Abstract
Natural killer (NK) cells play a vital role in the detection and destruction of virally infected and tumor cells during innate immune responses. The highly polymorphic Ly49 family of NK receptors regulates NK cell function by sensing major histocompatibility complex class I (MHC-I) molecules on target cells. Despite the determination of two Ly49-MHC-I complex structures, the molecular features of Ly49 receptors that confer specificity for particular MHC-I alleles have not been identified. To understand the functional architecture of Ly49-binding sites, we determined the crystal structures of Ly49C and Ly49G and completed refinement of the Ly49C-H-2Kb complex. This information, combined with mutational analysis of Ly49A, permitted a structure-based classification of Ly49s that we used to dissect the binding site into three distinct regions, each having different roles in MHC recognition. One region, located at the center of the binding site, has a similar structure across the Ly49 family and mediates conserved interactions with MHC-I that contribute most to binding. However, the preference of individual Ly49s for particular MHC-I molecules is governed by two regions that flank the central region and are structurally more variable. One of the flanking regions divides Ly49s into those that recognize both H-2D and H-2K versus only H-2D ligands, whereas the other discriminates among H-2D or H-2K alleles. The modular design of Ly49-binding sites provides a framework for predicting the MHC-binding specificity of Ly49s that have not been characterized experimentally.
Natural killer (NK)3 cells are an essential component of innate immunity against tumors and virally infected cells (1-3). The cytolytic activity of NK cells is regulated by a delicate balance of activating and inhibitory signals mediated through distinct classes of receptors found on their surface. The dominant signal received by an NK cell through its interaction with normal levels of major histocompatibility complex class I (MHC-I) molecules on target cells is inhibitory. If the level of MHC-I is reduced through tumorigenic or infectious processes, this inhibitory signal is attenuated, and the NK cell is activated (1-3). In this way, cells with abnormal MHC-I expression become targets of NK lytic activity.
Several receptor families have been identified on primate and rodent NK cells that monitor MHC-I expression on surrounding cells (3-5). These include the killer immunoglobulin-like receptors, members of the Ly49 family (Ly49s), and the CD94-NKG2 family of receptors. Additionally, the activating receptor NKG2D recognizes MHC-like molecules, such as MICA, that are up-regulated in stressed tissues (6). The mouse Ly49 family includes at least 23 expressed or potential members (Ly49A-W), along with about 15 allelic variants (2, 7). These highly polymorphic receptors constitute the main MHC-monitoring molecules on rodent NK cells. Although most Ly49s inhibit NK cell-mediated cytolysis after binding to MHC-I ligands, some are activating. Ly49s recognize one or more H-2D or H-2K alleles, independently of the MHC-bound peptide (2, 3). Their binding properties range from the broad recognition of MHC-I molecules by Ly49C to the allelic specificity of Ly49A. Recently, crucial roles for Ly49 receptors in antiviral immunity have been discovered. In one case, the m157 gene product of mouse cytomegalovirus (MCMV) was shown to interact directly with an inhibitory Ly49 (Ly49I) in a susceptible mouse strain and with an activating Ly49 (Ly49H) in a resistant one (8, 9). In another case, the activating receptor Ly49P was found to confer resistance to MCMV by interacting with H-2D molecules on MCMV-infected cells (10).
Ly49 receptors belong to the C-type lectin-like family of proteins, which also includes CD94-NKG2 and NKG2D (4, 5). Ly49s are homodimeric type II glycoproteins, with each chain composed of a C-type lectin-like domain, termed the natural killer receptor domain (NKD), connected by a stalk of ∼70 residues to the transmembrane and cytoplasmic domains. Crystal structures have been reported for Ly49A bound to H-2Dd (11) and for Ly49C bound to H-2Kb (12). These structures showed that both Ly49s engage MHC-I at a broad cavity beneath the peptide-binding platform formed by the heavy chain α1α2 and α3 domains and β2-microglobulin (β2m). However, the complexes differ considerably in the detailed architecture of their interfaces, raising the question of whether the different contacts in the two structures result from differences in the Ly49 receptors or the MHC-I ligands. Also unknown is the extent to which the Ly49A-H-2Dd and Ly49C-H-2Kb complexes are representative of MHC-I recognition by other Ly49s, which currently number over 20. Therefore, does each Ly49 engage MHC-I through a mostly unique set of interactions, as do Ly49A and Ly49C, or can Ly49s be sorted into a few clearly defined groups within which most interactions with MHC-I are conserved? Finally, is it possible to identify spatially separate regions within the Ly49-binding site that confer specificity for particular H-2D or H-2K alleles, while maintaining varying degrees of MHC cross-reactivity?
To better understand MHC recognition by Ly49 receptors, we determined the crystal structures of Ly49C and Ly49G, completed refinement of the Ly49C-H-2Kb complex, and engineered variants of Ly49A with altered MHC specificity. This information permitted a structure-based classification of Ly49s that provides new insights into the molecular architecture of their binding sites, which we show can be divided into distinct structural elements that together account for the ability of Ly49s to bind multiple MHC-I ligands, yet discriminate between individual alleles.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The entire extracellular portion of Ly49C from mouse strain B6 (residues 67-262), comprising both NKD and stalk regions, was cloned into the expression vector pT7-7 (Novagen). The protein was expressed in Escherichia coli as inclusion bodies, solubilized in 6 m guanidine, and folded in vitro by dilution into 0.4 m arginine, 3 mm reduced glutathione, and 0.8 mm oxidized glutathione, as described (12). Folded Ly49C was purified by size exclusion with a Superdex 75 HR column (Amersham Biosciences), followed by anion exchange using a Mono S column (Amersham Biosciences). The NKD of Ly49G from mouse strain BALB/c (residues 134-262) was expressed and purified similarly.
Three mutants of Ly49A were generated by PCR to introduce variations in the L3 loop (residues 211-231) as follows: Ly49Amut1, Ly49Amut2, and Ly49Amut3, in which residues 211-231, 218-226, and 227-231, respectively, were exchanged with the corresponding residues of Ly49C. The extracellular regions of wild-type and mutant Ly49A receptors (residues 67-262) were expressed and purified in the same way as Ly49C. H-2Dd was folded in vitro in the presence of β2m and the synthetic peptide AGPARAAAL as described (13); H-2Kb was similarly assembled with β2m and the peptide SIINFEKL (12).
Crystallization and Data Collection—Crystals of free Ly49C were grown in hanging drops at 4 °C in 2.0 m ammonium sulfate, 2% (volume/volume) PEG 400, and 0.1 m HEPES (pH 7.5). These conditions are identical to those for the Ly49C-H-2Kb complex (12). For data collection, free Ly49C crystals were cryoprotected by soaking in mother liquor containing 20% (volume/volume) ethylene glycol before being flash-cooled. Diffraction data were collected to 2.6 Å resolution at 100 K on beamline X29 of the Brookhaven National Synchrotron Light Source. The data were processed using the program HKL2000 (14). For the Ly49C-H-2Kb complex, data collection to 2.9 Å was described previously (12). Crystals of free Ly49C belong to space group P212121 with four Ly49C monomers per asymmetric unit. The Ly49C-H-2Kb complex crystallizes in space group P43212; the asymmetric unit contains one Ly49C monomer and one H-2Kb molecule. Data collection statistics are presented in Table 1.
TABLE 1.
X-ray crystallographic statistics
| Ly49C | Ly49C-H-2Kb | Ly49G | |
|---|---|---|---|
| Data processing statistics | |||
| Resolution limit (Å)a | 50.0-2.60 (2.66-2.60) | 50.0-2.90 (2.95-2.90) | 50.0-2.6 (2.67-2.60) |
| Space group | P212121 | P43212 | C2 |
| Cell dimensions (Å) | a = 99.1, b = 94.9, c = 104.2 | a = b = 152.0, c = 64.7 | a = 133.4, b = 56.2, c = 33.8, β = 100.3° |
| Unique reflectionsa | 21,005 (2076) | 16,931 (840) | 7989 (583) |
| Completeness (%)a | 96.4 (97.4) | 97.5 (97.9) | 90.8 (62.1) |
|
Rmerge
(%)a,b |
5.8 (35.7)
|
6.2 (32.5)
|
6.2 (15.7)
|
| Refinement statistics | |||
| Rwork (%)c | 19.8 | 20.0 | 22.7 |
| Rfree (%)c | 26.3 | 26.3 | 28.6 |
| Mean B values (Å2) | 60.1 | 74.4 | 55.0 |
| Main chains | 60.2 | 75.3 | 54.6 |
| Side chains | 60.5 | 75.4 | 55.4 |
| H2O | 52.8 | 59.6 | 54.8 |
| r.m.s. deviations from ideality | |||
| Bond lengths (Å) | 0.012 | 0.015 | 0.018 |
| Bond angles (°) | 1.416 | 1.543 | 1.777 |
| Ramachandran plot statistics | |||
| Most favored (%) | 85.9 | 87.2 | 73.2 |
| Additionally allowed (%) | 12.3 | 11.0 | 22.4 |
| Generously allowed (%) | 1.8 | 1.8 | 4.4 |
Values in the parentheses are statistics of the highest resolution shell.
Rmerge = ∑|Ij - <I>|/∑Ij where Ij is the intensity of an individual reflection and <I> is the average intensity of that reflection.
Rwork = ∑||Fo| - |Fc||/∑|Fo|, where Fc is the calculated structure factor. Rfree is as for Rwork but calculated for a randomly selected 5.0% of reflections not included in the refinement.
Crystals of Ly49G NKD grew at room temperature in 30% (weight/volume) PEG 4000, 0.1 mm Tris-HCl (pH 8.5), and 0.2 m MgCl2. For data collection, crystals were cryoprotected by brief soaking in reservoir solution containing 15% (volume/volume) glycerol and frozen in liquid nitrogen. X-ray diffraction data were first collected to 3.0 Å resolution on a Rigaku R-axis IV++ image plate detector using CuKα radiation from a Micromax-007 rotating anode generator. Higher resolution data (2.6 Å) were collected at beamline X25, Brookhaven National Synchrotron Light Source, and were processed using HKL2000 (14). The data showed severe anisotropy and scaled anisotropically using XPREP (15). Crystals of Ly49G NKD belong to space group C2 with two molecules per asymmetric unit (Table 1).
Structure Determination and Refinement—For free Ly49C, orientational and positional parameters of molecules in the unit cell were determined by molecular replacement with Phaser (16). The search model consisted of the Ly49C monomer from the Ly49C-H-2Kb structure (Protein Data Bank code 1P4L) (12). Refinement was carried out with REFMAC5 (17). Each cycle of refinement was followed by manual building. Noncrystallographic symmetry restraints were applied to the four Ly49C monomers in the asymmetric unit in the initial refinement; σA-weighted phases were used to calculate 2Fo - Fc and Fo - Fc electron density maps with XtalView (18) to aid model building. Solvent molecules were included in the final stages of refinement. Residues 67-132 of the Ly49C stalk region were not observed in density maps for any of the monomers.
For the Ly49C-H-2Kb complex, direct rigid body refinement was carried out with REFMAC5 (17) using the original structure (12) as a starting model. Residues 218-230 of the Ly49C NKD were omitted in the initial refinement. These residues were readily built with helical architecture, based on 2Fo - Fc and Fo - Fc maps after maximum likelihood refinement. The resulting electron density also clearly showed C-terminal residues 260-262 of Ly49C, missing from the original model. Waters were added in the final stages of refinement. Final refinement statistics for Ly49C in free form and bound to H-2Kb are given in Table 1. Model geometry was examined using PROCHECK (19).
The structure of Ly49G NKD was solved by molecular replacement with the program MOLREP in the CCP4 suite (20). A homology modeled structure of the Ly49G NKD based on the Ly49C monomer was used as a search model. The rotational and translation search found two unambiguous solutions corresponding to two Ly49G monomers in the asymmetric unit and resulted in a correlation coefficient of 0.37 and Rcryst = 0.507 at a resolution range of 30.0 to 3.0 Å. Initial refinement was performed with CNS (21). Manual model rebuilding was carried out iteratively in XtalView (18) based on σA-weighted 2Fo - Fc and Fo - Fc maps. After CNS refinement converged, the structure was further refined using REFMAC5 (17). The final refinement statistics are shown in Table 1.
Affinity Measurements—The interaction of wild-type and mutant Ly49s with MHC-I was measured by surface plasmon resonance (SPR). All binding assays were performed at 25 °C using a BIAcore T100 biosensor in a running buffer containing 150 mm NaCl, 3.5 mm EDTA, 0.05% surfactant P-20, and 10 mm HEPES (pH 7.4). The extracellular regions of Ly49s (Ly49A, Ly49A mutants, and Ly49C) were covalently coupled to CM5 sensor chips via primary amine groups with an amine coupling kit (BIAcore). Serially diluted H-2Dd and H-2Kb ligands were then injected over the immobilized Ly49s. Equilibrium affinity measurements were performed at a flow rate of 10 μl/min. The results were analyzed using BIAevaluation 4.1 software. Specific binding data were fitted with a 1:1 Langmuir binding model so that dissociation constants (KD) could be calculated.
Protein Data Bank Accession Codes—Coordinates and structure factors for Ly49C, Ly49G, and the Ly49C-H-2Kb complex have been deposited under accession codes 3C8J, 3CAD, and 3C8K, respectively.
RESULTS AND DISCUSSION
Structure of Unbound Ly49C—We expressed the entire extracellular portion of Ly49C (residues 67-262), comprising both NKD and stalk regions, by in vitro folding from bacterial inclusion bodies. The structure of Ly49C in free form was determined by molecular replacement to 2.6 Å resolution (Table 1; Fig. 1A). The four monomers (designated A-D) in the asymmetric unit, which are related by 222 point group symmetry, form two homodimers (AB and CD) that are positioned orthogonally to each other in the crystal. In none of the monomers could we detect electron density for residues 67-132, which comprise the stalk region connecting the Ly49C NKD to the transmembrane region. Because SDS-PAGE analysis of dissolved crystals confirmed the presence of an intact stalk (data not shown), with no evidence of proteolysis, we concluded that the disorder in the crystal lattice indicates flexibility of the stalk, which secondary structure predictions suggest is a long coiled coil interspersed with flexible loops. This mobility is presumably required to permit binding of Ly49 receptors on the NK cell to MHC-I on an opposing target (trans interactions), as well as to MHC-I on the NK cell itself (cis interactions) (22-24).
FIGURE 1.
Structures of Ly49C and Ly49G. A, ribbon diagram of the Ly49C homodimer. Secondary structure elements are labeled. α-Helices are colored in gold, β-strands in rose, and loops in gray. B, composite omit electron density map (dark green, contoured at 2σ) of Ly49C at 2.60 Å resolution, showing residues 218-226 of helix α3, is overlapped with the anomalous difference map (magenta, contoured at 3σ), showing the anomalous signal of the sulfur atom of Met-225. C, structure of the Ly49G homodimer. D, electron density from the final 2Fo - Fc map (contoured at 1.2σ) of Ly49G at 2.60 Å resolution showing residues 217-227 of loop L3.
The root mean square (r.m.s.) deviation in main-chain atom positions for the four Ly49C monomers in the asymmetric unit ranges between 0.4 and 0.5 Å, indicating close similarity. The only significant differences occur at the solvent-exposed N termini, and at residue Ser-229 of monomer D, whose unusual conformation compared with that in monomers A-C is caused by lattice contacts. The following description of the Ly49C NKD is based on monomer A and that of the Ly49C homodimer on monomers A and B (Fig. 1A), unless stated otherwise.
The Ly49C NKD adopts a fold similar to those of the NKDs of Ly49A (11) and NKG2D (25), and to that of the carbohydrate recognition domain of mannose-binding protein A (26), a C-type animal lectin. However, in addition to the highly conserved secondary structural features of C-type lectin-like domains (CTLDs) (27), including two anti-parallel β-sheets (β0, β1, β5 and β2, β2′, β3, and β4) and two α-helices (α1 and α2), Ly49C bears a third α-helix (α3), comprising residues 218-226 (Fig. 1, A and B). The α3 helix breaks the long loop (L3; residues 211-231 in Ly49A) that normally connects β-strands β2′ and β3 in other CTLD superfamily members into two shorter loops, L3a and L3b (Fig. 1A). Indeed, this helix is unique to Ly49C among all CTLDs of known structure, including Ly49A (11) and NKG2D (25). It is involved in multiple interactions with MHC-I in the Ly49C-H-2Kb complex, as described below.
At the cell surface, Ly49C, like other Ly49s, exists as a homodimer (2, 4). In the crystal, each Ly49C homodimer (AB and CD) shows a similar subunit arrangement as bound Ly49C (see below), which we term the “open” conformation, because the α2 helices of the NKDs make no contacts across the dimer interface (Fig. 1A). In another crystal form of free Ly49C (space group P1), we observed the same mode of association (data not shown), indicating that crystal packing does not influence Ly49C dimerization. This open conformation also resembles that of free Ly49A in solution determined by NMR (28). However, it is distinct from the “closed” conformation of MHC-bound Ly49A determined by x-ray crystallography (11), in which the α2 helices of the Ly49A dimer are juxtaposed (not shown). Thus, at least some Ly49 receptors can adopt both open and closed states, which may correlate with trans and cis binding to MHC-I (24).
Structure of the Ly49C-H-2Kb Complex—The identification of helix α3 in the free form of Ly49C prompted us to re-examine the original Ly49C-H-2Kb structure, in which residues 218-226 of bound Ly49C were modeled as a loop and residues 226-231 were reported to be disordered (12). To determine whether engagement of MHC-I actually induced conformational changes in residues 218-231, we rerefined the Ly49C-H-2Kb complex to 2.9 Å resolution. Indeed, the newly refined structure exhibited well defined electron density in this region (Fig. 2B); residues 218-226 were easily built as a helix (α3). Furthermore, the refinement statistics improved dramatically from those reported earlier (12); Rwork was reduced from 26.9 to 20.0% and Rfree from 29.5 to 26.3% (Table 1). The overall structure of the complex, however, is maintained.
FIGURE 2.
Structure of the Ly49C-H-2Kb complex. A, ribbon diagram of the Ly49C-H-2Kb complex. Domains are labeled. The α1, α2, and α3 domains of the MHC-I heavy chain are green; β2mis orange; the MHC-bound peptide in ball-and-stick representation is gray; the Ly49C dimer is rose. B, composite omit electron density map (dark green, contoured at 1.5σ) of Ly49C-H-2Kb at 2.90 Å resolution, showing residues 218-226 of Ly49C helix α3. C, structural rearrangements in Ly49C induced by binding to MHC-I. Bound Ly49C is rose; unbound Ly49C is gold; H-2Kb is green. Salt bridges are indicated by solid lines.
In the complex, the Ly49C dimer engages H-2Kb bivalently, such that each NKD makes identical interactions with MHC-I at a concave surface beneath the peptide-binding platform formed by the MHC-I α1α2 and α3 domains and β2m (Fig. 2A). In this view, the complex is oriented as if the two H-2Kb molecules stand on the target cell surface at the bottom and the Ly49C homodimer reaches H-2Kb from an opposing NK cell above, to which it is tethered through 66-residue stalk regions projecting down to the N termini of the NKDs (Ly49s are type II transmembrane proteins).
The MHC-bound Ly49C retains nearly the same conformation as the free receptor; superposition of the two homodimers gave an r.m.s. deviation of 0.8 Å for all atoms of residues 138-262. No substantial differences in main-chain conformation are observed in the MHC-binding site, except a displacement of 1.7 Åin the α-carbon position of Lys-228, which enables this residue to form salt bridges with H-2Kb Asp-30 and Asp-212 (Fig. 2C). To accommodate the shift in Lys-228, Ser-229 is displaced by 4.9 Å in its α-carbon position, accompanied by a side-chain rotation of 180°C about the Cα-Cβ axis.
The Ly49C-H-2Kb interface is very hydrophilic and is dominated by polar interactions, including 13 hydrogen bonds and 12 salt bridges (Table 2). The newly refined structure differs most from that determined earlier (12) in the interactions made by loop L3, which is now completely ordered and includes helix α3. Although this region, comprising residues 211-231, was not observed to contact MHC-I in the previous structure, it makes numerous interactions, including two hydrogen bonds and six salt bridges, in the current structure (Table 2; Fig. 3A). Additionally, the L3/α3 region accounts for 33% (785 Å2) of the total solvent-accessible surface buried in the Ly49C-H-2Kb interface (2392 Å2), suggesting an important contribution to complex stabilization.
TABLE 2.
Interactions between Ly49 and MHC-I molecules in the Ly49C-H-2Kb and Ly49A-H-2Dd complexes
Residues of H-2Kb or H-2Dd forming hydrogen bonds or salt bridges (bold letters) with Ly49C or Ly49A are in red. In parentheses is shown the number of hydrogen bonds or salt bridges between two residues, if more than one. Residues of H-2Kb or H-2Dd making van der Waals contacts with these Ly49s are in black. Hydrogen bonds and salt bridges were calculated using cutoff distances of 3.4 and 4.0 Å, respectively. The cutoff distance for van der Waals contacts was 4.0 Å. Residues from the L3/α3 region of Ly49C and Ly49A are in blue; residues from the L6 loop are in green.
FIGURE 3.
Comparison of Ly49-MHC-I interfaces. A, Ly49C-H-2Kb interface, highlighting interactions made by residues 211-231 of Ly49C. B, Ly49A-H-2Dd complex, showing interactions made by the corresponding region of Ly49A. Domains are labeled. The side chains of interacting residues are drawn in ball-and-stick representation, with carbon atoms in rose (Ly49C), cyan (Ly49A), green (H-2Kb or H-2Dd), or orange (β2m), and oxygen atoms in red, nitrogen atoms in blue, and sulfur in yellow. Salt bridges and hydrogen bonds are represented by solid and dotted lines, respectively.
Comparison of the Ly49C-H-2Kb and Ly49A-H-2Dd Complexes—Although the general features of the Ly49C-H-2Kb and Ly49A-H-2Dd complexes are similar, insofar as both Ly49s engage MHC at a site underneath the peptide-binding platform (24), the complexes differ markedly in the detailed interactions at their interfaces (Table 2). These differences are largely attributable to differences in the binding sites of the Ly49 receptors, rather than to differences in the MHC ligands, because H-2Kb and H-2Dd contact the receptors through β2m and mostly nonpolymorphic residues of the α1α2 and α3 domains.
A comparison of the MHC-binding sites of Ly49C and Ly49A showed that they differ most in the conformation of loop L3, whereas other regions of the binding sites (β3, β4, L5, and L6) display very similar main-chain conformations (Fig. 4A). Indeed, the only hydrogen bonds or salt bridges common to both the Ly49C-H-2Kb and Ly49A-H-2Dd complexes are mediated by residues in L5 (Ser-236, Ala/Thr-238, and Arg-239) or β4 (Glu/Asp-241 and Asp/Asn-242) (Table 2). By contrast, the L3 regions, because of their unique structures, mediate a completely different set of interactions in each complex (Fig. 3). Notably, Met-225 and Asn-226 in the signature α3 helix of Ly49C make numerous van der Waals contacts with H-2Kb Leu-110, Arg-111, and Glu-128, whereas Ly49C Lys-221, also in α3, forms a salt bridge with H-2Kb Glu-128. Hence, the Ly49A-H-2Dd and Ly49C-H-2Kb complexes define two distinct binding modes for MHC-I. To assess the extent to which these binding modes might apply to other Ly49s, we sought to better define structural diversity within the Ly49 family. Accordingly, we determined the crystal structure of a third Ly49 receptor, Ly49G, to 2.6 Å resolution (Table 1).
FIGURE 4.
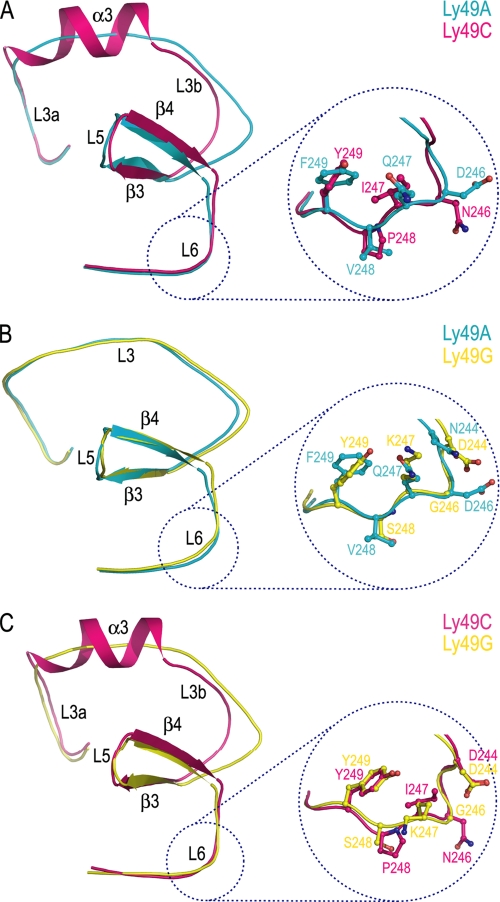
Comparison of the MHC-binding sites of Ly49 receptors. A, superposition of the MHC-binding regions of Ly49C (group I) and Ly49A (group II). Secondary structure elements are labeled according to Fig. 1. The inset shows MHC-contacting residues in loop L6 of Ly49C superposed on the corresponding residues of Ly49A. B, superposition of Ly49G and Ly49A (both group II). The inset shows MHC-contacting residues in L6 of Ly49A superposed on the corresponding residues of Ly49G. C, superposition of Ly49G (group II) and Ly49C (group I). The inset shows putative MHC-contacting residues in L6 of Ly49G superposed on the corresponding residues of Ly49C.
Structure of Ly49G—Like the Ly49A and Ly49C NKDs, the Ly49G NKD adopts a fold consisting of two α-helices and two anti-parallel β-sheets, including four intrachain disulfide bonds (Fig. 1, C and D). The two Ly49G monomers in the asymmetric unit (designated A and B) form an open dimer that closely resembles the Ly49C dimer in its subunit arrangement (Fig. 1, A and C). The r.m.s. difference in α-carbon positions between the two monomers is 1.1 for 117 α-carbon pairs, indicating significant flexibility in the Ly49G NKD. For simplicity, however, all subsequent analysis is based on monomer A of the Ly49G dimer. When the Ly49G NKD is superposed on the Ly49A and Ly49C NKDs, r.m.s. differences of 1.0 and 2.2 Å, respectively, are obtained. Hence, Ly49G is more closely related to Ly49A than to Ly49C. Consistent with this comparison, Ly49G, like Ly49A, recognizes only H-2D alleles (29), whereas Ly49C recognizes both H-2D and H-2K alleles (12).
The main structural difference between Ly49G and Ly49C is found in loop L3, which contacts MHC-I in both the Ly49A-H-2Dd and Ly49C-H-2Kb complexes (Fig. 3). This loop is continuous in Ly49G (and Ly49A) but is interrupted by helix α3 in Ly49C (Fig. 1, A and C). Thus, the r.m.s. deviation in α-carbon positions between the L3 loops of Ly49G and Ly49A is 1.1 Å for 21 α-carbon pairs, compared with an r.m.s deviation of 4.6 Å between the L3 loops of Ly49G and Ly49C.
A Structural Classification of Ly49 Receptors—Based on these results, we carried out sequence comparisons and secondary structure predictions of Ly49s using the program GOR. This analysis revealed that the Ly49 family can be neatly divided into four groups (I-IV), based on the known or predicted structure of the L3 region (Fig. 5A). Group I, which includes Ly49C, has a high probability of α-helix in the middle of L3, between residues 218 and 226, in excellent agreement with the crystal structure of Ly49C (Fig. 1A). In group II, which includes Ly49A and Ly49G, the probability of α-helix is low, and L3 is predicted to be random coil, consistent with the structures of Ly49A (11) and Ly49G (Fig. 1C) showing continuous loops. Importantly, a similar partitioning of Ly49s into two main groups was observed in a protein phylogenetic tree constructed by multiple sequence alignment of 21 Ly49 NKDs (residues 136-262), which permitted a further division into four subgroups (Ia, Ib; IIa, IIb) (Fig. 5B). Groups III and IV contain only single members, Ly49B and Ly49Q, respectively (Fig. 5A). Although the L3 region of Ly49B, like that of group II members, is predicted as random coil, it has a distinct amino acid sequence whose conformation remains to be defined. The L3 region of Ly49Q is also predicted as random coil, but phylogenetic analysis of the entire NKD sequence showed that Ly49Q, like Ly49B, is separate from group I or II Ly49s (Fig. 5B).
FIGURE 5.
Structural classification of Ly49 NK receptors. A, structure-based sequence alignment of the MHC-binding regions of Ly49 NKDs. Secondary structure elements for Ly49C and Ly49A are denoted by loops (α-helices) and arrows (β-strands). These, and the loop regions, are numbered according to Fig. 1. The classification into group I or II is based on the known, or predicted, presence (I) or absence (II) of helix α3 within loop L3. The classification into subgroups (Ia, Ib; IIa, IIb), and into group III or IV, is based on the dendrogram in B. White characters on a red background show MHC-contacting residues that are strictly, or mostly, conserved in group I or II, based on the Ly49C-H-2Kb or Ly49A-H-2Dd structure. MHC-contacting residues that differ are drawn in white and framed in blue. The remaining residues are black. Sequences were retrieved from GenBank™ or SwissProt using the following accession numbers: Ly49A, M25812; Ly49I, AF237686; Ly49B, U10304; Ly49C, U56404; Ly49D, L78247; Ly49E, Q60652; Ly49F, Q60653; Ly49G, S78689; Ly49H, U12889; Ly49L, AF213453; Ly49M, AF028134; Ly49O, AF146571; Ly49P, AF146570; Ly49R, AF288377; Ly49S, AF288378; Ly49T, AF288379; Ly49U, AF288380; Ly49V, AF288380; Ly49W, AF283250. Ly49K and Ly49N were not included because only partial sequences are available. B, dendrogram of Ly49 NK receptors. The sequences of the 21 Ly49 NKDs in A, corresponding to residues 136-262, were aligned and a dendrogram was generated using ClustalW 1.83.
Although no structure is available for Ly49G bound to an MHC-I ligand, it is likely from structural considerations and mutagenesis data that Ly49G, along with other group II members, engage MHC-I very similarly to Ly49A. Most notably, Ly49A and Ly49G, which have similar MHC specificities (29), are identical at 18 of 25 MHC-contacting positions identified in the Ly49A-H-2Dd complex (11), including 6 of 7 in loop L3, 4 of 4 in loop L5, and 3 of 3 strand β4 (Fig. 5A). By contrast, Ly49G differs from Ly49C (group I) at 16 of 21 MHC-contacting positions in the newly refined Ly49C-H-2Kb structure (Table 2). Ly49G differs most from Ly49A in loop L6, in which none of five MHC-contacting residues is conserved.
Previous mutational analysis of Ly49A residues in contact with MHC-I in the Ly49A-H-2Dd complex identified five residues (Asp-229, Ser-236, Thr-238, Arg-239, and Asp-241) whose replacement by alanine greatly reduced, or abolished, binding in an SPR assay (13). Significantly, all these critical residues are strictly conserved in Ly49G, as well as in all other group II Ly49s (Fig. 5A). In the Ly49A and Ly49G structures, these residues are located in L3 (Asp-229), L5 (Ser-236, Thr-238, and Arg-239), or β4 (Asp-241). Moreover, this region of the binding site is nearly superimposable in the two NKDs (Fig. 4B), with several small differences in side-chain conformation likely attributable to differences in crystal lattice environment or to MHC binding in the case of Ly49A. By contrast, mutagenesis of MHC-contacting residues in loop L6 of Ly49A (residues 244-249) had considerably less effect on affinity for MHC-I (13). Although L6 has similar main-chain conformations in Ly49A and Ly49G (Fig. 4B), and probably across group II, this element differs at all five MHC-contacting positions (Asn/Asp-244, Asp/Gly-246, Gln/Lys-247, Val/Ser-248, and Phe/Tyr-249). Indeed, except for position 223 in L3, sequence variability in the MHC-binding site of group II Ly49s is confined entirely to L6 (Fig. 5A).
This analysis suggests that MHC-I recognition by group II Ly49s is mediated primarily by a small number of invariant residues (229, 236, 238, 239, and 241) in highly conserved regions of these receptors (L3, L5, and β4) that together contribute the bulk of the binding free energy. However, the observed preference of individual group II Ly49s for particular MHC-I alleles is governed by the variable L6 loop, which modulates the affinity of the overall interaction.
This interpretation is supported by the finding that Ly49G from mouse strain BALB/c (the allele studied here) recognizes both H-2Dd and H-2Dk, whereas Ly49G from mouse strain B6 recognizes only H-2Dd (29). Remarkably, the MHC-binding sites of BALB/c and B6 Ly49G differ by only a single amino acid (Gly/Asp-246), which must therefore explain the observed specificity difference. This residue is located in L6 of the Ly49G NKD (Fig. 4B). The importance of L6 in determining MHC allele specificity is further supported by mutational analysis of Ly49P (group IIa) and Ly49W (group IIb), which recognize H-2Dd, or H-2Dd, and H-2Dk, respectively (30). The specificity for H-2Dk could be transferred from Ly49W to Ly49P by replacing three residues in loop L6 of Ly49P with the corresponding residues in Ly49W. Conversely, replacement of these same residues in Ly49W by the corresponding residues in Ly49P eliminated H-2Dk recognition, while preserving H-2Dd recognition (30). It should also be noted that Ly49A and Ly49D (group IIa), which closely resemble Ly49P in poorly recognizing H-2Dk relative to H-2Dd (30, 31), have the same L6 sequences as Ly49P (Fig. 5A). Likewise, BALB/c Ly49G (group IIb) and Ly49W, which recognize both H-2Dd and H-2Dk (29, 30), share identical L6 sequences.
Unlike group II Ly49s, which probably all bind MHC-I in the manner of Ly49A, structural considerations suggest that group I members engage MHC-I very similarly to Ly49C. Thus, group I Ly49s are identical at 11 of 21 MHC-contacting positions in the newly refined Ly49C-H-2Kb complex (Table 2), including 3 of 5 in the L3/α3 region, 4 of 4 in loop L5, and 2 of 3 in strand β4 (Fig. 5A). Among the invariant residues are Lys-221, Met-225, and Asn-226, all located in the unique α3 helix that distinguishes group I from group II Ly49s (Fig. 4, A and C). By comparison, Ly49C differs from Ly49A at 17 of 24 MHC-contacting positions in the Ly49A-H-2Dd structure (Table 2). Nevertheless, it is noteworthy that residues 236, 239, and 241, which were identified as hot spots for Ly49A binding to H-2Dd (see above) (13), are conserved, or conservatively substituted, in groups I and II (Fig. 5A). Moreover, these residues, which make similar interactions with nonpolymorphic MHC-I residues Arg-111 and Asp-122 in the Ly49C-H-2Kb and Ly49A-H-2Dd complexes (Table 2), are found in a region of the binding site, comprising L5 and β4, that is nearly superimposable in Ly49C and Ly49A (or Ly49G) (Fig. 4, A and C).
As in the case of group II Ly49s, group I members differ most in loop L6, where all four MHC-contacting residues show variability (Fig. 5A). This again implicates L6 in regulating MHC allele specificity. Thus, Ly49C and Ly49I (group Ia), which have identical L6 sequences, recognize the same MHC-I molecules (H-2Kb, H-2Kd, and H-2Dd) (32). By contrast, although the specificities of Ly49F, Ly49S and Ly49E (group Ib) are uncertain, these Ly49s clearly differ from Ly49C and Ly49I in terms of MHC recognition (32), which may reflect sequence differences in the L6 region of group Ia versus Ib receptors (Fig. 5A).
Role of L3 in MHC-I Recognition—The MHC specificities of Ly49 receptors have been analyzed by cell-cell adhesion and MHC-I tetramer binding assays, functional tests of NK cell cytolytic activity, and SPR to measure Ly49-MHC-I affinities (3). Although there are some discrepancies in MHC reactivities obtained by these diverse methods, and the specificity of most Ly49s remains unknown, the results are generally consistent and reveal that Ly49s fall into the following two broad categories: those that recognize both H-2D and H-2K alleles (Ly49C and Ly49I) and those that recognize only H-2D alleles (Ly49A, Ly49D, Ly49G, Ly49P, and Ly49W) (2, 29, 30, 32). Significantly, this functional dichotomy correlates exactly with our structure-based classification of Ly49s into groups I or II, respectively (Fig. 5A). Because these groups are distinguished by the structure of L3 (Fig. 4, A and C), we asked whether a group II Ly49 could be converted into a group I Ly49 by simply grafting L3 regions. Three mutants were constructed, in which residues 211-231 (Ly49Amut1), 218-226 (Ly49Amut2), or 227-231 (Ly49Amut3) of Ly49A (group II) were exchanged with the corresponding residues of Ly49C (group I) to completely (Ly49mut1) or partially (Ly49Amut2, Ly49Amut3) replace L3.
As measured by SPR (Fig. 6), wild-type Ly49A bound to H-2Dd with a KD of 4.4 μm (Table 3), comparable with the previously reported value of 1.8 μm (13), but it displayed no detectable binding to H-2Kb, even at the highest H-2Kb concentration tested (32 μm). By contrast, Ly49C recognized both H-2Dd and H-2Kb, with KD of 136 and 102 μm, respectively. As predicted from our analysis of Ly49-binding sites, Ly49Amut1, which contains the entire L3 of Ly49C, acquired the capacity to recognize H-2Kb (KD = 167 μm) (Fig. 6). At the same time, Ly49Amut1 retained binding to H-2Dd (KD = 28 μm), albeit with ∼6-fold lower affinity than wild-type Ly49A. However, partial replacement of L3 in Ly49Amut2 and Ly49Amut3 did not confer specificity for H-2Kb, even though both mutants still bound H-2Dd (Table 3). Hence, the gain of function exhibited by Ly49Amut1, but not Ly49Amut2 or Ly49Amut3, demonstrates that the L3 region, in its entirety, regulates the ability of Ly49s to recognize both H-2D and H-2K alleles, compared with only H-2D alleles.
FIGURE 6.
Equilibrium binding of Ly49C, Ly49A, and mutants of Ly49A to H-2Dd and H-2Kb. H-2Dd and H-2Kb flowed at five different concentrations ranging from 1 to 16 μm for H-2Dd and from 2 to 32 μm for H-2Kb, in 2-fold increments (red, 1 μm; gray, 2 μm; green, 4 μm; cyan, 8 μm; blue, 16 μm for H-2Dd, and red, 2 μm; gray, 4 μm; green, 8 μm; cyan, 16 μm; blue, 32 μm for H-2Kb) over immobilized Ly49s (Ly49A, 3000 RU; Ly49Amut1 3000 RU; Ly49Amut2 3000 RU; Ly49Amut3 3000 RU; Ly49C 4000 RU). Inset plots show nonlinear steady-state affinity analysis for each reaction.
TABLE 3.
Dissociation constants (× 10−6 M) of Ly49-MHC-I interactions
Equilibrium dissociation constants (KD) were derived by nonlinear steady-state affinity analysis of SPR data (Fig. 6). NB indicates no binding detectable.
| Ly49A | Ly49Amut1 | Ly49Amut2 | Ly49Amut3 | Ly49C | |
|---|---|---|---|---|---|
| H-2Kb | NB | 167 | NB | NB | 102 |
| H-2Dd | 4.4 | 28 | 23 | 26 | 136 |
Conclusions—Our structural analysis of Ly49s in free and MHC-bound forms has revealed the modular design of Ly49-binding sites. These polymorphic sites, which are formed by NKD residues 211-249, include three basic components as follows: 1) an N-terminal region (residues 211-231) consisting of loop L3 that may also include helix α3 (residues 218-226); 2) a central region (residues 232-243) composed of strand β3, loop L5, and strand β4; and 3) a C-terminal region (residues 244-249) composed of loop L6 (Fig. 4). These regions play distinct roles in MHC recognition that together account for the ability of Ly49s to bind multiple MHC-I molecules, while conferring preferences for particular alleles.
Based on the presence or absence of α3 within L3, we have assigned nearly all (19 of 21) Ly49s to groups I or II, respectively (Fig. 5). The significance of this classification is that it defines two distinct binding modes, in which the variable L3 element makes entirely different contacts with MHC-I in the two groups (Fig. 3). Moreover, these binding modes establish the functional division of Ly49s into broadly (H-2D and H-2K) or narrowly (H-2D) cross-reactive receptors, as we showed by engineering Ly49 variants with altered MHC-binding properties. In sharp contrast to L3, the portion of the binding site formed by β3, L5, and β4 has very similar structure in Ly49s from groups I and II. This region contains residues that mediate conserved interactions with MHC-I and that are energetic hot spots for MHC binding (13). Overlaid onto these primary interactions are secondary ones involving L6, which displays considerable sequence variability within each group. As demonstrated by mutational analysis (30), L6 regulates the specificity of Ly49s for particular H-2D or H-2K alleles, most likely by modifying the affinity derived from conserved interactions. However, it is not apparent which allele-specific features of MHC-I are detected by L6. In the Ly49A-H-2Dd structure, all MHC-I residues within 10 Å of L6 are identical between H-2Dd and H-2Dk, yet amino acid changes in L6 alone suffice to alter the specificity of Ly49s for these alleles (29, 30). Because H-2Dd and H-2Dk differ at only three positions in the binding interface (Ser/Pro-2, Glu/Gly-104, and Glu/Asp-224), none of which is closer than 13 Å to L6, long range effects must explain the ability of Ly49s to discriminate between H-2D alleles. Such communication between distant sites in proteins has been observed in many systems (33, 34), and it appears to involve long range perturbations in electrostatic fields or vibrational modes that are still poorly understood.
Determining the MHC specificity of Ly49 receptors has been complicated by a number of factors (3). For example, cell-cell adhesion assays using Ly49 and MHC-I transfectants, as well as direct binding assays using soluble recombinant proteins, may not detect low affinity interactions that are nevertheless biologically relevant. In addition, cis interactions between Ly49 and MHC-I molecules on the same NK cell may prevent trans interactions between these molecules on opposing cells (22-24). As a result, the MHC specificities of most Ly49s are unknown, or ambiguous at best (2, 3). Our structure-based classification of Ly49-binding sites (Fig. 5), whose overall validity is supported by biological data (2, 29-32), provides a guide for predicting the specificity of receptors that have not yet been characterized functionally. For example, Ly49J and Ly49U should resemble Ly49C in binding both H-2D and H-2K alleles, whereas Ly49O and Ly49R, like Ly49A, should be restricted to H-2D alleles, in particular H-2Dd. Such information can be used to analyze the overall repertoire of MHC specificities expressed by NK cells and to design tests of NK cell development and self-tolerance.
Acknowledgments
We thank Stephen Anderson (NCI, National Institutes of Health) for the Ly49G cDNA and Howard Robinson (Brookhaven National Synchrotron Light Source) for x-ray data collection.
The atomic coordinates and structure factors (codes 3C8J, 3CAD, and 3C8K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant AI047990. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NK, natural killer; NKD, natural killer receptor domain; MHC-I, major histocompatibility complex class I; MCMV, mouse cytomegalovirus; β2m, β2-microglobulin; SPR, surface plasmon resonance; CTLD, C-type lectin-like domain; r.m.s., root mean square.
References
- 1.McQueen, K. L., and Parham, P. (2002) Curr. Opin. Immunol. 14 615-621 [DOI] [PubMed] [Google Scholar]
- 2.Plougastel, B. F. M., and Yokoyama, W. M. (2003) Nat. Rev. Immunol. 3 304-316 [DOI] [PubMed] [Google Scholar]
- 3.Lanier, L. L. (2005) Annu. Rev. Immunol. 23 225-274 [DOI] [PubMed] [Google Scholar]
- 4.Natarajan, K., Dimasi, N., Wang, J., Mariuzza, R. A., and Margulies, D. H. (2002) Annu. Rev. Immunol. 20 853-885 [DOI] [PubMed] [Google Scholar]
- 5.Deng, L., and Mariuzza, R. A. (2006) Semin. Immunol. 18 159-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vivier, E., Tomasello, E., and Paul, P. (2002) Curr. Opin. Immunol. 14 306-311 [DOI] [PubMed] [Google Scholar]
- 7.Anderson, S. K., Ortaldo, J. R., and McVicar, D. W. (2001) Immunol. Rev. 181 79-89 [DOI] [PubMed] [Google Scholar]
- 8.Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B., and Lanier, L. L. (2002) Science 296 1323-1329 [DOI] [PubMed] [Google Scholar]
- 9.Smith, H. R., Heusel, J. W., Mehta, I. K., Kim, S., Dorner, B. G., Naidenko, O. V., Iizuka, K., Furukawa, H., Beckman, D. L., Pingel, J. T., Scalzo, A. A., Fremont, D. H., and Yokoyama, W. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8826-8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derosiers, M.-P., Kielczewska, A., Loredi-Osti, J.-C., Adam, S. G., Makrigiannis, A. P., Lemiex, S., Pham, T., Lodoen, M. B., Morgan, K., Lanier, L. L., and Vidal, S. M. (2005) Nat. Genet. 37 593-599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tormo, J., Natarajan, K., Margulies, D. H., and Mariuzza, R. A. (1999) Nature 402 623-631 [DOI] [PubMed] [Google Scholar]
- 12.Dam, J., Guan, R., Natarajan, K., Dimasi, N., Chlewicki, L. K., Kranz, D. M., Schuck, P., Margulies, D. H., and Mariuzza, R. A. (2003) Nat. Immunol. 4 1213-1222 [DOI] [PubMed] [Google Scholar]
- 13.Wang, J., Whitman, M. C., Natarajan, K., Tormo, J., Mariuzza, R. A., and Margulies, D. H. (2002) J. Biol. Chem. 277 1433-1442 [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 15.Sheldrick, G. M. (2002) XPREP, version 6.10, Brucker-AXS, Madison, WI
- 16.Storoni, L. C., McCoy, A. J., and Read, R. J. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 432-438 [DOI] [PubMed] [Google Scholar]
- 17.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 18.McRee, D. E. (1999) J. Struct. Biol. 125 156-165 [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 20.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 240-255 [Google Scholar]
- 21.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 22.Doucey, M. A., Scarpellino, L., Zimmer, J., Guillaume, P., Luescher, I. F., Dron, C., and Held, W. (2004) Nat. Immunol. 5 328-336 [DOI] [PubMed] [Google Scholar]
- 23.Back, J., Chalifour, A., Scarpellino, L., and Held, W. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3978-3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Held, W., and Mariuzza, R. A. (2008) Nat. Rev. Immunol. 8 269-278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, P., Morris, D. L., Willcox, B. E., Steinle, A., Spies, T., and Strong, R. K. (2001) Nat. Immunol. 2 443-451 [DOI] [PubMed] [Google Scholar]
- 26.Weis, W. I., Kahn, R., Fourme, R., Drickamer, K., and Hendrickson, W. A. (1991) Science 254 1608-1615 [DOI] [PubMed] [Google Scholar]
- 27.Drickamer, K. (1999) Curr. Opin. Struct. Biol. 9 585-590 [DOI] [PubMed] [Google Scholar]
- 28.Dam, J., Baber, J., Grishaev, A., Malchiodi, E. L., Schuck, P., Bax, A., and Mariuzza, R. A. (2006) J. Mol. Biol. 362 102-113 [DOI] [PubMed] [Google Scholar]
- 29.Silver, E. T., Lavender, K. J., Gong, D. E., Hazes, B., and Kane, K. P. (2002) J. Immunol. 169 4752-4760 [DOI] [PubMed] [Google Scholar]
- 30.Ma, B. J., Silver, E. T., Hazes, B., and Kane, K. P. (2003) J. Immunol. 171 5337-5344 [DOI] [PubMed] [Google Scholar]
- 31.Brennan, J., Mahon, G., Mager, D. L., Jeffries, W. A., and Takei, F. (1996) J. Exp. Med. 183 1553-1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanke, T., Takizawa, H., McMahon, C. W., Busch, D. H., Pamer, E. G., Miller, J. D., Altman, J. D., Liu, Y., Cado, D., Lemonnier, F. A., Bjorkman, P. J., and Raulet, D. H. (1999) Immunity 11 67-77 [DOI] [PubMed] [Google Scholar]
- 33.Süel, G. M., Lockless, S. W., Wall, M. A., and Ranganathan, R. (2003) Nat. Struct. Biol. 10 59-69 [DOI] [PubMed] [Google Scholar]
- 34.Moza, B., Buonpane, R. A., Zhu, P., Herfst, C. A., Nur-ur Rahman, A. K. M., McCormick, J. K., Kranz, D. M., and Sundberg, E. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9867-9872 [DOI] [PMC free article] [PubMed] [Google Scholar]