Abstract
Exon IV of SLC2A1, a multiple facilitator superfamily (MFS) transporter gene, is particularly susceptible to mutations that cause GLUT1 deficiency syndrome, a human encephalopathy that results from decreased glucose flux through the blood-brain barrier. Genotyping of 100 patients revealed that in a third of them who harbor missense mutations in the GLUT1 transporter, transmembrane domain 4 (TM4), encoded by SLC2A1 exon IV, contains mutant residues that have the periodicity of one face of a kinked α-helix. Arg-126, located at the amino terminus of TM4, is the locus for most of the mutations followed by other arginine and glycine residues located elsewhere in the transporter but conserved among MFS proteins. The Arg-126 mutants were constructed and assayed for protein expression, targeting, and transport capacity in Xenopus oocytes. The role of charge at position 126, as well as its accessibility, was investigated in R126H by determining its activity as a function of extracellular pH. The results indicate that intracellular charges at the MFS TM2–3 and TM8–9 signature loops and flanking TMs 3, 5, and 6 are critical for the structure of GLUT1 as are TM glycines and that TM4, located at the catalytic core of MFS proteins, forms a helix that surfaces into the extracellular solution where another proton facilitates transport.
Molecules that belong to the major facilitator superfamily (MFS)2 class of transporters mediate the translocation of substrates through cell membranes. Transporters exposed to extracellular or intracellular solutions make available recognition sites for the binding of nutrients, neurotransmitters, ions, and other molecules and then allow the emergence and release of bound substrate at the opposite side of the membrane. In the case of facilitative transporters, binding to either side of the transporter initiates translocation, but net flow across an ensemble of transporters over a period of time is driven by substrate concentration gradients across the cell independently of other cellular energy sources.
MFS proteins are loosely related in primary structure, but they probably fold similarly (1, 2). The structure of three MFS transporters has been solved: LacY (transporter classification (TC) 2.A.1.5.1) at 3.5-Å resolution (3), GltP (TC 2.A.1.4.3) at 3.3-Å (4), and OxlT (TC 2.A.1.11.1) at 6.5-Å (5) (for the TC system, see Ref. 6 and the Transport Classification Database (7)). Their architectures are superimposable: the transporters contain 12 transmembrane helices arranged in two six-helical quasisymmetrical domains situated around a transport pathway normal to the plane of the membrane. A set of the helices lines a central catalytic cavity. In the case of LacY (or lactose permease, a lactose/H+ cotransporter), the helices located at the catalytic core interact with water-avid substrate to provide a stabilizing network of hydrogen bonds, van der Waals interactions, and hydrophobic residues capable of decreasing substrate dissociation after binding (3). This transient substrate stabilization then probably mobilizes adjacent helices, facilitating translocation across the membrane.
GLUT1 (TC 2.A.1.1.28), encoded by the SLC2A1 gene, transports glucose in and out of many normal tissues and tumors, including erythrocytes (RBCs) and cerebral endothelial cells that form the blood-brain barrier (8). GLUT1 is densely expressed in the RBC membrane where it self-associates and exhibits modulated kinetics (9). Blood-brain barrier GLUT1, which mediates the entry of glucose into the nervous system and thus supplies most of the carbon and energy that the brain consumes, is less well characterized kinetically (10). A topological model based on hydropathy analysis (11) has been validated by the study of glycosylation mutants, establishing GLUT1 as a canonical 12-transmembrane domain MFS class member (12). GLUT1 is about 80% α-helical as also established by circular dichroism and Fourier transform infrared spectroscopy (13, 14), and the pattern of reactivity of substituted cysteines in several transmembrane domains is consistent with helical folding (15). Thus, the experimental evidence converges on the prediction that the structure of the transmembrane core of GLUT1 closely resembles that of MFS (LacY, GltP, and OxlT) proteins.
The study of pathological mutants offers a complementary window to the identification of residues and regions of unrecognized relevance. Amino acid-replacing (missense) mutants that retain correct folding and targeting are of particular value as they single out residues important for function. Homozygous SLC2A1 mutation causing GLUT1 absence is incompatible with embryonic survival because of the central role of GLUT1-mediated glucose transport during development (16). In contrast, heterozygous mutations cause encephalopathy (17). Patients afflicted by classic, severe GLUT1 deficiency (18) manifest varying degrees of epilepsy, mental retardation, and other neurological abnormalities and share two characteristic indicators of GLUT1 transporter dysfunction: diminished cerebrospinal fluid glucose (hypoglycorrhachia) and reduced glucose uptake by the cerebrum detectable by positron emission tomography (19, 20).
We set out to first identify residues essential for transport by identifying and genotyping 100 GLUT1 deficiency syndrome patients. A third of them (33 of 100) carried missense mutations that were distributed among any one of the 10 SLC2A1 exons with the peculiarity that, of these, nearly a third of patients (13 of 33) harbored mutations in exon IV. The rest of the patients (67 of 100) exhibited a broad variety of sequence variations not associated with single amino acid replacement and therefore were less informative from a structural point of view. Exon IV is particularly notable because it encodes the fourth transmembrane segment (TM4) of GLUT1, an important domain of MFS proteins given its proximity to the transport pathway that, in the case of LacY, interacts with substrate (3). The data support the notion that GLUT1 TM4 exists as an α-helix because it exhibits a periodic mutation pattern that is consistent with this folding, and we infer that the side of the helix where the mutations are located provides a surface that is essential for catalysis.
To probe the topology of TM4 relative to the membrane, we then tested the susceptibility of R126H, a naturally occurring mutant located near the amino end of TM4, to protonation from the extracellular solution and also analyzed other charged and neutral side chain substitutes of this residue. We found that Arg-126 is exposed at the extracellular aspect of the transporter and that a charge at this position facilitates glucose transport.
Additional missense mutations eliminated conserved glycines and replaced charged intracellular residues delimiting TMs 3, 4, 5, and 6 and arginines situated in the TM2–3 and TM8–9 loops, which are part of the two RXGRR signature motifs present in the MFS (21, 22). These findings support the folding of TM4 as an α-helix and highlight the essential function of protein charges located at transporter-membrane boundaries.
EXPERIMENTAL PROCEDURES
Patient Identification, RBC Glucose Uptake, and Genotyping— All contacts established with and information obtained about human subjects followed procedures approved by the Institutional Review Board of Columbia University and were voluntarily provided. Patients were identified on the basis of clinical and analytical criteria including infantile epilepsy and hypoglycorrhachia with diminished lactic acid concentration in the cerebrospinal fluid among other symptoms and parameters compatible with the diagnosis of GLUT1 deficiency (23).
RBC glucose uptake assays were performed in suspected patients using two control blood samples, which were sometimes obtained from the proband's normal parents, and were simultaneously drawn to avoid assay variability, a phenomenon that occurs when blood is subjected to shipping or storage (24). Blood was collected in sodium heparin or citrate-phosphatedextrose tubes. Zero-trans uptake of a fixed concentration of glucose below the apparent transport Km over time used 14C-labeled 3-O-methyl-d-glucose (3OMG; 0.5 mm, 1 μCi/ml) and was measured at 4 °C and pH 7.4 in duplicate. Influx was terminated at 5-s intervals starting at 0 s (no label added) and ending at 30 s. Washed cells were lysed, and retained radiolabel was quantitated by liquid scintillation counting. The slope of the plot of ln(1 - Ct/Ceq) over time, where Ct represents red cell label and Ceq represents isotope specific activity, was used to estimate uptake. Uptake of a series of glucose concentrations (0, 0.6, 1, 1.33, 2, 4, and 6 mm) during 15 s was similarly measured in quartets of identical samples. Typically the S.E. of the radioactive counts obtained from repeat samples did not exceed 6%. Apparent Km and Vmax values were obtained from Lineweaver-Burke plots with regression coefficients r generally greater than 0.96.
Genomic DNA was obtained from each patient and control blood sample and sequenced over the entire SLC2A1 coding region over all introns except 1 (which spans 15,312 bp) and over all intron-exon boundaries. Sequencing was performed on both DNA strands. All patients carried one mutation in one allele except where noted, and control blood from relatives was devoid of mutation except where indicated. After confirmatory RBC assay and genetic analysis of at least one proband per family, the rest of the familial cases sharing typical symptomatology were confirmed only by genotyping.
Recombinant DNA and Oocyte Culture—Subcloning of human GLUT1 in a custom plasmid, GLUT1/pM, was achieved by cloning a 1913-base pair Eco47III and HindIII GLUT1 cDNA restriction fragment derived from pcDNA3 (Invitrogen) into a vector (pM) containing fragments of the 5′- and 3′-untranslated regions of the Xenopus β-globin gene to enhance expression. Substitutions R126H, R126K, R126E, R126L, and R126C were introduced in GLUT1/pM by PCR performed on template plasmid cDNA using mutagenic oligonucleotides and from which a DNA segment was excised and then replaced by ligation using unique endonuclease sites. All constructs were subjected to restriction analysis and DNA sequencing extending over mutant and ligation areas. Capped runoff cRNA transcripts labeled with [3H]UTP were prepared after plasmid linearization with NotI using an mMessage mMachine kit (Ambion, Austin, TX), verified by denaturing agarose gel electrophoresis, quantitated by liquid scintillation counting, and stored in water at -80 °C.
Xenopus laevis (Nasco, Fort Atkinson, WI) ovarian lobes were collected following procedures approved by the Columbia University Institutional Animal Care and Use Committee. Stage V and VI oocytes were dissociated using collagenase and injected with 50 nl of diluted cRNA (0.40 μg/μl for all mutants). This dosage assured that the transport capacity of wild type-expressing oocytes remained below saturation. Control oocytes were injected with 50 nl of water. Cells were maintained in Barth's solution (containing 88 mm NaCl, 2.4 mm NaHCO3, 1 mm KCl, 0.33 mm Ca(NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 10 mm HEPES (178 mosm, pH 7.5)) at 18 °C for 2 days before use.
GLUT1 Transport and Expression Assays—Zero-trans influx of 3OMG into oocytes was measured as described previously (25, 26) with minor modifications using groups of oocytes placed into 0.6 ml of Barth's solution containing 10 μCi of 14C-labeled 3OMG (5 Ci/mmol; PerkinElmer Life Sciences) and 2 mm unlabeled 3OMG in the wells of a 24-well plate. For each data point, six to eight oocytes were used; each point was then replicated an additional one to three times except where noted. After incubation for 10 min, the oocytes were washed four times with ice-cold Barth's solution containing 0.2 mm phloretin to block GLUT1 activity. Individual oocytes were then placed into vials, and 0.2 ml of 0.2 n NaOH plus 0.2% SDS were added and mixed. After cellular dissolution, 5 ml of scintillation fluid was placed into each vial for disintegration detection. Zero-trans efflux of 3OMG was performed by injecting different amounts of 14C-labeled 3OMG together with unlabeled 3OMG to reach calculated concentrations of 1–50 mm in oocytes previously expressing wild type and mutant GLUT1 (assuming an average oocyte volume of 0.8 μl). After injection, immediately before the efflux assay, oocytes were rapidly washed three times in 2 ml of Barth's solution and transferred to a scintillation vial containing 0.5 ml of Barth's solution. After 2 min, the oocyte was transferred to another scintillation vial, dissolved, and counted as above. The measured amount of labeled glucose in both vials was used to estimate intracellular glucose concentration for efflux measurements. Basal transport rates from water-injected oocytes were subtracted from transport by oocytes expressing mutant and wild type GLUT1. Water-injected oocyte transport rates typically reached ∼10% of GLUT1-injected oocytes. For R126H protonation experiments, the pH was lowered to 6.5, and methanesulfonic acid (Sigma) replaced HEPES. The results were expressed as pmol of 3OMG uptake/10 min/oocyte. Transport kinetics were analyzed by fitting uptake data to the Michaelis-Menten equation utilizing Origin (OriginLab Software, Inc., Northampton, MA).
Confocal immunofluorescence microscopy permitted detection of GLUT1 in frozen oocyte sections as described previously (26) using affinity-purified rabbit primary antibody (Glut-101AP; FabGennix Inc., Frisco, TX) and donkey anti-rabbit IgG labeled with Alexa Fluor 488 (Molecular Probes, Eugene, OR) as secondary antibody, both diluted 1:200. Confocal microscopy was performed at the Optical Microscopy Facility at Columbia University.
For protein assay, total cell membranes were prepared as described previously (25) with the following modifications: after disruption of defolliculated oocytes by pipetting, cells were washed free of yolk. Oocyte ghosts were then suspended in freshly made homogenization buffer (50 μl/oocyte) containing 10 mm HEPES, 83 mm NaCl, 1.0 mm MgCl2, 0.5 mm phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin A, 5 μg/ml soybean trypsin inhibitor, 5 μg/ml leupeptin, pH 7.9, and then stroked 25 times in a glass-Teflon homogenizer. The homogenates were centrifuged at 1000 × g for 10 min at 4 °C three times, saving the supernatant. 15 μl of oocyte purified total membrane samples containing 2 μg of protein were subjected to 4–20% SDS-polyacrylamide gradient gel electrophoresis and probed with purified rabbit Glut-101AP primary antibody at 1:5000 dilution and secondary antibody-horseradish peroxidase (Santa Cruz Biotechnology Inc., Santa Cruz, CA) also at 1:5000 dilution. Under these conditions, GLUT1-expressing oocytes yielded a predominant band of about 55 kDa, corresponding to the monomeric, glycosylated transporter. A smaller, ∼45-kDa band was less abundant and reflected differential glycosylation as the ∼55-kDa fraction was susceptible to enzymatic deglycosylation and conversion to ∼45 kDa (not shown (4)). Additional ∼90–100-kDa bands and higher mass species were also detectable and possibly represented higher order GLUT1 aggregates.
The fractionation procedure for the collection of a plasma membrane-rich fraction followed the method of Finidori-Lepicard et al. (27). In brief, because oocytes are large cells (1-mm diameter) densely filled with vacuoles and with a small plasma membrane to total cell material ratio, it is impractical to purify plasma membrane-associated GLUT1. Instead we isolated a cell fraction known to be enriched in plasma membrane protein as estimated by 5′-nucleotidase (a membrane enzyme) activity assay (27, 28). When using this procedure, the P-1000 cell fraction is known to retain about 75% of the total cellular 5′-nucleotidase activity, and this fraction was used for Western blot analysis of predominantly plasma membrane material using the antibodies and procedures described above. Under these conditions, the molecular species detectable in the immunoblot resembled those identified in the whole-oocyte membrane extract except for a predominance of the monomeric transporter and an additional ∼25-kDa band that may represent a degradation product. GLUT1 signals were quantified digitally using ImageJ software (W. S. Rasband, National Institutes of Health, Bethesda, MD). The densities of wild type GLUT1 signals in purified oocyte membranes were used to normalize mutant GLUT1 densities.
GLUT1 Modeling—A human GLUT1 molecular model was constructed with the homology modeling algorithm Modeler 8v2 (29) based on the crystal structures of LacY (Protein Data Bank code 2CFQ) (3) and GlpT (Protein Data Bank code 1PW4) (4). The sequence alignment used in the modeling was based on (a) sequence-structure alignments of the human GLUT1 sequence and the LacY and GlpT structures with FUGUE (30) and (b) a pairwise structural alignment of LacY and GlpT calculated by Structal (31) excluding the amino and carboxyl termini, which diverge significantly among MFS. The initial sequence alignment was further refined and validated by comparing the accessible residue positions detected in GLUT1 so far by the substituted cysteine accessibility method (32) by Mueckler and Makepeace (15) with the solvent-accessible surface of both LacY and GlpT calculated by InsightII (Accelrys, San Diego, CA). The long cytoplasmic loop between TM6 and TM7 and the loop between TM1 and TM2 were not modeled.
RESULTS
Distribution of GLUT1 Mutations—Of 100 GLUT1-deficient patients identified by clinical and analytical criteria, 91 carried a mutation in one SLC2A1 allele. One patient, reported previously (33), was a compound heterozygote and was clinically severely affected. The remainder eight patients did not exhibit sequence variants associated with abnormal polypeptide encoding or other obvious pathogenic mechanisms and thus may carry undetected mutations in either the promotor region or intron I of SLC2A1 (neither was sequenced) or in another unidentified gene that can also cause GLUT1 deficiency.
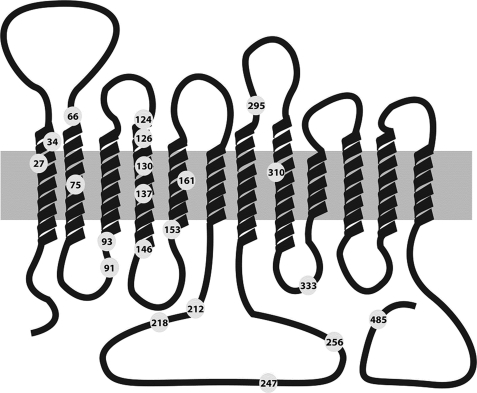
A total of 21 independent amino acid loci were susceptible to missense mutation (Fig. 1 and Table 1). Several patients shared missense mutations involving one of six individual residues: three patients at Asn-34, three at Gly-91 (reported in Ref. 34), three at Arg-93, nine at Arg-126, three at Arg-153, and four at Arg-333. The most frequent substitution was R126H, a heritable mutation (25) found in five patients (35). The rest of the mutations in SLC2A1 included a variety of sequence variations that followed no discernible pattern when mapped on the gene structure (not shown).
FIGURE 1.
GLUT1 topological model. GLUT1 is represented in the context of the plasma membrane (gray rectangle) and comprising cytoplasmic amino- (left side) and carboxyl-terminal (right side) domains. The extracellular side of the membrane is situated on the top part of the schematic. The relative length of TMs follows the crystal structure of LacY, and the length of the extramembranous amino and carboxyl termini and interhelical loops is also drawn approximately to scale. Numbered circles indicating residue number identify the location of missense mutations.
TABLE 1.
Genetic and kinetic characteristics of missense GLUT1 mutations NR, not reported; ND, not determined. Normal Km = 1.6 ± 0.2 mm. Normal Vmax values varied across groups of samples but were consistently similar (within 10%) in samples subjected to the same processing (see “Results”), ranging from 907 to 2417 fmol × 106 RBC × s–1.
| Mutation | Number of patients | SLC2A1 exon | Km | Vmax |
|---|---|---|---|---|
| mm | % of normal | |||
| G27V | 1 | II | 0.9 | 60 |
| N34I | 1 | II | 1.6 | 49 |
| N34K | 1 | II | 1.6 | 71 |
| N34Sa | 2 | II | 1.6 | 55 |
| S66F | 1 | III | 0.7 | 45 |
| G75Wa | 1 | III | NR | NR |
| G91Db | 3 | IV | ND | ND |
| R93Q | 2 | IV | 1.4 ± 0.1 | 67 ± 5 |
| R93W | 1 | IV | 2.7 | 65 |
| L124R | 1 | IV | 2.0 | 84 |
| R126Lc | 1 | IV | 1.4 | 43 |
| R126H | 5 | IV | 1.6 ± 0.3 | 74 ± 12 |
| R126C | 3 | IV | 1.5 ± 0.3 | 43 ± 4 |
| G130S | 1 | IV | 1.6 | 75 |
| T137I | 1 | IV | 2.2 | 53 |
| E146K | 1 | IV | 1.8 | 50 |
| R153L | 2 | IV | 0.9 ± 0.1 | 50 ± 0 |
| R153C | 1 | IV | 0.9 | 52 |
| Q161H | 1 | IV | ND | ND |
| R212Ha | 1 | V | NR | NR |
| R218Ha | 1 | V | NR | NR |
| E247D | 1 | VI | 2.2 | 47 |
| K256Vc | 1 | VI | 1.4 | 43 |
| T295M | 1 | VII | ND | ND |
| T310I | 1 | VII | 2.2 | 53 |
| R333Wd | 4 | VIII | 1.7 ± 0.1 | 44 ± 13 |
| P485L | 1 | X | 1.8 | 55 |
Indicates one patient reported by Klepper et al. (66) (Km and Vmax not included in the report)
Indicates three patients reported by Klepper et al. (34) (Km and Vmax not included in the report)
Mutants occurring simultaneously in both alleles of one patient (33)
Data obtained only from two patients
Of note, TM4 included mutations at residues 124, 126, 130, 137, and 146. Interestingly Pro-141, located a third of the way from the extracellular end of TM4 and thus located near its cytoplasmic end (Table 2), can be predicted to break the secondary structure of the segment. Mutation of Arg-333 disrupts the RXGRR signature of MFS transporters located in the loop between TM8 and TM9 (21). Three patients exhibited mutations at Arg-93, which flanks the cytoplasmic end of TM3 and is also part of the TM2-TM3 RXGRR MFS signature (36). Another three patients carried mutations at Arg-153, a residue predicted to lie at an equivalent, membrane-flanking location in TM5. Other charge-substituting variants involved Arg-212 at the carboxyl end of TM6, Arg-218, Glu-247, and Lys-256 (Fig. 1 and Table 1). In summary, of the 21 mutant loci, 13 involved charged amino acids.
TABLE 2.
TM4 of GLUT1 and related MFS transporters Membrane helices 4 of GLUT1 (G1), LacY (LY), and GlpT (GT) were manually aligned. Conserved proline residues are bold. Outside and inside delimit the extracellular and intracellular boundaries of TM4, respectively.
| G1 | LY | GT |
|---|---|---|
| Outside | ||
| 122 | 105 | 121 |
| Val | Ala | |
| Gly | Val | |
| Leu | Ser | Met |
| Ile | Ile | Phe |
| Leua | Val | Val |
| Gly | Gly | Leu |
| Arga | Glyb | Leu |
| Phe | Ile | Phe |
| Ile | Tyr | Leu |
| Ile | Leu | Cys |
| Glya | Gly | Gly |
| Val | Phe | Trp |
| Tyr | Cys | Phe |
| Cys | Phe | Gln |
| Gly | Asn | Gly |
| Leu | Ala | Met |
| Thr | Gly | Gly |
| Thra | Alab | Trp |
| Gly | Pro | Pro |
| Phe | Ala | Pro |
| Val | Val | Cys |
| Pro | Glub | Gly |
| Met | Ala | Arg |
| Tyr | Phe | Thr |
| Val | Ile | Met |
| Gly | Glu | Val |
| Glua | Lys | His |
| Val | Val | |
| Ser | Ser | |
| Pro | Arg | |
| 149 | 134 | 148 |
| Inside |
Missense mutant residues in GLUT1 (124, 126, 130, 137, and 146)
Important structural features in LacY (Gly-111 and Pro-123 as helix breakers and Glu-126, a residue essential for transport (3))
Function of GLUT1 Mutants in RBC—RBC clinical indexes (morphology, hemoglobin contents, and volume) were normal in all patients. Nevertheless GLUT1 constitutes at least 5% of the RBC plasma membrane protein (37) and can transport water (38, 39), which in turn may change cell volume and thus alter glucose flux during the transport assay. Therefore, the potential contribution of water permeability to the transport assay was assessed to ensure that RBC volume and hence glucose distribution volume in RBC were preserved during the assay. Hematological osmotic fragility assays (40) were performed in normal RBC and select mutants to compare water permeability and membrane integrity. The fragility assays included three GLUT1 mutants that caused profound decreases in RBC glucose uptake due to truncation of the transporter, a mutation type associated with haploinsufficiency (41). A normal osmotic fragility was observed for all of the mutants, indicating that any potential changes in RBC membrane structure and cell volume resulting from impaired GLUT1 production or function did not significantly influence the transport assay of GLUT1 mutants.
All GLUT1 mutants exhibited diminished RBC glucose transport (Fig. 2 and Table 1). Changes in both Vmax (reductions to 43–84% of normal Vmax) and Km (modest increases or decreases by less than 2-fold) were observed. The Vmax changes were variable and did not always amount to the 50% reduction expected of mutants that behaved as pure hemizygous traits, probably indicating residual transport by these mutants (see below). The observed changes in Km, on the other hand, were inconsistent with the notions of the mutations simply diminishing the population of carriers and of the carriers working independently of one another. Instead the measured Km probably reflects a more complex transport kinetics influenced by factors such as subunit cooperativity arising from transporter polymerization (42), a phenomenon not taken into consideration in our analysis. An alternative possibility that could also explain the relatively high (greater than 50% of normal) residual Vmax observed in some mutants is that the normal SLC2A1 allele compensated for the loss of GLUT1 function by increasing transcription or translation. In this context, our transport assays would overestimate flux through mutant transporters. Still in this setting, the results would also imply that the magnitude of such compensation would be mutation-dependent. This possibility, however, is unlikely because Western blot of 26 RBC samples harboring a variety of mutation types failed to demonstrate any up-regulation of GLUT1 contents over normal RBC levels (not shown). These experiments included mutants devoid of transport capacity but capable of targeting to the plasma membrane based on oocyte studies (26). Additionally allele hemizygosity, the most severe mutation, also failed to induce an increase in the GLUT1 RBC protein produced by the only residual (normal) allele (see below), arguing against any significant compensation by the normal allele.
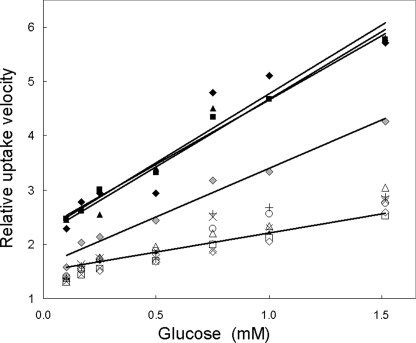
FIGURE 2.
Glucose influx into human RBC. Zero-trans velocity of influx of glucose into normal and patient RBCs was normalized to normal (control) maximum velocity (Vmax) obtained from Lineweaver-Burke plots of uptake and expressed as relative uptake velocity as a function of extracellular glucose concentration. The lower set of symbols (open symbols and crosses) represents uptake measured in seven normal samples. The fitted line represents a fitted Lineweaver-Burke relationship using a larger set of normal samples (n = 30). Filled symbols reflect uptake by mutant RBCs associated with complete loss of function of one allele (▪, deletion of nucleotides 616–617 causing a frameshift; ▴, R330X, a truncating mutation; ♦, R126C, a missense mutant). Three fitted lines (upper set) indicate Lineweaver-Burke relationships for these mutants. Gray diamonds (♦) indicate transport by G130S; data are fit by a similar linear relationship.
TM4 mutants at residues 124, 126, and 130 transported glucose with a Vmax intermediate between normal RBC and one-half of normal (the uptake expected of functionally hemizygous mutations), suggesting that they express functional transporters (Fig. 2). These sequence variants constitute the most informative set among all GLUT1 mutants because they probably fold and are targeted correctly, exposing residues potentially critical for transport. R126C was associated with a diminished Vmax that approached hemizygous magnitude. RBC Western blot, however, demonstrated no significant change in R126C protein expression (100 ± 1% of normal, n = 2) in contrast with RBCs from two hemizygous patients who carried large scale deletions that comprised the entire SLC2A1 allele and whose protein expression levels were 54 and 56%. Similarly mutation R126H, at the same locus, transported inefficiently but expressed 91 ± 10% (n = 3) of protein relative to normal RBC. Mutation R126L was associated with the lowest measured Vmax (43%), but this mutation occurred in a compound heterozygote patient who also carried K256V in the trans allele.
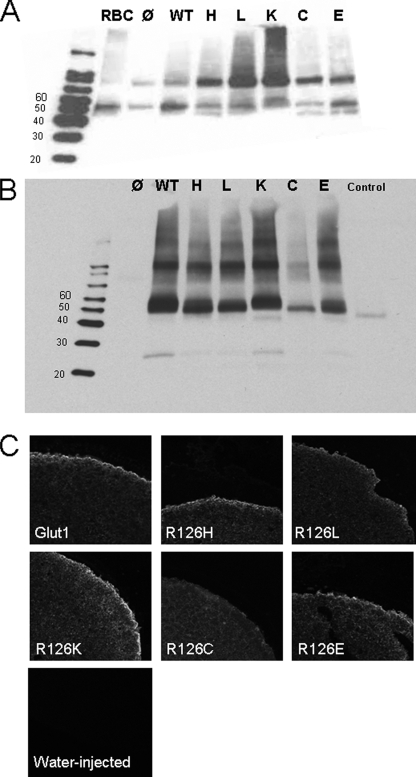
Expression of Arg-126 Mutants in Oocytes—Functional studies of Arg-126 in oocytes were of particular interest because (a) Arg-126 is the locus most often mutated in GLUT1 deficiency; (b) it bears a positive charge and was predicted to lie near the extracellular end of TM4 (11), a substrate-interacting region in LacY (3); and (c) substitution R126H, the most frequent mutation in GLUT1 deficiency, may be susceptible to protonation. Arg-126 transitions His, Lys, Glu, Leu, and Cys were associated (in decreasing order) with diminished plasma membrane expression by Western blot. A similar pattern was also detected by confocal microscopy (Fig. 3). In all cases, the amount of mutant GLUT1 detectable by immunoblot of oocyte plasma membrane-rich fractions was within 20% of wild type except for R126C, which was reduced by about 75%. Thus, at least in part, all mutants were targeted to the cell membrane. A fraction of each mutant protein was retained in vesicles or in the cytoplasm (Fig. 3).
FIGURE 3.
Expression and targeting of GLUT1 and Arg-126 mutants in oocytes. A, total normal human RBC membranes (RBC, leftmost lane) and oocyte membrane extracts after reaction with the same antibodies. Additional water-injected oocyte and purified commercial GLUT1 controls were added and labeled as in B. B, purified oocyte plasma membrane-enriched extract stained with GLUT1 antibody. Water-injected oocyte membranes (Ø, leftmost lane) and purified GLUT1 (Control, rightmost lane, 45–49 kDa, obtained from FabGennix, Inc.) were added in parallel as controls to GLUT1-injected (wild type (WT)) and mutant-injected oocyte membranes. C, confocal microscopy of oocyte sections using GLUT1 primary antibody. Plasma membrane targeting qualitatively similar to normal GLUT1 was observed for charged substitutions Lys (K), Glu (E), His (H), and, to a lesser extent, Leu (L), whereas mutant Cys (C) was associated with diminished membrane signal relative to cytoplasmic signal. Water-injected oocytes did not exhibit appreciable staining. For both A and B, molecular mass markers are labeled in kDa. The gel in A represents one of seven experiments, and B represents one of two independent experiments, all of which yielded similar results.
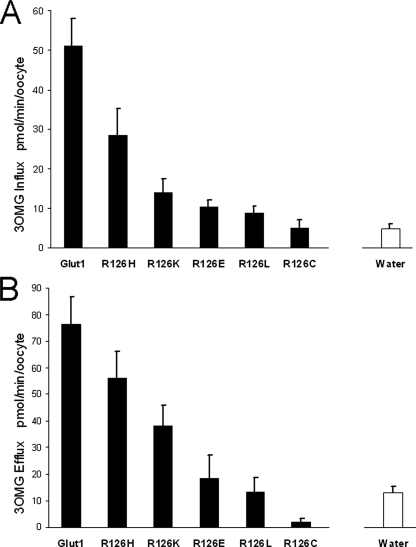
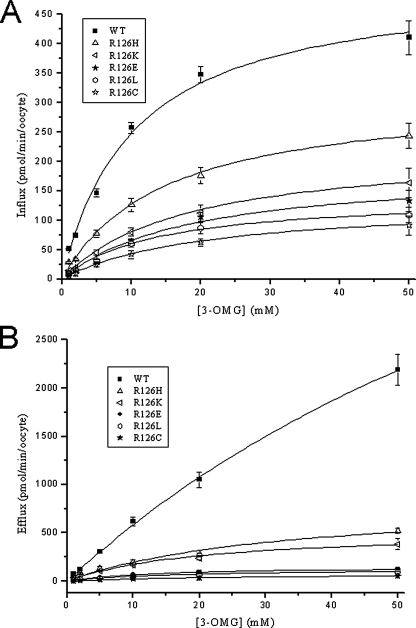
Fig. 4 illustrates zero-trans influx into and efflux from oocytes. Under these experimental conditions, efflux proceeded slowly (almost linearly) for wild type GLUT1 (Fig. 5). All of the mutations decreased efflux more than influx. Efflux was nearly abolished by most substitutions except His and Lys, which retained 15–20% of normal transport velocity. Nevertheless it was particularly difficult to quantitate efflux because of the potential contribution of intracellular vesicular transport revealed by confocal microscopy (Fig. 3). Simple calculations illustrate this limitation: if the ratio between total vesicular membrane surface and vesicular volume were small and the surface of the oocyte was large, efflux might be rate-limited by vesicular depletion of glucose. With a 0.5-mm radius, a typical oocyte surface of π mm2 can be estimated. However, capacitance measurements from oocytes by voltage clamp steps assuming a universal membrane capacitance of 1 microfarad/cm2 (43, 44) indicated that the plasma membrane surface is much larger than expected, typically measuring ∼18 mm2 (not shown). Thus, efflux kinetics from organelle-rich oocytes were only qualitatively interpretable.
FIGURE 4.
Transport capacity of GLUT1 and Arg-126 mutants expressed in oocytes. Each solid bar represents 3OMG flux through oocytes injected with GLUT1 or mutant transporters after subtraction of water-injected oocyte 3OMG flux. The open bar in each panel represents the actual 3OMG flux through water-injected oocytes used for subtraction. A, influx was measured in cells exposed to 1 mm extracellular 3OMG in near zero-trans conditions. B, efflux was determined in cells injected with concentrated 3OMG to achieve ∼1 mm intracellular 3OMG. Measurements were obtained from a least three independent batches of oocytes with n ≥ 15. Error bars represent S.D.
FIGURE 5.
Concentration dependence of 3OMG influx and efflux. A, concentration dependence of 3OMG influx into oocytes expressing GLUT1 and Arg-126 mutants. B, 3OMG efflux measured from oocytes injected with 3OMG. Transport isotherms were obtained by fitting a Michaelis-Menten equation to the efflux and influx data. Measurements were obtained from a least three independent batches of oocytes with n ≥ 15. Error bars represent S.D. WT, wild type.
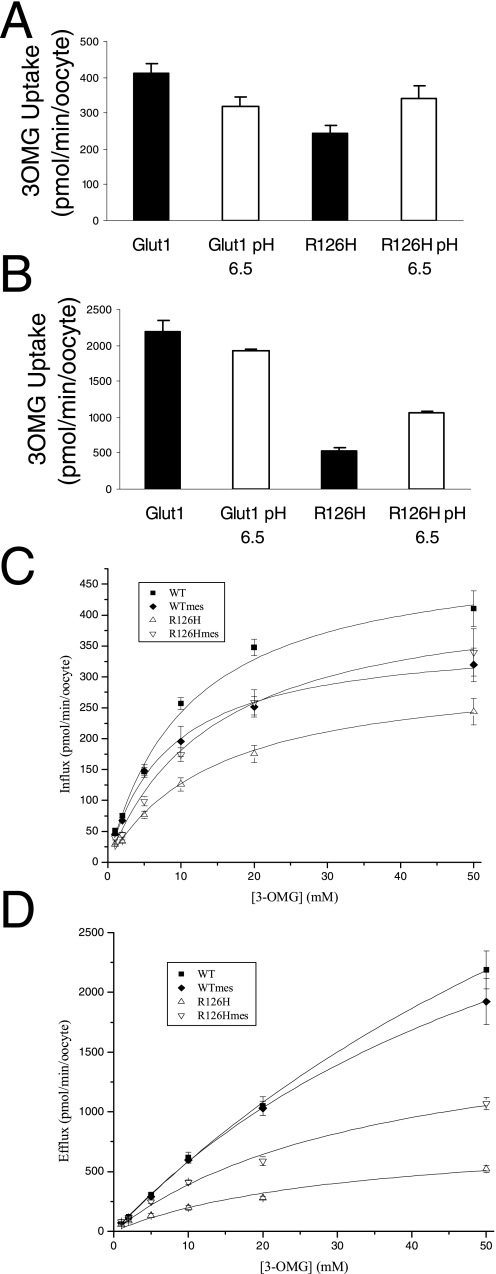
Modification of R126H—To probe the topology of R126H, a substitution that replaces an imidazole for the guanidinium group of the native arginine, the pH of the extracellular solution was decreased to 6.5. Although the pKa of histidine in solution is 6.0, it is not feasible to predict the pKa of R126H given its location in an unknown protein environment possibly subject to intrinsic electrostatic and membrane potentials (45). Additionally the predicted α-helical folding of TM4 would generate a helix dipole near Arg-126 that would decrease the pKa. Nevertheless we reasoned that if the presence of a positive charge were important for transport and if the R126H imidazole was titratable with a proton and this resulted in an enhanced glucose flux, Arg-126 would necessarily be exposed to and accessible from the extracellular solution. The ability of pH to modulate transport kinetics by protonation of titratable residues has been documented in disease states (46), as a tool to investigate membrane protein topology (47), and as a regulatory mechanism of physiological relevance (48).
Decreasing extracellular pH below 6.5 had deleterious effects on oocyte integrity and glucose transport. However, a pH of 6.5 exerted a modest inhibitory effect on GLUT1, whereas R126H transport function was augmented (Fig. 6), indicating that Arg-126 resides in a location accessible to protons and that, at this residue, charge rather than side chain size (as shown by the transport-defective R126L mutant) facilitates substrate translocation.
FIGURE 6.
Effect of pH 6.5 on 3OMG transport through GLUT1 and R126H. A and B, efflux and influx were assayed as in Fig. 4. C and D, concentration dependence of influx and efflux, respectively. Error bars represent S.D. WT, wild type; mes, methanesulfonic acid.
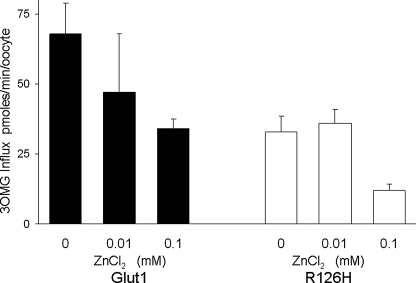
To further probe accessibility, the effect of ZnCl2 was tested on R126H (Fig. 7). At both 10 and 100 μm, ZnCl2 caused a concentration-related block of both wild type and R126H of similar magnitude, probably indicating that the R126H mutation does not create an accessible metal binding site (Fig. 7). The lack of a significant effect of ZnCl2 on R126H despite the effects of protons at this site suggests either that Arg-126 is not exposed into a large, water-filled space, such as a vestibule at the extracellular mouth of GLUT1, and/or that a single His residue is insufficient for the coordination of the metal ion due to the absence of a neighboring binding partner.
FIGURE 7.
Inhibition of 3OMG transport by ZnCl2 in GLUT1- (filled bars) and R126H (open bars)-expressing oocytes. Cells were preincubated in freshly dissolved ZnCl2 for 10 min prior to the assay and maintained in the same blocker concentration in the presence of 50 mm glucose during the assay. The efficacy of ZnCl2 as a blocker of GLUT1 and R126H was not significantly different (t test p > 0.1) under 10 and 100 μm ZnCl2. n ≥ 4 oocytes per assay. Error bars represent S.D.
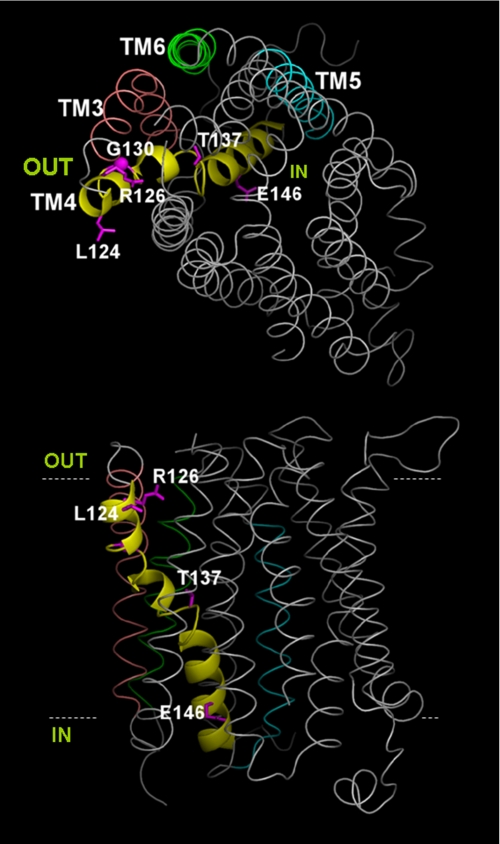
TM4 in the Context of the GLUT1 Model—The GLUT1 model illustrates the topology of helices relative to one another and to the plasma membrane (Fig. 8). As established by the crystal structures of LacY and GlpT, the transporting (catalytic) core of GLUT1 is formed by a set of the 12 transmembrane helices. It is only within this transmembrane core that some homology exists among the MFS proteins. In the GLUT1 model, helical length, curvature, kinks, and tilt angle vary considerably across the 12 TMs, thus decreasing the number of potential helix-to-helix contacts and preventing dense packing. Although loose packing may render the molecule more prone to large conformational changes such as those proposed for LacY, our model offers too many degrees of conformational freedom to be useful for dynamic predictions even accounting for the relatively short loops between TM1 through TM6 and between TM7 through TM12. On the other hand, crystallization of stabilized transporter in preferred conformations may make not large but rare movements apparent. Therefore, our model (comparable in this regard to the crystal structure) offers only a framework that limits some potential interactions and suggests others on the basis of TM packing and connecting loop length.
FIGURE 8.
GLUT1 model. TM helices are represented in gray except for TM4, indicated by a yellow ribbon, and select helices, labeled and shown in color. The side chains of residues subject to missense mutation are depicted in pink and numbered. Top panel, view normal to the membrane from the extracellular aspect of the transporter. Bottom panel, view parallel to the plane of the membrane. Dotted lines represent the putative boundaries of the plasma membrane and have been arbitrarily placed. IN and OUT denote the extracellular and intracellular spaces, respectively.
In this context, TM4 was modeled as a kinked helix (at residues Gly-130 and Pro-141) situated at the catalytic core and in close apposition to TM1 and TM5. It was not possible to identify a potential pairing charge in the vicinity of Arg-126, suggesting that the residue may increase the electrostatic potential in the vicinity of the transporter and probably interacts with a water shell.
DISCUSSION
We undertook the identification of residues essential for GLUT1 function by analyzing human SLC2A1 missense mutations that cause GLUT1 deficiency. The rest of the (non-missense) mutations identified in SLC2A1 included a variety of sequence variations that followed no discernible pattern when mapped on the gene structure. These mutations may cause GLUT1 haploinsufficiency through a variety of plausible mechanisms: (a) hemizygosity resulting from large scale deletion of an entire allele in chromosome 1 (17), (b) primary or secondary stop codon insertion (the latter arising from frameshift, insertion, or deletion mutations) resulting in polypeptide truncation and obligatory complete loss of function (23), (c) aberrant targeting of a normally folded protein, (d) inefficient catalysis due to active site mutation or conformational restrictions (26), and (e) inadequate RNA processing due to splice site mutations (33). In contrast, missense mutations highlight residues potentially important for folding, processing, subunit interactions, and catalysis. Of these, residues involved in establishing protein interactions and/or transport may be inferred from the fraction of mutants that retain glucose transport capacity in RBC. Structure-function studies and comparative topological models based on proteins of known structure can then be used to discern the subset of amino acids involved in catalysis.
A modest degree of homology and a similar function indicate that GLUT1 must fold like other MFS proteins (2, 21). Three MFS crystal structures are available (3–5), providing a framework for the interpretation of GLUT1 mutational analyses. Although these structures represent one (the predominant) state of the stabilized transporter, substantial evidence from work on LacY and GLUT1 has indicated that both large scale conformational changes and small atomic exchanges occur incessantly (49, 50), many of which affect transport and would have been difficult to predict from the crystal structure alone. On the other hand, the functional analysis of site-directed mutants often is not directly interpretable in structural terms except in three special instances: (a) when a reactive site is introduced and then perturbed to yield information about the local environment (51), (b) when antibody epitopes are inserted in the protein to probe their location relative to the membrane (12), and (c) when transport is disrupted by mutation or chemical modification of residues that are located at short periodic intervals in the peptide chain, allowing for the inference of ordered or disordered secondary structures (32, 52). Yet in these experimental approaches, as in others, functional inferences are subordinate to (or at least necessitate) complementary modeling of the rest of the molecule, a subject of intense, ongoing efforts in the case of GLUT1 (53–58).
Several GLUT1 mutations revealed a pattern consistent with α-helical folding: residues 124, 126, 130, 13,7 and 146, all located in TM4, can be modeled as lining one side of a helix. In LacY, TM4 constitutes one of the longest helices, extending into the cytoplasm as a kinked, tilted rod. Kinks occur at LacY positions Gly-111 and Pro-123, demarcating three approximately equal length segments in TM4. In the ligand-free LacY, Glu-126 is part of a helix-to-helix charge pair advanced into the vicinity of the pore by the kinked configuration of TM4 imposed by Pro-123 on the cytoplasmic third segment of the helix. LacY Glu-126 dissociates from its paired state in the occupied permease to allow interaction with the substrate via water molecules. The rest of the membrane-spanning part of the helix not facing the substrate translocation pathway is embedded in the protein core in close proximity to TMs 1, 2, and 3 (3). In GlpT, a glycerol 3-phosphate and phosphate exchanger, Arg-45 and Arg-269, located in TM1 and TM7, respectively, offer unpaired charges that may also participate in phosphate binding as the substrate induces a conformational change. GLUT1 Arg-126, the residue most frequently mutated in GLUT1 deficiency, is the only charge in TM4 (Table 2). Our GLUT1 model (Fig. 8) did not identify any potential charge-pairing, acidic residues in the vicinity of Arg-126, suggesting that Arg-126 may exist in free ionized form rather than neutralized by a neighboring charge or embedded in the protein core.
To test the latter possibility, we expressed and probed access to substitution R126H. The results supported the hypothesis: first, the histidine replacement affected glucose influx and efflux but was compatible with the expression of functional transporters (as were other substitutions at this locus; Fig. 4 and Table 1), indicating that an arginine at 126 is not required for function, and second, the residue is accessible to protons (and possibly not to zinc) from the extracellular solution. Thus, Arg-126 may be positioned at a location near the opening of the transporter from which the branched guanidinium group is capable of offering two hydrogen bonds to substrate oxygen atoms and of contributing to the intrinsic electrostatic potential at the extracellular aspect of the transporter. The rest of the transmembrane core of GLUT1 is devoid of charged residues, except for His-337 in TM9, a peripheral helix in both LacY and GlpT, which is therefore unlikely to contribute to substrate binding directly, and for His-160 in TM5 and Glu-380 in TM10. Although it is not known whether GLUT1 His-160 is ionized, the latter two residues are located at catalytically important regions of LacY, suggesting that they may participate, together with Arg-126, in glucose binding to GLUT1. In particular, LacY Arg-144, located in TM5, also interacts with substrate via hydrogen bonding (3). GLUT1 H160C is not accessible to extracellular p-chloromercuribenzenesulfonate, as no other residue in the cytoplasmic half of TM5 is (perhaps because of hindered penetration of the reagent), but mutation to cysteine is associated with reduced transport capacity (59), whereas Glu-380 reacts with p-chloromercuribenzenesulfonate under similar conditions (60). These results are compatible with the potential role of these residues in substrate binding. No cysteine-substituted mutant in the GLUT1 TM4 region spanning residues 127–148 reacted appreciably with p-chloromercuribenzenesulfonate (15), possibly indicating tight packing of the helix below position 127. However, mutation of residues 130, 134, 143, and 146, located on the side of the helix identified as functionally important (see “Results”), abolished transporter function, expanding the inference made from the location of the human mutations. Residues 145 and 148, situated near the carboxyl end of TM4, can be bonded with TM8 residues by membrane-permeant cross-linkers, consistent with a cytoplasm-immersed end of a helical TM4 that offers another side of the helix to the vicinity of TM8 (61).
The majority of the remaining GLUT1 mutations involved TM-associated glycines and charged intracellular residues that were previously recognized as structural signatures on the basis of sequence conservation among many MFS transporters and other related membrane carriers (62). Some of these signature residues have been subjected to mutational analysis, and thus their function is known. TM glycines can induce helical breaks and are also a conserved feature of numerous membrane helices (62). In LacY, TM4 Gly-111 causes a break in secondary structure as do Gly-150 and Gly-268 in other substrate-interacting TMs, allowing the direct exposure of many of the residues directly involved in catalysis to the vicinity of the substrate (3). Gly-27 (TM1), Gly-75 (TM2), and Gly-91 (TM2–3 loop) are transporter signatures mutated in GLUT1 deficiency and thus may be essential for substrate docking with the recognition site even if they do not contact the glucose molecule.
The last category of mutations involved charged residues flanking membrane domains, which generally serve to anchor TMs to the membrane particularly when membrane helices are curved (4). Among these, Arg-93 (at the cytoplasmic end of TM2 or TM2–3 loop), Arg-153 (at the intracellular end of TM5 or TM4–5 loop), and Arg-333 (TM8–TM9 loop) are also conserved across MFS transporters. GLUT4 Arg-153, aligned with GLUT1 Arg-153, diminishes transport capacity and eliminates cytochalasin B binding to its endofacial aspect while reducing recognition of 2-N4-(1-azi-2,2,2-trifluoroethyl)benzoyl-1,3-bis(d-mannosyloxy)-2-propylamine at its exofacial site to a lesser degree (63). These effects are consistent with an altered transporter structure incapable of undergoing normal conformational changes. Arg-93 and Arg-333 are part of RXGRR loop motifs situated at homologous positions in both halves of the transporter (TM1–TM6 and TM7–TM12) and probably reflect evolutionary gene duplication (22). Charge reductions in any of the two RXGRR loops lead to the production of misinserted transporters devoid of activity (36). In GLUT4, mutation of Arg-92 in the first loop motif reduces function but does not impair cytochalasin B binding, whereas combined substitution of R333L and R334A also reduces transport without affecting cytochalasin B or 2-N4-(1-azi-2,2,2-trifluoroethyl)benzoyl-1,3-bis(d-mannosyloxy)-2-propylamine binding, indicating that, in addition to their role in folding, the charged loops participate in the conformational changes necessary for substrate translocation (63). Of note, Arg-333 also participates in the regulation of GLUT1 kinetics by ATP (64), a phenomenon of probable physiological and pathological relevance under conditions of impaired oxidative phosphorylation (65).
Based on the frequency of GLUT1 polymorphisms observed in normal individuals,3 we conclude that many of the transporter residues are probably replaceable without noticeable functional consequences. Only a set of conserved amino acids is essential. These include glycine, capable of inducing helical breaks, and charged residues mainly involved in the stabilization of membrane helices required for conformational changes. The results, however, do not imply that these or other pathogenic mutations (in TM4 and elsewhere) alter substrate flow through the pore directly, a feature incorporated in some structural models (57, 58). Rather in light of the available MFS structures, they identify a membrane helix (TM4) ideally situated as a transducer of substrate binding. We conclude that TM4 may present a surface of interaction necessary for the intramolecular rearrangement of this and other helices as the substrate is translocated.
Acknowledgments
We are grateful to Arthur Karlin and James Stull for critically reviewing parts of the manuscript, to Philip Thomas for advice on deglycosylation, and to Sujit Sheth for supervising the osmotic fragility assays.
This work was supported, in whole or in part, by National Institutes of Health Grants NS037949 and NS001698 (to J. M. P., D. W., and D. C. D.). This work was also supported by the Colleen Giblin Foundation and the Will Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MFS, multiple facilitator superfamily; TM, transmembrane domain; TC, transporter classification; RBC, red blood cell; 3OMG, 3-O-methyl-d-glucose.
J. M. Pascual, D. Wang, H. Yang, and D. C. De Vivo, personal observations.
References
- 1.Abramson, J., Kaback, H. R., and Iwata, S. (2004) Curr. Opin. Struct. Biol. 14 413-419 [DOI] [PubMed] [Google Scholar]
- 2.Vardy, E., Arkin, I. T., Gottschalk, K. E., Kaback, H. R., and Schuldiner, S. (2004) Protein Sci. 13 1832-1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H. R., and Iwata, S. (2003) Science 301 610-615 [DOI] [PubMed] [Google Scholar]
- 4.Huang, Y., Lemieux, M. J., Song, J., Auer, M., and Wang, D. N. (2003) Science 301 616-620 [DOI] [PubMed] [Google Scholar]
- 5.Hirai, T., Heymann, J. A., Shi, D., Sarker, R., Maloney, P. C., and Subramaniam, S. (2002) Nat. Struct. Biol. 9 597-600 [DOI] [PubMed] [Google Scholar]
- 6.Busch, W., and Saier, M. H., Jr. (2002) CRC Crit. Rev. Biochem. Mol. Biol. 37 287-337 [DOI] [PubMed] [Google Scholar]
- 7.Saier, M. H., Jr., Tran, C. V., and Barabote, R. D. (2006) Nucleic Acids Res. 34 D181-D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uldry, M., and Thorens, B. (2004) Pfluegers Arch. Eur. J. Physiol. 447 480-489 [DOI] [PubMed] [Google Scholar]
- 9.Hebert, D. N., and Carruthers, A. (1992) J. Biol. Chem. 267 23829-23838 [PubMed] [Google Scholar]
- 10.Vannucci, S. J., Maher, F., and Simpson, I. A. (1997) Glia 21 2-21 [DOI] [PubMed] [Google Scholar]
- 11.Mueckler, M., Caruso, C., Baldwin, S. A., Panico, M., Blench, I., Morris, H. R., Allard, W. J., Lienhard, G. E., and Lodish, H. F. (1985) Science 229 941-945 [DOI] [PubMed] [Google Scholar]
- 12.Hresko, R. C., Kruse, M., Strube, M., and Mueckler, M. (1994) J. Biol. Chem. 269 20482-20488 [PubMed] [Google Scholar]
- 13.Chin, J. J., Jung, E. K., Chen, V., and Jung, C. Y. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 4113-4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin, J. J., Jung, E. K., and Jung, C. Y. (1986) J. Biol. Chem. 261 7101-7104 [PubMed] [Google Scholar]
- 15.Mueckler, M., and Makepeace, C. (2005) J. Biol. Chem. 280 39562-39568 [DOI] [PubMed] [Google Scholar]
- 16.Wang, D., Pascual, J. M., Yang, H., Engelstad, K., Mao, X., Cheng, J., Yoo, J., Noebels, J. L., and De Vivo, D. C. (2006) Hum. Mol. Genet. 15 1169-1179 [DOI] [PubMed] [Google Scholar]
- 17.Seidner, G., Alvarez, M. G., Yeh, J. I., O'Driscoll, K. R., Klepper, J., Stump, T. S., Wang, D., Spinner, N. B., Birnbaum, M. J., and De Vivo, D. C. (1998) Nat. Genet. 18 188-191 [DOI] [PubMed] [Google Scholar]
- 18.Pascual, J. M., Wang, D., Hinton, V., Engelstad, K., Saxena, C. M., Van Heertum, R. L., and De Vivo, D. C. (2007) Arch. Neurol. 64 507-513 [DOI] [PubMed] [Google Scholar]
- 19.Pascual, J. M., Wang, D., Lecumberri, B., Yang, H., Mao, X., Yang, R., and De Vivo, D. C. (2004) Eur. J. Endocrinol. 150 627-633 [DOI] [PubMed] [Google Scholar]
- 20.Pascual, J. M., Van Heertum, R. L., Wang, D., Engelstad, K., and De Vivo, D. C. (2002) Ann. Neurol. 52 458-464 [DOI] [PubMed] [Google Scholar]
- 21.Pao, S. S., Paulsen, I. T., and Saier, M. H., Jr. (1998) Microbiol. Mol. Biol. Rev. 62 1-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiden, M. C., Davis, E. O., Baldwin, S. A., Moore, D. C., and Henderson, P. J. (1987) Nature 325 641-643 [DOI] [PubMed] [Google Scholar]
- 23.Wang, D., Pascual, J. M., Yang, H., Engelstad, K., Jhung, S., Sun, R. P., and De Vivo, D. C. (2005) Ann. Neurol. 57 111-118 [DOI] [PubMed] [Google Scholar]
- 24.Klepper, J., Garcia-Alvarez, M., O'Driscoll, K. R., Parides, M. K., Wang, D., Ho, Y. Y., and De Vivo, D. C. (1999) J. Clin. Lab. Anal. 13 116-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brockmann, K., Wang, D., Korenke, C. G., von Moers, A., Ho, Y. Y., Pascual, J. M., Kuang, K., Yang, H., Ma, L., Kranz-Eble, P., Fischbarg, J., Hanefeld, F., and De Vivo, D. C. (2001) Ann. Neurol. 50 476-485 [DOI] [PubMed] [Google Scholar]
- 26.Wang, D., Pascual, J. M., Iserovich, P., Yang, H., Ma, L., Kuang, K., Zuniga, F. A., Sun, R. P., Swaroop, K. M., Fischbarg, J., and De Vivo, D. C. (2003) J. Biol. Chem. 278 49015-49021 [DOI] [PubMed] [Google Scholar]
- 27.Finidori-Lepicard, J., Schorderet-Slatkine, S., Hanoune, J., and Baulieu, E. E. (1981) Nature 292 255-257 [DOI] [PubMed] [Google Scholar]
- 28.Finidori, J., Hanoune, J., and Baulieu, E. E. (1982) Mol. Cell. Endocrinol. 28 211-227 [DOI] [PubMed] [Google Scholar]
- 29.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779-815 [DOI] [PubMed] [Google Scholar]
- 30.Shi, J., Blundell, T. L., and Mizuguchi, K. (2001) J. Mol. Biol. 310 243-257 [DOI] [PubMed] [Google Scholar]
- 31.Gerstein, M., and Levitt, M. (1998) Protein Sci. 7 445-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlin, A., and Akabas, M. H. (1998) Methods Enzymol. 293 123-145 [DOI] [PubMed] [Google Scholar]
- 33.Wang, D., Pascual, J. M., Ho, Y. Y., Engelstad, K., Jhung, S., Miller, C., Kranz-Eble, P., and De Vivo, D. C. (2001) Ann. Neurol. 50 S125 (Abstr. P89) [Google Scholar]
- 34.Klepper, J., Monden, I., Guertsen, E., Voit, T., Willemsen, M., and Keller, K. (2001) FEBS Lett. 498 104-109 [DOI] [PubMed] [Google Scholar]
- 35.Wang, D., Kranz-Eble, P., and De Vivo, D. C. (2000) Hum. Mutat. 16 224-231 [DOI] [PubMed] [Google Scholar]
- 36.Sato, M., and Mueckler, M. (1999) J. Biol. Chem. 274 24721-24725 [DOI] [PubMed] [Google Scholar]
- 37.Sogin, D. C., and Hinkle, P. C. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 5725-5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeidel, M. L., Albalak, A., Grossman, E., and Carruthers, A. (1992) Biochemistry 31 589-596 [DOI] [PubMed] [Google Scholar]
- 39.Iserovich, P., Wang, D., Ma, L., Yang, H., Zuniga, F. A., Pascual, J. M., Kuang, K., De Vivo, D. C., and Fischbarg, J. (2002) J. Biol. Chem. 277 30991-30997 [DOI] [PubMed] [Google Scholar]
- 40.Godal, H. C., Elde, A. T., Nyborg, N., and Brosstad, F. (1980) Scand. J. Haematol. 25 107-112 [DOI] [PubMed] [Google Scholar]
- 41.Oka, Y., Asano, T., Shibasaki, Y., Lin, J. L., Tsukuda, K., Katagiri, H., Akanuma, Y., and Takaku, F. (1990) Nature 345 550-553 [DOI] [PubMed] [Google Scholar]
- 42.Coderre, P. E., Cloherty, E. K., Zottola, R. J., and Carruthers, A. (1995) Biochemistry 34 9762-9773 [DOI] [PubMed] [Google Scholar]
- 43.Curtis, H. J., and Cole, K. S. (1938) J. Gen. Physiol. 21 1980-1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumgartner, W., Islas, L., and Sigworth, F. J. (1999) Biophys. J. 77 1980-1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pascual, J. M., and Karlin, A. (1998) J. Gen. Physiol. 111 717-739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulte, U., Hahn, H., Konrad, M., Jeck, N., Derst, C., Wild, K., Weidemann, S., Ruppersberg, J. P., Fakler, B., and Ludwig, J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 15298-15303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starace, D. M., and Bezanilla, F. (2001) J. Gen. Physiol. 117 469-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkins, M. E., Hosie, A. M., and Smart, T. G. (2002) J. Neurosci. 22 5328-5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan, L., and Kaback, H. R. (2006) Annu. Rev. Biophys. Biomol. Struct. 35 67-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung, E. K., Chin, J. J., and Jung, C. Y. (1986) J. Biol. Chem. 261 9155-9160 [PubMed] [Google Scholar]
- 51.Voss, J., Wu, J., Hubbell, W. L., Jacques, V., Meares, C. F., and Kaback, H. R. (2001) Biochemistry 40 3184-3188 [DOI] [PubMed] [Google Scholar]
- 52.Pascual, J. M., Shieh, C. C., Kirsch, G. E., and Brown, A. M. (1995) Neuron 14 1055-1063 [DOI] [PubMed] [Google Scholar]
- 53.Fischbarg, J., Cheung, M., Czegledy, F., Li, J., Iserovich, P., Kuang, K., Hubbard, J., Garner, M., Rosen, O. M., Golde, D. W., and Vera, J. C. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 11658-11662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng, H., Parthasarathy, R., Rampal, A. L., and Jung, C. Y. (1996) Biophys. J. 70 14-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwyer, D. S. (2001) Proteins 42 531-541 [PubMed] [Google Scholar]
- 56.Zuniga, F. A., Shi, G., Haller, J. F., Rubashkin, A., Flynn, D. R., Iserovich, P., and Fischbarg, J. (2001) J. Biol. Chem. 276 44970-44975 [DOI] [PubMed] [Google Scholar]
- 57.Salas-Burgos, A., Iserovich, P., Zuniga, F., Vera, J. C., and Fischbarg, J. (2004) Biophys. J. 87 2990-2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunningham, P., Afzal-Ahmed, I., and Naftalin, R. J. (2006) J. Biol. Chem. 281 5797-5803 [DOI] [PubMed] [Google Scholar]
- 59.Mueckler, M., and Makepeace, C. (1999) J. Biol. Chem. 274 10923-10926 [DOI] [PubMed] [Google Scholar]
- 60.Mueckler, M., and Makepeace, C. (2002) J. Biol. Chem. 277 3498-3503 [DOI] [PubMed] [Google Scholar]
- 61.Alisio, A., and Mueckler, M. (2004) J. Biol. Chem. 279 26540-26545 [DOI] [PubMed] [Google Scholar]
- 62.Joost, H. G., and Thorens, B. (2001) Mol. Membr. Biol. 18 247-256 [DOI] [PubMed] [Google Scholar]
- 63.Schurmann, A., Doege, H., Ohnimus, H., Monser, V., Buchs, A., and Joost, H. G. (1997) Biochemistry 36 12897-12902 [DOI] [PubMed] [Google Scholar]
- 64.Levine, K. B., Cloherty, E. K., Hamill, S., and Carruthers, A. (2002) Biochemistry 41 12629-12638 [DOI] [PubMed] [Google Scholar]
- 65.Shi, Y., Liu, H., Vanderburg, G., Samuel, S. J., Ismail-Beigi, F., and Jung, C. Y. (1995) J. Biol. Chem. 270 21772-21778 [DOI] [PubMed] [Google Scholar]
- 66.Klepper, J., Scheffer, H., Leiendecker, B., Gertsen, E., Binder, S., Leferink, M., Hertzberg, C., Nake, A., Voit, T., and Willemsen, M. A. (2005) Neuropediatrics 36 302-308 [DOI] [PubMed] [Google Scholar]