Abstract
Excitable cells express a variety of ion channels that allow rapid exchange of ions with the extracellular space. Opening of Na+ channels in excitable cells results in influx of Na+ and cellular depolarization. The function of Nav1.5, an Na+ channel expressed in the heart, brain, and gastrointestinal tract, is altered by interacting proteins. The pore-forming α-subunit of this channel is encoded by SCN5A. Genetic perturbations in SCN5A cause type 3 long QT syndrome and type 1 Brugada syndrome, two distinct heritable arrhythmia syndromes. Mutations in SCN5A are also associated with increased prevalence of gastrointestinal symptoms, suggesting that the Na+ channel plays a role in normal gastrointestinal physiology and that alterations in its function may cause disease. We collected blood from patients with intestinal pseudo-obstruction (a disease associated with abnormal motility in the gut) and screened for mutations in SCN5A and ion channel-interacting proteins. A 42-year-old male patient was found to have a mutation in the gene TCAP, encoding for the small protein telethonin. Telethonin was found to be expressed in the human gastrointestinal smooth muscle, co-localized with Nav1.5, and co-immunoprecipitated with sodium channels. Expression of mutated telethonin, when co-expressed with SCN5A in HEK 293 cells, altered steady state activation kinetics of SCN5A, resulting in a doubling of the window current. These results suggest a new role for telethonin, namely that telethonin is a sodium channel-interacting protein. Also, mutations in telethonin can alter Nav1.5 kinetics and may play a role in intestinal pseudo-obstruction.
Sodium channel function is altered by a number of interacting proteins known as either SCIPs (sodium channel-interacting proteins) or sodium ChIPs (channel-interacting proteins). SCIPs/ChIPs do not conduct ions themselves but alter the kinetics of the channel with which they interact (1). Nav1.5 is a tetrodotoxin-resistant sodium channel expressed in heart (2), human gastrointestinal tract (3), and brain (4). In the human gastrointestinal tract, Nav1.5 is expressed in smooth muscle and interstitial cells of Cajal; the latter generate the electrical signal that paces smooth muscle. In these cells, the native Na+ current is mechanosensitive (5), and mechanosensitivity is dependent on an intact cytoskeleton (6).
Given the recent identification of Nav1.5 in the gut (5) and a positive gastrointestinal phenotype observed in patients with heritable arrhythmia syndromes due to defective Nav1.5 channels (7), we hypothesized that gastrointestinal dysmotility, particularly intestinal pseudo-obstruction, might stem from a defect in the sodium channel macromolecular complex. Intestinal pseudo-obstruction is a gastrointestinal motility disorder characterized by abnormal gastrointestinal motility together with symptoms of bowel obstruction in the absence of any lesions occluding the gut lumen. Secondary intestinal pseudo-obstruction is due to a systemic disease process affecting enteric nerves, interstitial cells of Cajal, or smooth muscle cells, whereas primary intestinal pseudo-obstruction has no known inciting cause (8, 9).
Therefore, we conducted comprehensive mutational analysis of SCN5A, six established SCIPs (SCN1B, SCN2B, SCN3B, SCN4B, CAV3, and SNTA1), and TCAP-encoded telethonin, which is known to interact with ion channels (10) and to act as a stretch sensor (11–13).
During screening, we identified a rare, novel missense mutation in telethonin in one of the patients. No other mutations were found. Telethonin is a small protein (19 kDa) expressed mainly in striated muscle (14) that binds to and is phosphorylated by both titin kinase (15) and protein kinase D (16). It is known to interact with several proteins: MLP (13), Ankrd2 (11), myostatin (17), MURF-1 (18), and the FATZ/calsarcin/myozenin family (17, 19) as well as MinK, the β subunit of the slow activating component of the delayed rectifier potassium current channel (10). Among its varied functions, telethonin works as a stretch sensor in the heart (13), links the sarcomere to a K+ channel subunit (10), and interacts with titin (20) and Ankrd2 (11), a member of the MARP (muscle ankyrin repeat proteins) family of proteins that have also been proposed to act as stress sensors in muscle.
Here, we demonstrate that telethonin is expressed in the circular smooth muscle layer of the human jejunum. Electrophysiological study of the patient's mutation in telethonin suggests that telethonin is a Na+ channel interacting protein and that mutations in telethonin can alter Nav1.5 kinetics, resulting in altered regulation of Na+ entry. The data also suggest a similar function in the heart.
EXPERIMENTAL PROCEDURES
Mutational Analysis of Telethonin (TCAP)—Informed written consent was obtained in accordance with study protocols approved by the Mayo Foundation Institutional Review Board. Genomic DNA for 20 unrelated patients with idiopathic intestinal pseudo-obstruction was extracted from peripheral blood lymphocytes using the Purgene DNA Isolation kit (Gentra Systems). Mutational analysis of both translated exons of TCAP was performed using PCR, denaturing high performance liquid chromatography (HPLC),2 and direct DNA sequencing utilizing a 3500HT DNA Fragment Analysis System (WAVETM, Transgenomic Inc.), as previously described (21). Briefly, 5 μl of PCR product is loaded onto a preheated DNAsep HT cartridge (part number DNA-99-3710; Transgenomic Inc.) and is eluted under partially denaturing conditions with a linear acetonitrile gradient composed of buffers A (0.1 m triethylammonium acetate) and B (0.1 m triethylammonium acetate and 25% acetonitrile). A gradient of 5% buffer B/min at a flow rate of 1.5 ml/min in WAVETM 3500 HT rapid DNA mode allows mutation detection at temperatures optimized for each exon. Eluted DNA fragments are detected by UV absorption at a wavelength of 260 nm, and resulting chromatograms are examined for abnormal elution profiles. Primer sequences, PCR conditions, and denaturing HPLC conditions are shown in Table 1.
TABLE 1.
PCR and dHPLC conditions for mutational analysis of TCAP PCRs for amplifying TCAP were performed in 20-μl volumes using 50 ng of DNA, 16 pmol of each primer, 200 μm each dNTP, 50 mmol/liter KCl, 10 mmol/liter Tris-HCl (pH 8.3), 1.5 mmol/liter MgCl2, and 1.0 unit of Amplitaq Gold (Applied Biosystems). Thermocycling conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, and a final extension of 72 °C for 10 min. Denaturing HPLC was performed using a 5% buffer B/min gradient. The start and stop percentage of buffer B (%B) is indicated, followed by the temperature (Temp) at which the gradient was performed.
| Exon | Template | Forward primer (5′–3′) | Reverse primer (5′–3′) | Size | Gradient 1 (%B; Temp) |
|---|---|---|---|---|---|
| bp | |||||
| 1 | 1 | GGGCTATTTAAAGGGCCTG | TCATGGCTCAGTGAGGGTG | 222 | 52–62%; 64 °C |
| 2 | 2A | TGCCCAGAGAGCAACAGCT | TTGCTGACAGGCACCACAG | 385 | 54–64%; 67 °C |
| 2 | 2B | GGAGGTGGCTGAGATCACA | CACAGGTCCTAGCCAGGAA | 180 | 52–62%; 65 °C |
To be considered a possible pseudo-obstruction-predisposing mutation, the genetic variant had to (i) be a nonsynonymous variant (synonymous single nucleotide polymorphisms were excluded from consideration), (ii) involve a highly conserved residue, (iii) be absent among 400 reference alleles from 100 healthy white and 100 healthy black control subjects, and (iv) result in a functionally altered cellular phenotype. Control genomic DNA was obtained from the Human Genetic Cell Repository sponsored by NIGMS, National Institutes of Health, and the Coriell Institute for Medical Research (Camden, NJ).
Dissection of Human Jejunal Smooth Muscle Strips and Isolation of Smooth Muscle Cells—The Institutional Review Board approved the use of human jejunal tissue, obtained as surgical waste tissue from elective gastric bypass operations performed for morbid obesity in otherwise healthy subjects. The Institutional Animal Use and Care Committee approved the use of mouse tissue. Both longitudinal and circular smooth muscle strips from human jejunum were isolated by dissection, as described previously (22, 23). Briefly, a 2.5–8-cm-long segment of proximal jejunum was cut along the mesentery and pinned mucosa side up onto a Sylgard-coated dish filled with ice-cold Krebs solution. The mucosa was removed using a sharp scissors and discarded. The remaining tissue was repinned serosal side up, and the longitudinal muscle layer was teased away from the circular muscle layer with a combination of forceps and scalpel. The tissue obtained was used for protein extraction and Western blot experiments. Alternatively, circular smooth muscle cells were isolated from muscle strips using a combination of enzymatic digestion and centrifugation, as previously described (23) and used for RT-PCR.
RT-PCR—All RT-PCR amplifications were performed using GeneAmp Gold RNA PCR kit systems (Applied Biosystems) following standard procedures. For the RT-PCR, RNA was extracted using the RNA-Bee solution (Tel-Test Inc.), according to the manufacturer's instructions, and the quality and quantity of the extracted RNA were determined spectrophotometrically by checking the 260 nm/280 nm absorbance ratios. Alternatively, cells were collected as described above, and RNA was extracted from a pool of about 3–4 cells as previously described (24). RT was performed using a mixture of random hexamers and oligo(dT) primers, and the reaction protocol consisted of annealing at 25 °C for 10 min, followed by one cycle at 42 °C for 20 min. The product of the RT reaction was then amplified using gene-specific primers (Table 2). The amplification protocol consisted of two successive rounds of PCR amplification with the following standard protocol: 95 °C for 8 min, followed by cycles at 94 °C for 20 s, 58–60 °C for 30 s, and 72 °C for 30–60 s, ending with 72 °C for 7 min. The annealing temperature and extension period varied, depending on the primers used and the size of products. The first round of amplification consisted of 25 cycles, and the PCR mixture was then used for a second round of 35 cycles. The second amplification protocol was performed as described above, using primers internal to the ones used for the first protocol. The PCR products were separated on a 1.5% agarose gel. Fragments of the expected size were purified from the gel using a standard procedure (WIZARD SV gel and PCR clean up system; Promega) and sent to the Mayo Molecular Core Facility for automated DNA sequencing to confirm the identity of the amplicons.
TABLE 2.
Sequences of the primers used for the RT-PCR
| Primer | Sequence |
|---|---|
| Telethonin primers | |
| Forward external | 5′–CACCGGAGGGGAGAGAGAATGAGG |
| Forward internal | 5′–CTGGGCAGAATGGAAGGAT |
|
Reverse
|
5′–TCAGCCTCTCTGTGCTTCCT
|
| Glyceraldehyde-3-phosphate dehydrogenase primers | |
| Forward | 5′–CCATCACCATCTTCCAGGAG |
| Reverse | 5′–CCTGCTTCACCACCTTCTTG |
Telethonin Antibody Production—Human purified His-tagged full-length telethonin protein was used to immunize BALB/c female mice. Injections to immunize mice for the generation of polyclonal antibody were made at intervals at least 3 weeks apart. Freund's incomplete adjuvant (100 μl) and 15–50 μg of His-tagged telethonin protein in PBS (100 μl) were mixed together at a ratio of 1:1, and the mice were injected intraperitoneally with 200 μl. After the third injection, the mice were bled from the submandibular artery (∼50–100 μl of blood/mouse). The collected blood was allowed to clot for 60 min at 37 °C or overnight at 4 °C and then centrifuged at 10,000 × g for 10 min at 4 °C to separate the serum. Sodium azide (final concentration of 0.02%) was added to the sera. The sera were stored at 4 °C and tested by Western blot analysis to detect telethonin in human skeletal muscle extracts. Similar individual serum samples were pooled, tested, and stored at -20 °C. The use of animals in these experiments was approved by the Italian Ministry of Health.
Immunoprecipitation and Immunoblotting—Mice were killed by CO2 asphyxiation. Then total homogenates were prepared from mouse heart by lysis in a buffer containing 20 mm Tris/HCl, pH 7.5, 0.32 m sucrose, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 10 mm NEM, and a protease inhibitor mixture (protease inhibitor mixture III; Calbiochem). Tissue was homogenized using a Polytron homogenizer for 1 min and centrifuged at 1000 × g for 10 min. The pellet was resuspended using a Teflon/glass homogenizer and centrifuged again in the same condition. The total homogenates were then incubated overnight by rotation at 4 °C with a polyclonal pan-sodium channel antibody (SP19; Chemicon). After the addition of protein-A-Sepharose beads (Amersham Biosciences), incubation followed for 2 h. After washing of the beads, the solutions were then subjected to SDS-PAGE on a polyacrylamide gel and transferred to Immobilon-P polyvinylidene difluoride membrane (Millipore). Membranes were then probed for the presence of telethonin (see above). After washing, horseradish peroxidase-conjugated secondary antibodies were used, and the immunoreactive bands were visualized by ECL according to the manufacturer's instructions (Amersham Biosciences). For the depletion experiments, the Nav1.5 antibody was incubated with the immunogenic peptide, following the manufacturer's instructions.
Immunohistochemistry—Mice were killed by CO2 asphyxiation, and heart tissue was flash-frozen in liquid nitrogen. 12-μm sections were cut, and after air-drying, they were fixed for 10 min in acetone and rinsed in phosphate-buffered saline (PBS). Sections were then incubated with 10% normal donkey serum and 0.3% Triton X-100 for 1 h to block nonspecific absorption sites and then incubated overnight at 4 °C with specific primary antibodies (diluted at the proper concentration in 5% normal donkey serum, 0.3% Triton X-100 in PBS). After several rinses in PBS, the sections were incubated for 1 h with the specific secondary antibody (diluted at the proper concentration in 2.5% normal donkey serum, 0.3% Triton X-100 in PBS), rinsed in PBS, and mounted with Slowfade Gold (Invitrogen). Control experiments were performed by omitting the primary antibodies. Rabbit anti-rat polyclonal antibody to Nav1.5 (Alomone Laboratories) was used to a final dilution of 1:200; mouse polyclonal antibody against human telethonin, made as described above, was used at a dilution of 1:100.
Construction of Expression Vectors and Transfection of HEK293 Cells—The pcDNA3 expression vector (Invitrogen) containing the full-length human SCN5A (H558/Q1077) was used in the Na+ channel expression experiments, as previously described (24). The cDNA for telethonin was produced by RT-PCR and inserted into the pCMV-SPORT vector (Invitrogen) using NotI and SpeI. All constructs were verified by sequencing. Lipofectamine™ 2000 reagent (Invitrogen) was used to transiently co-transfect vectors containing green fluorescent protein pEGFP-C1 (Clontech), SCN5A, and wild-type or R76C telethonin into HEK293 cells (ATCC). Transfected cells were identified by fluorescence microscopy prior to patch clamping.
Site-directed Mutagenesis—The R76C mutation was recreated in the telethonin expression vector using the QuikChange® II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The appropriate nucleotide changes for R76C were engineered into the pCMV-SPORT vector (Invitrogen) using the following nucleotide mutagenic sense and antisense primers: 5′-GGCATCCTCGGCTGTGGGCTGCAGG and 5′-CCTGCAGCCCACAGCCGAGGATGCC. The integrity of the constructs and the absence of undesired mutations were verified by sequencing.
Small Interfering RNA (siRNA) Transfection—The siRNAs used in this study were ON-TARGET SMARTtpools against human telethonin obtained from Dharmacon. Using siPORT Amine Reagent (Ambion), siRNAs (at a final concentration of 100 nm) were transiently transfected into HEK 293 cells stably expressing SCN5A according to the manufacturer's instructions. Cells in each experiment were treated with scrambled or TCAP siRNA. The cells were incubated for 4 days and then used for patch clamping to record activation and inactivation kinetics from scrambled or TCAP siRNA-treated cells (recordings carried out on the same day) and harvested for quantitative RT-PCR experiments.
Patch Clamp Recordings—Whole cell records were obtained by standard patch clamp techniques. Recording electrodes were pulled on a P-97 puller (Sutter Instruments) using Kimble KG-12 glass, coated with R6101 (Dow Corning), and fire-polished to a final resistance of 3–5 megaohms. Currents were amplified, digitized, and processed using an Axopatch 200B amplifier, Digidata 1322A, and pCLAMP 9 software (Axon Instruments). Whole cell records were sampled at 10 kHz and filtered at 4 kHz with an 8-pole Bessel filter. 70–85% series resistance compensation with a lag of 60 μs was applied during each recording. To determine various kinetic parameters, Na+ currents were recorded using pulse protocols described previously (24). For steady state activation, time to peak, and τ of inactivation, HEK cells were held at -100 mV and stepped from -80 to 35 mV in 5-mV intervals for 50 ms. The start-to-start time was 1 s. For steady state inactivation, cells were held at -100 mV, stepped from -130 to -60 mV in 5-mV intervals for 3 s to reach a steady state of inactivation and then briefly stepped to -110 mV for 0.1 or 0.2 ms to standardize transients and finally stepped to -40 mV. Current was measured at -40 mV and plotted against the 3 s prepulse potential. The start-to-start time was 4 s.
Drug and Solutions—Currents were recorded in solutions at room temperature (22 °C). The intracellular solution contained 130 mm Cs+, 125 mm methanesulfonate, 20 mm Cl-, 5 mm Na+, 5 mm Mg2+, 5 mm HEPES, 2 mm EGTA, 2.5 mm ATP, 0.1 mm GTP. The extracellular solution was Ringer's solution modified to contain 149.2 mm Na+, 159 mm Cl-, 4.74 mm K+, 2.54 mm Ca2+, 5 mm HEPES, and 5.5 mm glucose. Chemicals were purchased from Sigma. Intra- and extracellular solutions were equilibrated to pH 7.0 and 7.35 with CsOH or NaOH, respectively. Osmolality of either was 300 mosm/kg.
Data Analysis—Electrophysiological data were analyzed using Clampfit (Axon Instruments Inc.), Excel (Microsoft), GraphPad InStat (GraphPad Software Inc.), and SigmaPlot (SPSS Inc.). Voltages were adjusted for junction potentials. Statistical comparisons were made using a paired, two-tailed Student's t test or one-way analysis of variance. Statistical significance was accepted when p values were <0.05. Data are expressed as mean values ± S.E. The τ of inactivation was measured between the points at peak inward current and at 50 ms. Time to peak was measured as the difference between the time when the pulse started and the time at maximal peak inward current. τ of inactivation and time to peak were both fitted by a standard two-exponential decay curve: y = y0 + ae-bx. Steady state activation was calculated as the peak inward current of a single control trace normalized to 100 using the equation, |INORM| = 100·(IV – IMIN)/(IMAX – IMIN), where INORM represents normalized current, IV is measured current at a given voltage, IMIN is minimum peak inward current, and IMAX is maximum peak inward current. Steady state inactivation was calculated similarly. Steady state activation and inactivation curves were fitted with a three-parameter sigmoid (Boltzmann) function: y = 100/(1 + e–(x – x0)/b), where x0 is V½, the voltage at half-activation or inactivation, and b is the slope.
RESULTS
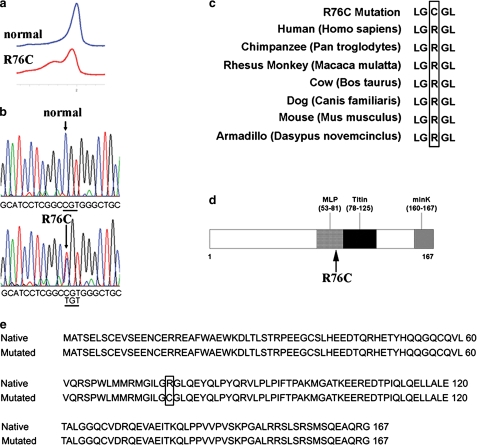
Identification of a Novel TCAP Mutation in a Pseudo-obstruction Patient—In a screen of 20 unrelated patients with primary intestinal pseudo-obstruction, we identified a patient with a heterozygous mutation, R76C, in telethonin by direct DNA sequencing. To our knowledge, this is the first description of such a missense mutation in telethonin involving this residue and the first description of a mutation in telethonin in a patient with intestinal pseudo-obstruction. A C to T base change at position 226 in exon 2 (Fig. 1a and b), involving a highly conserved residue (Fig. 1c), resulted in the substitution of cysteine (TGT) for arginine (CGT) in residue 76 (Fig. 1e). The mutation is located in the region of the telethonin where the protein has been shown to interact, in the heart, with sarcomeric proteins (muscle LIM protein and titin) (13, 21) (Fig. 1d). The R76C mutation in telethonin was found in a 42-year-old male diagnosed with intestinal pseudo-obstruction. There was no family history of intestinal pseudo-obstruction. The diagnosis was based on radiological studies and clinical picture several years before he was referred to the corresponding author for further management. Intestinal pseudo-obstruction was confirmed with panintestinal radionuclide transit tests, which showed normal gastric emptying with delayed small bowel and colon transit, and by gastroduodenal manometry, suggesting a mixed neuropathic/myopathic picture with decreased amplitude of contractions and an incomplete fed pattern. A small bowel x-ray and computed tomography scan of the abdomen did not show any evidence for obstruction. A small bowel aspirate showed small bowel bacterial overgrowth, consistent with his abnormal motility, that was treated with antibiotics. The clinical history was also significant for sleep apnea. Although extensive cardiac testing was not carried out, no overt cardiac phenotype was apparent by clinical examination and ECG. Specifically, the base-line ECG did not show either QT interval prolongation to suggest long QT syndrome or ST segment elevation in the right precordial leads to suggest Brugada syndrome.
FIGURE 1.
Molecular characterization of TCAP mutation. a, normal (blue) and mutated (red) TCAP denaturing HPLC. b, sequencing chromatograms showing a heterozygous C to T base change in position 226 in exon 2 of the telethonin gene, TCAP, in the patient compared with normal. c, amino acid conservation analysis showing the highly conserved Arg in position 76 in different species. d, graphic map of the telethonin gene showing the localization of the identified mutation in the area of interaction with muscle LIM protein and titin. e, alignment of the amino acid sequences of native and mutated telethonin shows the resulting R76C codon change in the protein sequence (box).
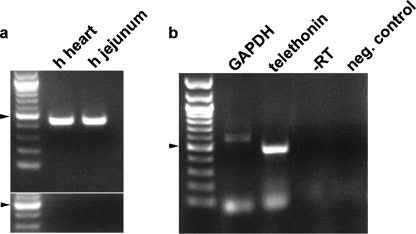
Telethonin mRNA Is Expressed in Human Circular Smooth Muscle Cells—Telethonin was detected in human intestinal muscle layers using RT-PCR on circular muscle strips separated from the jejunum mucosa and longitudinal muscle by dissection (Fig. 2a). Several cell types reside in the intestinal smooth muscle layers; therefore, RT-PCR on individually picked dissociated cells was used to localize telethonin. Human circular smooth muscle cells were identified by their spindle shape and ability to contract. 3–4 individual cells were collected in each tube. Telethonin was amplified with gene-specific primers designed to span an intron to exclude genomic DNA contaminations (Fig. 2b).
FIGURE 2.
Expression of telethonin mRNA in human jejunum smooth muscle. a, expression of telethonin mRNA in muscle strips from human jejunum. RT-PCR using primers designed against the human telethonin gene showed the presence of telethonin mRNA in the jejunal smooth muscle strips isolated by dissection. RNA extracted from heart tissue was used as a positive control. No bands were amplified in the -RT negative controls shown in the lower panel. b, RT-PCR on a pool of 3–4 human jejunal smooth muscle single cells showing the presence of a correct size band for TCAP. The identity of the band was confirmed by sequence analysis. There were no products in negative controls containing water bath solution (4 μl aspirated just above a smooth muscle cell) and in controls performed by omitting the reverse transcriptase enzyme during the RT reaction. Arrowhead, 500-bp size band. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
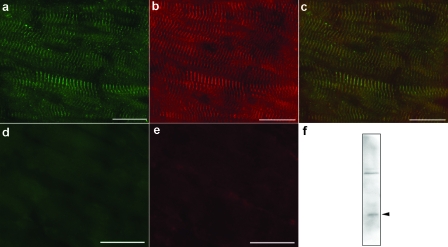
Co-localization of Nav1.5 and Telethonin Protein in Mouse Heart—To determine if telethonin is linked to the mechanosensitive sodium channels, we stained mouse heart with antibodies against telethonin and Nav1.5. Immunostaining for telethonin showed the striated pattern typical of sarcomeric proteins (Fig. 3a). Immunostaining for Nav1.5 unexpectedly also showed a striated distribution (Fig. 3b), and the two staining patterns almost completely overlapped (Fig. 3c). Negative controls carried out by omitting the primary antibodies showed an absence of staining (Fig. 3, d–e). To further test the co-expression of telethonin and a Na+ channel, immunoprecipitation experiments were carried out from mouse heart total homogenates. A 19 kDa band for telethonin was found in immunoprecipitates obtained using a pan-sodium channel antibody (SP19) (Fig. 3f). The identity of the band was confirmed by mass spectrometry analysis (data not shown).
FIGURE 3.
Expression of telethonin protein in mouse cardiac muscle. a, immunofluorescence on mouse heart showed the typical striated staining for telethonin (green). b, the staining for SCN5A (red) appeared to have a distribution similar to telethonin. c, merged image showing the overlapping distribution of the two proteins (yellow). Negative controls carried out by omitting the primary antibodies showed no staining. Immunofluorescence on mouse heart performed by omitting the primary antibodies showed an absence of the striated staining for telethonin (green) (d) and SCN5A (red) (e). Scale bars, 20 μm. f, immunoblotting using an antibody against telethonin showed the presence of this protein (arrowhead) in immunoprecipitates obtained from total homogenates of mouse heart tissue using a pan-sodium channel antibody (SP19).
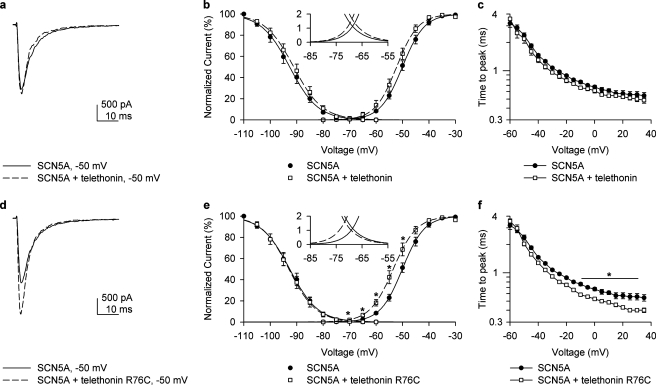
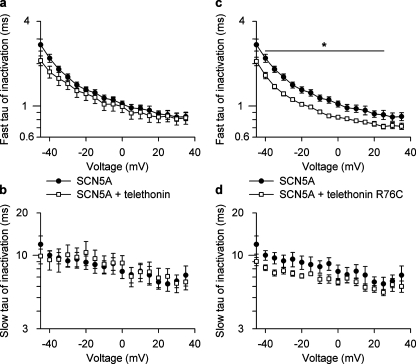
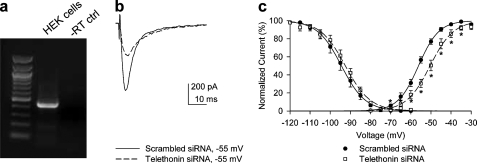
Telethonin Mutation R76C Alters Activation and Inactivation Kinetics of Nav1.5—To determine the potential effect of telethonin on Nav1.5 kinetics, whole cell patch clamp recordings were obtained from HEK 293 cells co-expressed either with SCN5A plus telethonin or SCN5A plus R76C telethonin (Fig. 4, a–f). Green fluorescent protein was used as a marker. Maximum inward peak currents were similar (Fig. 4, a and d). No change in steady state activation or inactivation kinetics was seen upon expression of native telethonin with SCN5A (Fig. 5, a and b). However co-expression of R76C telethonin with SCN5A resulted in a leftward shift in steady state activation kinetics of SCN5A (Fig. 4b, V½, SCN5A alone, -50.0 ± 0.1 mV; Fig. 4e, V½, SCN5A plus R76C telethonin, -53.5 ± 1.8 mV; n = 6, p < 0.05). The absolute voltage shift was 3.5 mV, resulting in the window current approximately doubling and shifting in the negative direction (Fig. 4e, inset). This would result in increased Na+ entry at the resting membrane potential, making the cell more depolarized. Time to peak current (activation) was not different between SCN5A and SCN5A plus telethonin (Fig. 4c). It was, however, faster for voltages ranging from -10 to 30 mV for SCN5A plus R76C telethonin compared with SCN5A alone (Fig. 4f). Inactivation fit well with two τ values (representing fast and slow inactivation). Significant differences were observed only in the fast τ of inactivation (n = 6, p > 0.05, Fig. 5, c and d), which was faster with the mutation. Since we found no difference in the kinetics of SCN5A when expressed in HEK 293 cells alone and when co-expressed with nonmutated telethonin, we carried out RT-PCR experiments on total RNA extracted from nontransfected HEK 293 cells to investigate the expression of endogenous telethonin in HEK cells. Message for telethonin was found in the HEK cells (Fig. 6a). Therefore, to examine the effect of telethonin on SCN5A kinetics, we knocked down endogenous telethonin in HEK cells using siRNA. The procedures for these experiments are described in detail under “Experimental Procedures.” Patch clamp experiments were carried out after 4 days of treatment of HEK cells stably transfected with SCN5A with siRNA against telethonin. Maximum inward peak currents were similar (Fig. 6b). There was a positive shift in the activation kinetics of SCN5A of 5.2 mV (Fig. 6c, TCAP siRNA V½, -51.2 ± 1, n = 11 compared with scrambled siRNA V½, -56.4 ± 1 mV, n = 10, p < 0.01). Knockdown of telethonin (mean knockdown of 70%) was confirmed by quantitative RT-PCR. No change in steady state inactivation kinetics were observed (Fig. 6c, TCAP siRNA V½, -92.8 ± 1, n = 11 compared with scrambled siRNA V½, -94.5 ± 2 mV, n = 10, p = 0.46). The data suggest that the R76C mutation in telethonin enhances the effect of telethonin on SCN5A.
FIGURE 4.
Telethonin mutation R76C alters activation and inactivation kinetics of SCN5A co-transfected in HEK cells. a, inward Na+ current at -50 mV recorded from HEK 293 cells co-transfected with SCN5A alone or with telethonin. b, steady state inactivation and activation curves for SCN5A alone or with telethonin, showing that telethonin did not alter activation or inactivation curves. c, telethonin did not delay the time to peak of activation of Na+ current in cells transfected with both SCN5A and telethonin compared with SCN5A alone. d, inward Na+ current at -50 mV for SCN5A alone or SCN5A with R76C telethonin. e, steady state inactivation and activation curves for SCN5A with R76C or SCN5A alone. Steady state activation of SCN5A with telethonin R76C shifted negative, resulting in a larger window current (inset) than SCN5A alone. f, telethonin R76C sped up the time to peak of activation of Na+ current in cells co-transfected with SCN5A as compared with those with SCN5A alone.
FIGURE 5.
Inactivation kinetics of SCN5A co-transfected in HEK cells in the presence of wild-type or mutant telethonin. Telethonin wild type did not affect the fast τ (a) or the slow τ (b) of inactivation of Na+ current in HEK cells co-transfected with SCN5A as compared with those with SCN5A alone. No significant differences were present at any voltages. Telethonin R76C significantly decreased the fast τ (c) but not the slow τ of inactivation (d).
FIGURE 6.
Expression and knockdown of endogenous telethonin in HEK 293 cells stably transfected with SCN5A. a, RT-PCR using primers designed against the human telethonin gene showed the presence of telethonin mRNA in HEK 293 cells. No bands were identified in the -RT negative controls (ctrl). b, representative current traces at -55 mV stepped from a holding potential of -100 mV, recorded from HEK 293 cells stably expressing SCN5A treated for 4 days with scrambled siRNA or with siRNA against telethonin. c, steady state inactivation and activation curves for sodium current in HEK 293 cells stably expressing SCN5A and transfected with either scrambled siRNA (n = 10) or TCAP siRNA (n = 11). Steady state activation of SCN5A after treatment with siRNA against TCAP was shifted positive with respect to cells treated with scrambled siRNA (*, p < 0.05).
DISCUSSION
The data presented in this study indicate that telethonin is expressed in tissues other than cardiac and skeletal muscle (14) and suggest a new role for telethonin, as a sodium channel-interacting protein. We also describe a novel telethonin mutation, R76C, found in a patient with intestinal pseudo-obstruction. The heterozygous mutation identified in a pseudo-obstruction patient is located in the second exon of the telethonin gene, further upstream of the mutations known to be responsible for the limb girdle muscular dystrophy type 2G (25), in an area of the telethonin gene that is highly conserved among the different species examined.
We also showed that mRNA for telethonin is found in human jejunal circular smooth muscle cells. Immunofluorescence and immunoprecipitation studies showed that telethonin and SCN5A appear to be co-expressed and interact, and analysis of the channels in a heterologous expression system (HEK 293 cells) revealed a functional interaction between the mutated telethonin and SCN5A. It is not known if telethonin is also expressed in other cell types in the gut, such as the interstitial cells of Cajal that also express Nav1.5 and regulate motility (26). The expression of SCN5A in the gut varies across species (23). There are currently no animal models that we can use to predict the effect of the observed shift in kinetics on motility. Altered Na+ entry may change gastrointestinal smooth muscle contractile behavior in several ways, including by affecting membrane potential, amplitude and duration of the gastrointestinal slow wave, the Na+/K+ pump, and the Na+/Ca2+ exchanger, all of which will impact the ability of smooth muscle to sustain regular contractile activity (3) required for normal function.
The mutation was only found in one patient of the 20 screened, suggesting that intestinal pseudo-obstruction is a heterogeneous disorder, not unexpected given the wide range of symptoms and clinical findings. An extensive cardiovascular testing of this patient was not possible due to protocol limitations on contacting the patient. It is known from the literature that telethonin interacts with ion channels in the heart. The C terminus of telethonin directly interacts with the MinK subunit of the slow activating component of the delayed rectifier potassium current channel, and this complex, through the interaction of telethonin with the N terminus of titin, connects the ion channel to the sarcomere (10). In cardiac myocytes, Nav1.5 channels are localized in two different pools: at the lateral membrane and in the intercalated disks (27). However, Gavillet et al. (28) conclude that only the pool of Nav1.5 present on the membrane of cardiac myocytes can interact with the dystrophin complex, because dystrophin is absent from the intercalated disks. They hypothesized that in the intercalated disks, Nav1.5 could be associated with another protein complex still to be determined. Recently, it has been suggested that macromolecular complexes, including N-RAP, MLP, telethonin, and titin, can be present at the intercalated disks in cardiomyocytes and function as stress sensors (29). The finding that the mechanosensitive sodium channel Nav1.5 is functionally coupled to telethonin is in line with these findings and suggests that Nav1.5 expressed in the intercalated disks may associate with these complexes through telethonin.
In conclusion, telethonin appears to function as a sodium channel-interacting protein. Expression of telethonin is not limited to cardiac and skeletal muscle as previously thought but is broader, with telethonin also found in smooth muscle. The telethonin mutation found in the patient with intestinal pseudo-obstruction provides evidence that telethonin can alter Nav1.5 function and raises the possibility for a similar effect in the heart.
Acknowledgments
We thank Kristy Zodrow for secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK52766 and 57061 (to G. F.). This work was also supported by Telethon Foundation-Italy Grant GGP04088 (to G. F.), COFIN 2004062155 and Fondazione Del Monte di Bologna e Ravenna,” Bologna, Italy (to R. De G.), RO1 HL71092 (to J. C. M.), and the Windland Smith Rice Comprehensive Sudden Cardiac Death Program (to M. J. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HPLC, high performance liquid chromatography; RT, reverse transcription; PBS, phosphate-buffered saline; siRNA, small interfering RNA.
References
- 1.Abriel, H., and Kass, R. S. (2005) Trends Cardiovasc. Med. 15 35-40 [DOI] [PubMed] [Google Scholar]
- 2.Gellens, M. E., George, A. L., Jr., Chen, L. Q., Chahine, M., Horn, R., Barchi, R. L., and Kallen, R. G. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 554-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holm, A. N., Rich, A., Miller, S. M., Strege, P., Ou, Y., Gibbons, S., Sarr, M. G., Szurszewski, J. H., Rae, J. L., and Farrugia, G. (2002) Gastroenterology 122 178-187 [DOI] [PubMed] [Google Scholar]
- 4.Hartmann, H. A., Colom, L. V., Sutherland, M. L., and Noebels, J. L. (1999) Nat. Neurosci. 2 593-595 [DOI] [PubMed] [Google Scholar]
- 5.Ou, Y., Gibbons, S. J., Miller, S. M., Strege, P. R., Rich, A., Distad, M. A., Ackerman, M. J., Rae, J. L., Szurszewski, J. H., and Farrugia, G. (2002) Neurogastroenterol. Motil. 14 477-486 [DOI] [PubMed] [Google Scholar]
- 6.Strege, P. R., Holm, A. N., Rich, A., Miller, S. M., Ou, Y., Sarr, M. G., and Farrugia, G. (2003) Am. J. Physiol. 284 C60-C66 [DOI] [PubMed] [Google Scholar]
- 7.Locke, G. R., III, Ackerman, M. J., Zinsmeister, A. R., Thapa, P., and Farrugia, G. (2006) Am. J. Gastroenterol. 101 1299-1304 [DOI] [PubMed] [Google Scholar]
- 8.Milla, P. J. (1994) in Gastrointestinal Transit: Pathophysiology and Pharmacology (Kamm, M. A., and Lennard-Jones, J. E., eds) Wrightson Biomedical Publishing, Petersfiend, UK
- 9.Schuffler, M. D. (1981) Med. Clin. N. Am. 65 1331-1358 [DOI] [PubMed] [Google Scholar]
- 10.Furukawa, T., Ono, Y., Tsuchiya, H., Katayama, Y., Bang, M. L., Labeit, D., Labeit, S., Inagaki, N., and Gregorio, C. C. (2001) J. Mol. Biol. 313 775-784 [DOI] [PubMed] [Google Scholar]
- 11.Kojic, S., Medeot, E., Guccione, E., Krmac, H., Zara, I., Martinelli, V., Valle, G., and Faulkner, G. (2004) J. Mol. Biol. 339 313-325 [DOI] [PubMed] [Google Scholar]
- 12.Miller, M. K., Bang, M. L., Witt, C. C., Labeit, D., Trombitas, C., Watanabe, K., Granzier, H., McElhinny, A. S., Gregorio, C. C., and Labeit, S. (2003) J. Mol. Biol. 333 951-964 [DOI] [PubMed] [Google Scholar]
- 13.Knoll, R., Hoshijima, M., Hoffman, H. M., Person, V., Lorenzen-Schmidt, I., Bang, M. L., Hayashi, T., Shiga, N., Yasukawa, H., Schaper, W., McKenna, W., Yokoyama, M., Schork, N. J., Omens, J. H., McCulloch, A. D., Kimura, A., Gregorio, C. C., Poller, W., Schaper, J., Schultheiss, H. P., and Chien, K. R. (2002) Cell 111 943-955 [DOI] [PubMed] [Google Scholar]
- 14.Valle, G., Faulkner, G., De Antoni, A., Pacchioni, B., Pallavicini, A., Pandolfo, D., Tiso, N., Toppo, S., Trevisan, S., and Lanfranchi, G. (1997) FEBS Lett. 415 163-168 [DOI] [PubMed] [Google Scholar]
- 15.Mayans, O., van der Ven, P. F., Wilm, M., Mues, A., Young, P., Furst, D. O., Wilmanns, M., and Gautel, M. (1998) Nature 395 863-869 [DOI] [PubMed] [Google Scholar]
- 16.Haworth, R. S., Cuello, F., Herron, T. J., Franzen, G., Kentish, J. C., Gautel, M., and Avkiran, M. (2004) Circ. Res. 95 1091-1099 [DOI] [PubMed] [Google Scholar]
- 17.Nicholas, G., Thomas, M., Langley, B., Somers, W., Patel, K., Kemp, C. F., Sharma, M., and Kambadur, R. (2002) J. Cell. Physiol. 193 120-131 [DOI] [PubMed] [Google Scholar]
- 18.Witt, S. H., Granzier, H., Witt, C. C., and Labeit, S. (2005) J. Mol. Biol. 350 713-722 [DOI] [PubMed] [Google Scholar]
- 19.Faulkner, G., Pallavicini, A., Comelli, A., Salamon, M., Bortoletto, G., Ievolella, C., Trevisan, S., Kojic, S., Dalla Vecchia, F., Laveder, P., Valle, G., and Lanfranchi, G. (2000) J. Biol. Chem. 275 41234-41242 [DOI] [PubMed] [Google Scholar]
- 20.Mues, A., van der Ven, P. F., Young, P., Furst, D. O., and Gautel, M. (1998) FEBS Lett. 428 111-114 [DOI] [PubMed] [Google Scholar]
- 21.Ackerman, M. J., Tester, D. J., Jones, G. S., Will, M. L., Burrow, C. R., and Curran, M. E. (2003) Mayo Clin. Proc. 78 1479-1487 [DOI] [PubMed] [Google Scholar]
- 22.Farrugia, G., Rae, J. L., Sarr, M. G., and Szurszewski, J. H. (1993) Am. J. Physiol. 265 G873-G879 [DOI] [PubMed] [Google Scholar]
- 23.Strege, P. R., Mazzone, A., Kraichely, R. E., Sha, L., Holm, A. N., Ou, Y., Lim, I., Gibbons, S. J., Sarr, M. G., and Farrugia, G. (2007) Neurogastroenterol. Motil. 19 135-143 [DOI] [PubMed] [Google Scholar]
- 24.Ou, Y., Strege, P., Miller, S. M., Makielski, J., Ackerman, M., Gibbons, S. J., and Farrugia, G. (2003) J. Biol. Chem. 278 1915-1923 [DOI] [PubMed] [Google Scholar]
- 25.Moreira, E. S., Wiltshire, T. J., Faulkner, G., Nilforoushan, A., Vainzof, M., Suzuki, O. T., Valle, G., Reeves, R., Zatz, M., Passos-Bueno, M. R., and Jenne, D. E. (2000) Nat. Genet. 24 163-166 [DOI] [PubMed] [Google Scholar]
- 26.Strege, P. R., Ou, Y., Sha, L., Rich, A., Gibbons, S. J., Szurszewski, J. H., Sarr, M. G., and Farrugia, G. (2003) Am. J. Physiol. 285 G1111-G1121 [DOI] [PubMed] [Google Scholar]
- 27.Mohler, P. J., Rivolta, I., Napolitano, C., LeMaillet, G., Lambert, S., Priori, S. G., and Bennett, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17533-17538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavillet, B., Rougier, J. S., Domenighetti, A. A., Behar, R., Boixel, C., Ruchat, P., Lehr, H. A., Pedrazzini, T., and Abriel, H. (2006) Circ. Res. 99 407-414 [DOI] [PubMed] [Google Scholar]
- 29.Hoshijima, M. (2006) Am. J. Physiol. 290 H1313-H1325 [DOI] [PMC free article] [PubMed] [Google Scholar]