ATF, were co-transfected with the luciferase reporter plasmid driven by the ZNF268 promoter into HEK293 cells. Promoter activities were determined by measuring the relative luciferase activity in transfected cells. Luciferase activities correspond to an average of at least three independent experiments, and the data are shown as mean values with S.E.. *, p < 0.05; **, p < 0.02, standard t test.
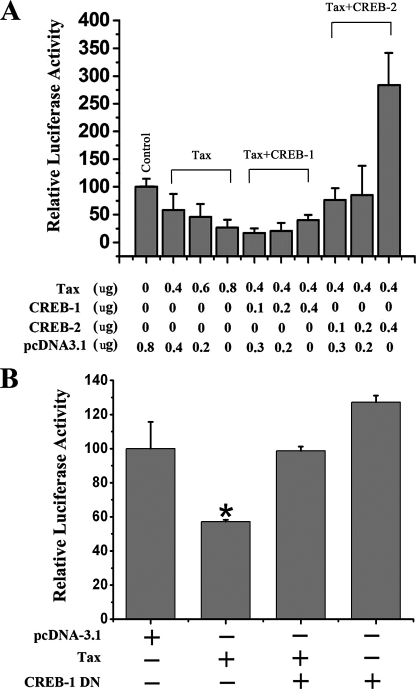
CREB-1 Plays a Role in the Repression of ZNF268 Expression by Tax—To identify whether CREB-1 or CREB-2 is involved in the repression of the ZNF268 gene by Tax, we co-transfected Tax expression plasmids for Tax, CREB-1, and CREB-2 into HEK293 cells with different concentrations and combinations together with the reporter DNA. The results indicated that the overexpression of CREB-1 stimulated the repression of ZNF268 by low levels of Tax, whereas the overexpression of CREB-2 eliminated the repression of ZNF268 by Tax (Fig. 7A).
FIGURE 7.
A role of CREB-1 in the repression of the ZNF268 promoter by Tax. A, the indicated amounts of expression plasmids for Tax, CREB-1, and CREB-2 were transfected into HEK293 cells together with the plasmid containing the luciferase reporter gene driven by the ZNF268 promoter. Promoter activities were determined as before. Note that overexpression of CREB-1 at low concentrations enhanced repression by low levels of Tax. B, the plasmids expressing Tax (0.3 μg in 24-well plates) and CREB-1 dominant negative (DN) (0.2 μg in 24-well plates) were transfected together with the plasmid containing the luciferase reporter gene driven by the ZNF268 promoter into HEK293 cells, respectively. Promoter activities were determined as before. *, p < 0.05; **, p < 0.02, standard t test. Note that the dominant negative CREB-1 inhibited the repression by Tax.
To further investigate whether CREB-1 is involved in the repression of the ZNF268 gene by Tax, we constructed an expression plasmid for a dominant negative mutant of CREB-1 (S133A), which can bind to the CRE, whereas it has no transcriptional activity (34). Co-transfection of cells with this dominant negative CREB-1 and Tax inhibited the repression of the ZNF268 promoter by Tax (Fig. 7B). (The lower level of Tax expression plasmid used here compared with that in Fig. 2 led to a lower level of repression.) In the absence of Tax, dominant negative CREB-1 had little effect (a small, nonsignificant increase in Fig. 7B) on the ZNF268 promoter, consistent with the lack of effect of CREB-1 on this promoter in HeLa cells (27) (see “Discussion” for a possible explanation). This result suggests that CREB-1 plays an important role in the repression of ZNF268 gene by Tax.
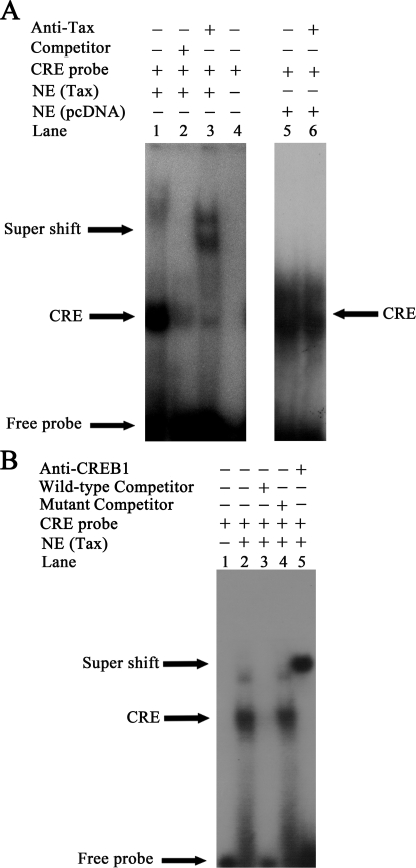
Corecruitment of Tax and CREB-1 to the CRE Site of the ZNF268 Promoter Represses ZNF268 Gene Expression—The CRE sequence +732 to +741 (5′-ATGACGCAAT-3′) of the ZNF268 promoter has a high level of similarity with the Tax-responsive element (35). Existing evidence suggests that Tax need not directly bind to DNA to accomplish its function but rather that it can act through binding to CREB-1 bound to the Tax-responsive element (22). To define the association between the complex of Tax·CREB-1 and ZNF268 promoter, we conducted an EMSA. HEK293 cells were transfected with a control plasmid (Fig. 8A, lanes 5 and 6) or a plasmid containing the tax gene (Fig. 8, A, lanes 1–3, and B, lanes 2–5). Nuclear extracts were prepared from the transfected cells, and EMSA was performed with 4 μg of nuclear extract and labeled CRE probe. A strong CRE complex(es) was observed (Fig. 8, A, lane 1, and B, lane 2). To ensure specific binding of transcription factors to the probe, unlabeled wild-type double-stranded oligonucleotide competitors (Fig. 8, A, lane 2, and B, lane 3) and unlabeled mutated double-stranded oligonucleotide competitors (Fig. 8B, lane 4) were added prior to the addition of labeled probe. To determine whether Tax protein or CREB-1 protein was bound to the promoter, anti-Tax monoclonal antibody (Tab172) or anti-CREB-1 monoclonal antibody was incubated with nuclear extracts before adding the binding buffer (Fig. 8, A, lane 3, and B, lane 5). The CRE complex(es) was supershifted by both antibodies, indicating the presence of Tax and CREB-1 in the CRE complex(es). When similar experiments were carried out with nuclear extracts from cells without Tax transfection, no bands supershifted by the anti-Tax antibody were observed (Fig. 8A, lane 6). It is interesting to note that the anti-Tax and anti-CREB-1 antibodies supershifted nearly all of the CRE complexes, suggesting that in the presence of Tax, Tax·CREB-1 was the predominant complex bound to the CRE, at least under our in vitro binding conditions. These results indicate that both Tax and CREB-1 bind the CRE in vitro.
FIGURE 8.
Both Tax and CREB-1 bind to the CRE in the ZNF268 promoter in vitro. A, EMSA was performed with nuclear extracts (NE) from HEK293 cells transfected with pcDNA-Tax (lanes 1–3) or with the empty vector pcDNA-3.1 (lanes 5 and 6) as a control. Labeled CRE probe was added to all reactions (lanes 1–6). Unlabeled wild-type double-stranded oligonucleotide competitors were added during preincubation prior to probe addition (lane 2). For supershift experiments, anti-Tax (lanes 3 and 6) was incubated with nuclear extracts before adding to the reaction. Free probe without any nuclear extracts or antibody (lane 4) was used as a negative control. Samples were electrophoresed on 5% nondenaturing polyacrylamide gel and visualized by autoradiography. B, EMSA was performed with nuclear extracts of HEK293 cells transfected with pcDNA-Tax (lanes 2–5). Labeled CRE probe was added to all reactions (lanes 1–5). Unlabeled wild-type double-stranded oligonucleotide competitors and mutated double-stranded oligonucleotide competitors were added during preincubation prior to the probe addition (lanes 3 and 4). For supershift experiments, anti-CREB-1 antibody (lane 5) was incubated with nuclear extracts before adding to the reaction. Free probe without any nuclear extracts or antibody (lane 1) was used as negative control. Samples were electrophoresed on a 5% nondenaturing polyacrylamide gel and visualized by autoradiography. Note that the anti-Tax and anti-CREB-1 antibodies supershifted nearly all of the CRE complexes. It is possible that in the presence of Tax, Tax·CREB-1 is the predominant complex binding to the CRE, whereas little CREB-2 is bound to the CRE, at least under our in vitro binding conditions. CRE, complexes containing the CRE probe. Supershift, complexes supershifted by the anti-Tax or anti-CREB-1 antibody. (Note that the CRE complex(es) formed with nuclear extract with or without Tax transfection had similar mobility (A, lanes 1 and 5). It is unclear why. However, the mobility of complexes in native gels is affected by many factors and is not simply related to the mass of the complexes. It is possible that Tax·CREB·CRE complexes had similar mobility as CREB·CRE complexes under our gel conditions.)
To further confirm Tax-ZNF268 promoter binding, a ChIP assay was performed. Chromatin fragments were prepared from HEK293 cells transfected with plasmid expressing Tax and immunoprecipitated with specific anti-Tax monoclonal antibody (Tab172) or anti-phospho-CREB-1. The immunoprecipitated DNA was amplified by PCR with primers PECS11/PECA (+594 to +925) (Table 1) for the promoter region containing the CRE. A fragment of the expected size of 332 bp was detected when anti-Tax or anti-phospho-CREB-1 antibody was used (Fig. 9A, lanes 2, 4, and 5), but no signal was detected when anti-GFP antibody was used for the ChIP assay (Fig. 9A, lane 8). In addition, no signal was detected for the anti-Tax antibody ChIP assay with the immunoprecipitated DNA from the cells transfected without any plasmid or with the empty vector pcDNA-3.1 (Fig. 9A, lanes 3 and 7). Furthermore, when no antibody was included in the ChIP assay as a negative control, no signal was detected (Fig. 9A, lane 6). Finally, no signal was detected when PCR amplification of the precipitated DNA was done for a negative control region with primers PU/PCT1A (–1790 to –1381) (Table 1) (Fig. 9B). These results indicated that both Tax and CREB-1 were bound to the CRE in the ZNF268 promoter, suggesting the formation of the Tax·CREB-1·CRE complex in vivo.
FIGURE 9.
ChIP assay shows that both Tax and CREB-1 are bound to the CRE region of the ZNF268 promoter in vivo. HEK293 cells transfected without any plasmid (lanes 3 and 5) or with empty vector pcDNA-3.1 (lane 7) or pcDNA-Tax (lanes 1, 2, 4, 6, and 8) were lysed and subjected to ChIP assay. Shown is PCR amplification of DNA precipitated with anti-Tax (lanes 2, 3, and 7), anti-phospho-CREB-1 (lanes 4 and 5), and anti-GFP (lane 8) by using primers for the ZNF268 promoter, PECS11 and PECA, flanking the CRE (+594 to +925) (A). PCR amplification of DNA precipitated without any antibody (lane 6) was used as the negative control. The input lane (lane 1) shows product after PCR amplification of chromatin DNA prior to immunoprecipitation. PCR with primers PU and PDT1A for an upstream region (–1790 to –1381) (B) of the ZNF268 promoter was done as a negative control.
DISCUSSION
The HTLV-1 Tax is crucial for viral replication and for initiating malignant cell transformation, leading to leukemogenesis (22). There are data demonstrating that Tax uses the CREB/ATF factors to repress the expression of genes such as cyclin A, cyclin D3, and DNA polymerase α. This CRE-dependent effect of Tax on such cellular genes may contribute to the initiation of an oncogenic process by impairing the cell and growth control (22). Many cellular genes contain in their promoters CREs and are regulated by signals that elevate the cellular cAMP level (22). However, the ability of Tax to regulate transcription via the CRE site is context-specific, since at many other CREB-binding sites, where Tax·CREB complex formation may occur, transcriptional regulation by Tax is not seen (35). Our previous studies suggested that ZNF268 played a role in the differentiation of blood cells during early human embryonic development and the pathogenesis of leukemia (25, 26, 32). Here, we first showed that ZNF268 mRNA was repressed in an HTLV-1-infected cell line and cells stably expressing Tax, suggesting that Tax is responsible for the repression of ZNF268 genes in HTLV-1-infected cells. We also found that Tax repressed the ZNF268 promoter in different cell lines. Finally, we showed that ZNF268 was transcriptionally regulated by the HTLV-1-encoded Tax through the CREB/ATF pathway and that both Tax and CREB-1 are bound to the CRE in the ZNF268 promoter in vitro and in vivo.
Our luciferase assays showed that Tax repressed the ZNF268 promoter in a dose-dependent manner. Semiquantitative RT-PCR and quantitative real time PCR confirmed this repression of ZNF268 at the mRNA level by Tax. There are at least three potential protein products encoded by the ZNF268 gene, ZNF268a, ZNF268b1, and ZNF268b2, due to alternative splicing (32). Since Tax represses the ZNF268 promoter, all three ZNF268 products are probably reduced in the presence of Tax, although further studies are needed to confirm this. Studies with mutants of the ZNF268 promoter and Tax revealed that Tax specifically recognized the CRE regulatory element in the ZNF268 promoter. Based on our results and previous findings, it is reasonable to suggest that Tax represses ZNF268 gene expression through the CREB/ATF pathway. Indeed, EMSA and ChIP assays confirmed the binding of both Tax and CREB-1 to the ZNF268 CRE site in vitro and in vivo, respectively. These results suggest the formation of a CREB-1·Tax complex at the CRE site of the promoter, which would be consistent with the luciferase assay showing that dominant negative CREB-1 inhibited the repression by Tax.
Historically, Tax was first characterized as a potent activator of gene expression. Among the genes are transcription factors (such as c-fos, c-jun, c-myc, egr-1, and egr-2), cytokines (such as IL-1α, IL-2, IL-4, IL-6, and IL-8), and cell cycle regulators (such as cyclin D1 and cyclin D2) (33). Recently, however, several prototypic transcription activators, such as p53, E2F, and E1a, have been shown to also function as potent transcriptional repressors (20). Thus, it may not be surprising to find that HTLV-1 Tax can also function as a repressor, as shown recently by several studies describing its repressive activity on genes such as cyclin A, lck, bax, and c-myb (20). Here, we have extended this repressive effect of Tax to a new target gene, ZNF268, and more importantly, revealed that Tax can repress through the CREB/ATF pathway.
Our previous in vitro and in vivo binding studies showed that in HeLa cells, CREB-2 but not CREB-1 binds to the CRE within the minimal promoter, and overexpression of CREB-2 but not CREB-1 dramatically enhances ZNF268 promoter activity (27). Interestingly, here we were able to detect CREB-1 binding to the CRE in the absence of Tax. It is possible that the binding of CREB-1 to the CRE is stronger in HEK293 cells than in HeLa cells. Alternatively, weak binding of CREB-1 in HeLa cells was not detected under the previous experimental conditions. Although the mechanism underlying the differences between CREB-1 and CREB-2 remains to be determined, the promoter context within the ZNF268 gene may also affect either the function or the binding of CREB-1 and -2 to the CRE in vivo. In addition, it remains possible that the levels of CREB-1 and -2 in HeLa and HEK293 cells with or without transfection may be different to account for the observed results. Finally, there may be a delicate balance of the CRE binding activity between CREB-1 and CREB-2. In normal cells in the absence of Tax, CREB-2 may be the main CREB bound to the CRE at ZNF268 promoter in the context of this gene and activates it by recruiting co-activators, which would be consistent with the lack of significant effects of overexpression of CREB-1 (27) or CREB-1 dominant negative (DN) on the ZNF268 promoter in the absence of Tax (Fig. 7B). In the presence of Tax, CREB-1·Tax complex may now be able to compete effectively against CREB-2 for binding to the promoter. For an unknown reason (e.g. due to the particular conformation of the complex at the promoter), both CREB-1 and Tax cannot activate the promoter, thus leading to repression. It is also possible that CREB-1·Tax may recruit other co-repressors in a complex to actively repress the promoter. Thus, the repression of ZNF268 by Tax may be due to a combination of two effects: the loss of the activation by CREB-2 and the additional repressive activity of Tax. Clearly, further studies, including a detailed quantitative analysis of CREB-1 and CREB-2 binding to the CRE in the presence and absence of Tax, are needed to determine the exact mechanism.
As a factor very likely to be important for blood cell and leukemia development, abnormal expression of ZNF268 is potentially an important event in cell transformation induced by Tax. The unexpected observation that Tax represses ZNF268 provides a further illustration of the intimate relationship between viruses and host factors. It additionally highlights the delicate balance between positive and negative events in maintaining cellular homeostasis.
Acknowledgments
The Tax, M22, and M47 expression plasmids were kind gifts from Warner C. Greene (Gladstone Institute of Virology and Immunology, University of California, San Francisco). The anti-Tax monoclonal antibody (Tab172) was a kind gift from John N. Brady (NCI, National Institutes of Health). We thank Dr. Xue Zhang for experimental technology.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program (NICHD). This research was also supported by National High Technology Research and Development Program of China (863 Program) Grant 2006AA02A306, Programme of Introducing Talents of Discipline to Universities Grant B06018, Key Project of the Ministry of Science and Technology of China (973 Program) Grant 2005CB522903, and National Natural Science Foundation of China Grant 30500266. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HTLV-1, human T-cell leukemia virus type 1; CREB, cyclic AMP-responsive element-binding protein; CRE, cyclic AMP-responsive element; RT, reverse transcription; EMSA, electrophoretic mobility shift assay; ChIP, chromatin immunoprecipitation; ATF, activating transcription factor; GFP, green fluorescent protein; DN, dominant negative; IL, interleukin.
References
- 1.Hinuma, Y., Nagata, K., Hanaoka, M., Nakai, M., Matsumoto, T., Kinoshita, K. I., Shirakawa, S., and Miyoshi, I. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 6476–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poiesz, B. J., Ruscetti, F. W., Mier, J. W., Woods, A. M., and Gallo, R. C. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 6815–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robek, M. D., and Ratner, L. (1999) J. Virol. 73 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grassmann, R., Berchtold, S., Radant, I., Alt, M., Fleckenstein, B., Sodroski, J. G., Haseltine, W. A., and Ramstedt, U. (1992) J. Virol. 66 4570–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Napolitano, M., Modi, W. S., Cevario, S. J., Gnarra, J. R., Seuanez, H. N., and Leonard, W. J. (1991) J. Biol. Chem. 266 17531–17536 [PubMed] [Google Scholar]
- 6.Tsukahara, T., Kannagi, M., Ohashi, T., Kato, H., Arai, M., Nunez, G., Iwanaga, Y., Yamamoto, N., Ohtani, K., Nakamura, M., and Fujii, M. (1999) J. Virol. 73 7981–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brauweiler, A., Garrus, J. E., Reed, J. C., and Nyborg, J. K. (1997) Virology 231 135–140 [DOI] [PubMed] [Google Scholar]
- 8.Zhao, L. J., and Giam, C. Z. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 7070–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, L. J., and Giam, C. Z. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 11445–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki, T., Fujisawa, J. I., Toita, M., and Yoshida, M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandre, C., and Verrier, B. (1991) Oncogene 6 543–551 [PubMed] [Google Scholar]
- 12.Fujii, M., Niki, T., Mori, T., Matsuda, T., Matsui, M., Nomura, N., and Seiki, M. (1991) Oncogene 6 1023–1029 [PubMed] [Google Scholar]
- 13.Fujii, M., Tsuchiya, H., Chuhjo, T., Akizawa, T., and Seiki, M. (1992) Genes Dev. 6 2066–2076 [DOI] [PubMed] [Google Scholar]
- 14.Trejo, S. R., Fahl, W. E., and Ratner, L. (1996) J. Biol. Chem. 271 14584–14590 [DOI] [PubMed] [Google Scholar]
- 15.Fu, W., Shah, S. R., Jiang, H., Hilt, D. C., Dave, H. P., and Joshi, J. B. (1997) J. Neurovirol. 3 16–27 [DOI] [PubMed] [Google Scholar]
- 16.Joshi, J. B., and Dave, H. P. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka, A., Takahashi, C., Yamaoka, S., Nosaka, T., Maki, M., and Hatanaka, M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishiguro, N., Abe, M., Seto, K., Sakurai, H., Ikeda, H., Wakisaka, A., Togashi, T., Tateno, M., and Yoshiki, T. (1992) J. Exp. Med. 176 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshino, H., Tanaka, H., Shimotohno, K., Miwa, M., Akatsuka, T., and Odaka, T. (1985) Virology 147 223–226 [DOI] [PubMed] [Google Scholar]
- 20.Kibler, K. V., and Jeang, K. T. (2001) J. Virol. 75 2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka, M. (2003) Oncogene 22 5131–5140 [DOI] [PubMed] [Google Scholar]
- 22.Azran, I., Schavinsky-Khrapunsky, Y., and Aboud, M. (2004) Retrovirology 1 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gou, D. M., Sun, Y., Gao, L., Chow, L. M., Huang, J., Feng, Y. D., Jiang, D. H., and Li, W. X. (2001) Biochim. Biophys. Acta 1518 306–310 [DOI] [PubMed] [Google Scholar]
- 24.Gebelein, B., and Urrutia, R. (2001) Mol. Cell. Biol. 21 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun, Y., Gou, D. M., Liu, H., Peng, X., and Li, W. X. (2003) IUBMB Life 55 127–131 [DOI] [PubMed] [Google Scholar]
- 26.Sun, Y., Shao, H., Li, Z., Liu, J., Gao, L., Peng, X., Meng, Y., and Li, W. (2004) Int. J. Mol. Med. 14 971–975 [PubMed] [Google Scholar]
- 27.Guo, M. X., Wang, D., Shao, H. J., Qiu, H. L., Xue, L., Zhao, Z. Z., Zhu, C. G., Shi, Y. B., and Li, W. X. (2006) J. Biol. Chem. 281 24623–24636 [DOI] [PubMed] [Google Scholar]
- 28.Krackhardt, A. M., Witzens, M., Harig, S., Hodi, F. S., Zauls, A. J., Chessia, M., Barrett, P., and Gribben, J. G. (2002) Blood 100 2123–2131 [DOI] [PubMed] [Google Scholar]
- 29.Smith, M. R., and Greene, W. C. (1990) Genes Dev. 4 1875–1885 [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, L. Q., Kopp, P., Martinson, F., Stanfield, K., Roth, S. I., and Jameson, J. L. (2000) Mol. Endocrinol. 14 1448–1461 [DOI] [PubMed] [Google Scholar]
- 31.Pathak, S. K., Bhattacharyya, A., Pathak, S., Basak, C., Mandal, D., Kundu, M., and Basu, J. (2004) J. Biol. Chem. 279 55127–55136 [DOI] [PubMed] [Google Scholar]
- 32.Shao, H., Zhu, C., Zhao, Z., Guo, M., Qiu, H., Liu, H., Wang, D., Xue, L., Gao, L., Sun, C., and Li, W. (2006) Int. J. Mol. Med. 18 457–463 [PubMed] [Google Scholar]
- 33.Wycuff, D. R., and Marriott, S. J. (2005) Front. Biosci. 10 620–642 [DOI] [PubMed] [Google Scholar]
- 34.Amorino, G. P., Mikkelsen, R. B., Valerie, K., and Schmidt-Ullrich, R. K. (2003) J. Biol. Chem. 278 29394–29399 [DOI] [PubMed] [Google Scholar]
- 35.Cox, J. M., Sloan, L. S., and Schepartz, A. (1995) Chem. Biol. 2 819–826 [DOI] [PubMed] [Google Scholar]