Abstract
α-Synuclein (α-syn) phosphorylation at serine 129 is characteristic of Parkinson disease (PD) and related α-synulceinopathies. However, whether phosphorylation promotes or inhibits α-syn aggregation and neurotoxicity in vivo remains unknown. This understanding is critical for elucidating the role of α-syn in the pathogenesis of PD and for development of therapeutic strategies for PD. To better understand the structural and molecular consequences of Ser-129 phosphorylation, we compared the biochemical, structural, and membrane binding properties of wild type α-syn to those of the phosphorylation mimics (S129E, S129D) as well as of in vitro phosphorylated α-syn using a battery of biophysical techniques. Our results demonstrate that phosphorylation at Ser-129 increases the conformational flexibility of α-syn and inhibits its fibrillogenesis in vitro but does not perturb its membrane-bound conformation. In addition, we show that the phosphorylation mimics (S129E/D) do not reproduce the effect of phosphorylation on the structural and aggregation properties of α-syn in vitro. Our findings have significant implications for current strategies to elucidate the role of phosphorylation in modulating protein structure and function in health and disease and provide novel insight into the underlying mechanisms that govern α-syn aggregation and toxicity in PD and related α-synulceinopathies.
Mounting evidence from pathologic, genetic, animal model, biochemical, and biophysical studies support the hypothesis that α-synuclein (α-syn)3 plays a central role in the pathogenesis of Parkinson disease (PD) and several other neurodegenerative diseases, including Alzheimer disease, multiple system atrophy, dementia with Lewy bodies, Down syndrome, and neurodegeneration with brain iron accumulation, collectively referred to as “synucleinopathies” (1). Although the exact function of α-syn remains poorly understood, it is thought to play a role in regulating dopamine neurotransmission (2), vesicular trafficking (3, 4), and modulating synaptic function and plasticity (5, 6). Increasing evidence suggests that phosphorylation may be an important regulator of α-syn oligomerization, fibrillogenesis, Lewy body (LB) formation, and neurotoxicity in vivo (7). Immunohistochemical and biochemical studies suggest that the majority of α-syn within LBs from patients with PD and related synucleinopathies (8-12) is phosphorylated at Ser-129 (Ser(P)-129). Proteinaceous inclusions formed in cellular and animal models overexpressing WT or mutant α-syn can also be stained with an antibody against Ser(P)-129.
A study by Fujiwara et al. (8) reported that in vitro phosphorylated α-syn (at Ser-129, using casein kinase II (CK2)) forms fibrils more readily than unmodified α-syn. Phosphorylation at Ser-129 was also reported to promote the formation of cytoplasmic inclusions in some cell culture models of synucleinopathies (13). Together, these findings suggested that phosphorylation at Ser-129 plays an important role in modulating α-syn aggregation, LB formation, and toxicity in vivo. However, in vivo studies by Feany and co-worker (7) suggest a lack of correlation between phosphorylation at Ser-129 and the level of α-synfibrillation. Overexpression of the phosphomimic S129D or coexpression of WT α-syn and G protein-coupled receptor kinase 2 (Gprk2), which phosphorylates α-syn specifically at Ser-129, in the Drosophila model of PD results in increased α-syn toxicity without an increase in the number of α-syn inclusions (compared with overexpression of WT α-syn). Interestingly, overexpression of S129A results in a significant increase (4×) in the number of inclusions and suppression of dopaminergic neuronal loss produced by expression of WT human α-syn.
We considered that a rigorous examination and comparison of the biochemical and biophysical properties of phosphorylation mimicking mutants (S129A and S129E/D), and WT α-syn may clarify the observations described above as well as the molecular mechanisms by which phosphorylation at Ser-129 may modulate α-syn aggregation and toxicity in vivo. In addition to the phosphorylation mimics, we prepared, purified, and characterized the in vitro phosphorylated Ser(P)-129 form of α-syn. To better understand the role of the phosphate group in modulating α-syn aggregation and membrane binding properties, we compared the structural, oligomerization, fibrillation, and membrane binding properties of WT α-syn to those of the phosphorylation mimics (S129E, S129D) as well as the purified in vitro phosphorylated form of α-syn using NMR, circular dichroism (CD), size exclusion chromatography (SEC), SDS-PAGE, thioflavin T (ThT), and transmission electron microscopy (TEM). Together our results demonstrate that phosphorylation at Ser-129 inhibits rather than promotes α-syn fibril formation in vitro. Equally important is our finding that the phosphorylation mimics (S129E/D) do not reproduce the effect of phosphorylation at this site on α-syn structure and aggregation properties in vitro. These findings have significant implications for modeling synulceinopathies in vivo and our understanding of the role of α-syn in the pathogenesis of PD and related disorders.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of α-Syn Variants—The S129E, S129D, S129A, and S87A α-syn mutants were generated using site-directed mutagenesis employing complementary internal mutagenic primers and two-step PCR. All constructed sequences were confirmed by DNA sequencing. All proteins used in these studies were expressed as previously described (14). For the NMR studies, expression and purification of unlabeled and 15N-labeled α-syn were performed as described previously (15, 16). To enable attachment of a spin label, a single cysteine was introduced into α-syn at position 18 (A18C). The nitroxide spin label chosen for reaction with the cysteine-containing mutant was 1-oxy-2, 2, 5, 5-tetramethyl-d-pyrroline-3-methyl)-methanethiosulfonate (MTSL, Toronto Research Chemicals, Toronto, Ontario, Canada). MTSL has already been shown to efficiently react with α-syn cysteine mutants, and the reaction was carried out as described previously (17).
In Vitro Phosphorylation of Recombinant α-Syn—WT or mutant α-syn was phosphorylated by CK1 (New England Biolabs) at a concentration of 1.44 mg/ml (100 μm) unless otherwise stated. The reaction was carried out in the presence of 1.09 mm ATP (Sigma), 1× reaction solution supplied with the enzyme, and 1200 units of CK1/145 μg of α-syn. The phosphorylation reaction was incubated at 30 °C for the stated time points, and the reaction was stopped with EDTA disodium salt (5 mm final concentration) (Axon Lab). The progress of the reaction was monitored by reverse phase HPLC and mass spectrometry.
Fibrillation Studies—To probe the effect of CK1 phosphorylation on the aggregation of α-syn, WT α-syn was phosphorylated for 24 h at 30 °C, and the reaction was stopped with EDTA disodium salt before the samples were subjected to fibrillation conditions at 37 °C with continuous shaking for the indicated time points. The unphosphorylated controls were subjected to the same conditions, but CK1 was not added. For fibrillation studies the proteins were dissolved in 20 mm Bis-Tris propane (Aldrich), 100 mm lithium chloride (Aldrich), pH 7.4, at a concentration of 70 μm, and fibril formation was monitored by ThT using an Analyst Fluorescence instrument (LJL Biosystems) as described previously (18).
Sedimentation and Gel-filtration Separation of Monomeric α-Syn from Fibrillar Material—The degree of α-syn oligomerization was measured by determining the amount of soluble monomeric and oligomeric α-syn in solution at the indicated time points during the aggregation process. 75-μl aliquots withdrawn at the indicated time points from a main α-syn sample incubating at 37 °C were centrifuged at 18,000 × g for 30 min at 4 °C to remove any fibrils. The supernatant was then separated from the pelleted fibrils, and 30 μl were injected on an analytical Waters 2795 system equipped with a Waters 996 photodiode array detector and using an analytic Superdex 200 PC 3.2/30 column (Amersham Biosciences). The supernatant was also run on SDS-PAGE gels and stained with Coomassie Blue staining to verify the stability of the protein (see below).
Gel Electrophoresis (SDS-PAGE) and Immunoblotting—α-Syn samples were diluted in loading buffer and separated on 12% SDS 1-mm gels. Gels were stained with Simply Blue safe stain (Invitrogen) or silver staining (Invitrogen) according to the manufacturer's instructions. For dot blot analysis, the proteins were deposited onto nitrocellulose membrane (Omnilab SA), and the membrane was blocked with Odyssey blocking buffer (Li-COR Biosciences GmbH) diluted 1:3 in PBS (Sigma) for 1 h at room temperature. The membrane was probed with the primary antibody (mouse monoclonal anti-α-syn (121-125) (211) at a dilution of 1:500 (Santa Cruz Biotechnology) or mouse anti-α-syn (15-123) at a dilution of 1:1000 (BD Transduction) or mouse monoclonal anti-phosphorylated α-syn Ser-129 at a dilution of 1:5000 (Wako) or rabbit polyclonal anti-phosphorylated α-syn Ser-87 at a dilution of 1:100) at room temperature for 1-2 h. After 4 PBST (PBS, 0.01% (v/v) Tween 20 (Fluka)) washes, the membrane was incubated with the secondary antibody (i.e. goat anti-mouse ALEXA Fluor 680) protected from light at room temperature for 1 h. The immunoblot was finally washed 4 times with PBST and 3 times with PBS and scanned in a Li-COR scanner at a wavelength of 700 nm.
TEM—For TEM studies, WT or mutant α-syn samples were deposited on Formvar-coated 200 mesh copper grids (Electron Microscopy Sciences) at a concentration of 25 μm. Grids were washed with 2 drops of water and stained with 2 drops of freshly prepared 0.75% (w/v) uranyl acetate (Electron Microscopy Sciences). Specimens were inspected on a Philip CIME 12 electron microscope, operated at 80 kV. Digitized photographs were recorded with a slow scan CCD camera (Gatan, Model 679).
Reverse Phase HPLC—Monitoring the phosphorylation reaction and purification of the various phosphorylated species of α-syn (i.e. monophosphorylated or diphosphorylated α-syn) was accomplished using reverse phase HPLC on a Waters 2795 instrument equipped with a Waters 996 photodiode array detector and using an analytical (4.6-mm inner diameter) or semi-preparative (10-mm inner diameter) C4 or C18-RP column (Vydac). A 35-50% linear gradient over (flow rate = 1 ml/min) (0.09% (v/v) trifluoroacetic acid (solution A), 90% (v/v) acetonitrile aqueous solution (solution B)) was applied, and the signal was monitored at 214 nm. In the case of the semi-preparative reverse phase HPLC, a 40-60% linear gradient over (flow rate = 5 ml/min) solution A, solution B was applied.
Preparation of Large Unilamellar Vesicles and Small Unilamellar Vesicles and α-Syn-Liposome Complexes—1-Palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt) (POPG) (Avanti Polar Lipids Inc.) was purchased in chloroform which was removed by evaporation and lyophilization. The residual phospholipid was hydrated with 50 mm HEPES (Fluka), 150 mm NaCl (Fluka), pH 7.4, solution, giving rise to a phospholipid suspension of 10 mg/ml. To increase the efficiency of large unilamellar vesicles formation, 10 cycles of freezing in dry ice and thawing at 37 °C water bath were carried out. Small unilamellar vesicles were prepared by extrusion through a 100-nm polycarbonate membrane (Avestin Inc.) according to the manufacturer's instructions. The small unilamellar vesicles were stored at 4 °C and used within 3-5 days. The appropriate amount and volume of α-syn in PBS or sodium phosphate, pH 7.5, was mixed with the appropriate volume of POPG to generate a mass ratio of α-syn:POPG of 1:20. The α-syn-liposome complex was incubated for 2 h at room temperature before CD spectroscopy.
Circular Dichroism—The average secondary structure of monomeric α-syn was determined by CD spectroscopy using a Jasco 810 Spectrometer. The Far UV-CD spectra (190-250 nm, integration time of 2 s for 0.2 nm) were collected at room temperature in a 1-mm path length quartz cuvette containing 0.1 mg/ml of α-syn in PBS or sodium phosphate buffer.
Matrix-assisted Laser Desorption Ionization Time-of-flight (MALDI-TOF) Mass Spectrometry—For two layer sample preparation, MALDI matrices and calibrants were from Sigma/Fluka (Schnelldorf, Germany), trifluoroacetic acid was from Pierce, and sinapinic acid and 2,5-dihydroxybenzoic acid were from Fluka. Cytochrome c and apomyoglobin were from Sigma. 2-μl aliquots of each collected peaks were reserved for the MALDI analysis. A two-layer sample preparation was selected for the molecular weight analysis. Two matrix solutions were prepared; a saturated solution of sinapinic acid in methanol (matrix solution 1) and a 14 mg/ml sinapinic acid solution in 0.1% trifluoroacetic acid, acetonitrile (1/1) (matrix solution 2). A thin matrix layer was first generated on the mirror-polished target using a gel loader tip with matrix solution 1. 1 μl of sample was mixed with 1 μl of sinapinic acid matrix solution, and 0.8 μl of this mixture was deposited on top of the thin layer and allowed to air dry. Samples were deposited twice and analyzed with a 4700 MALDI-TOF/TOF instrument (Applied Biosystems). 8000 laser shots were typically summed by random sampling of the surface to generate the spectra that were calibrated externally on the pseudomolecular peak of cytochrome c, and apomyoglobin was prepared with the same matrix and deposited close to the samples.
NMR—NMR samples of free α-syn contained ∼0.1 mm 15N-labeled WT or mutant α-syn in 90% H2O, 10% D2O, 50 mm phosphate buffer at pH 7.4, 100 mm NaCl. NMR experiments were acquired on Bruker Avance 600 and 700 MHz NMR spectrometers. The temperature was set to 15 °C. At higher temperatures, NMR resonances particularly in the N-terminal domain were broadened. NMR data were processed and analyzed using NMRPipe (19) and Sparky 3 (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) and NMR-View (20). Spectra were referenced indirectly to sodium 2,2-dimethyl-2-silapentane-5-sulfonate and ammonia using the known chemical shift of water. Tentative assignments for the spectra of mutant and phosphorylated proteins were obtained by transferring each previously assigned cross-peak in the 1H,15N heteronuclear single quantum coherence (HSQC) spectrum of the WT protein to the nearest unassigned cross-peak in each new spectrum. Subsequently, they were verified by three-dimensional HNHA and three-dimensional NOE-HSQC spectra. Normalized weighted average chemical shift differences for amide 1H and 15N chemical shifts were calculated according to Δav = [(Δδ2HN +Δδ2N/25)/2]1/2. 3J(HN,Hα) scalar couplings were measured in WT α-syn, phosphorylated S87A α-syn, and S129D α-syn using intensity modulated HSQC experiments. Pulse field gradient NMR experiments were acquired on unlabeled WT and mutant α-syn (200 μm) dissolved in 99.9% D2O, 50 mm phosphate buffer, pH 7.4, and containing dioxane (∼20 mm) as an internal radius standard and viscosity probe (21). Twenty one-dimensional 1H spectra were collected as a function of gradient amplitude employing the PG-SLED sequence (21, 22). Each experiment was repeated at least twice. The gradient strength was increased from 1.69 to 33.72 Gauss/cm in a linear manner. Each 1H spectrum comprised 32 scans plus 16 steady-state scans. 16,000 complex points were acquired with a spectral width of 6000 Hz. Signals corresponding to the aliphatic region of the 1H spectra (3.3-0.5 ppm) were integrated, and diffusion data (signal intensity versus gradient strength) were fitted to Gaussian functions using XWINNMR (Bruker Instruments, Karlsruhe, Germany). Stokes radii of α-syn were calculated from the apparent diffusion coefficients of α-syn and dioxane and the known Stokes radius of dioxane (22). Errors in stokes radii estimated from repeat measurements are about 0.3 Å. For WT and phosphorylated S87A α-syn, paramagnetic relaxation enhancement profiles were derived from the measurement of peak intensity ratios between two 1H,15N HSQC spectra in the presence (Ipar) and absence (Idia) of the nitroxide radical.
NMR for SDS Micelle-bound α-Syn—NMR experiments on SDS micelle-bound α-syn were performed on samples containing 120 μm α-syn. Lyophilized S87A, either unphosphorylated or phosphorylated, was dissolved in sample buffer (100 mm NaCl, 10 mm Na2HPO4, pH 7.4, in 90%, 10% H2O, D2O) with 40 mm SDS. Two-dimensional 1H,15N HSQC spectra were recorded on a Varian INOVA spectrometer operating at a 1H resonance frequency of 600MHz and a sample temperature of 40 °C for all samples.
RESULTS
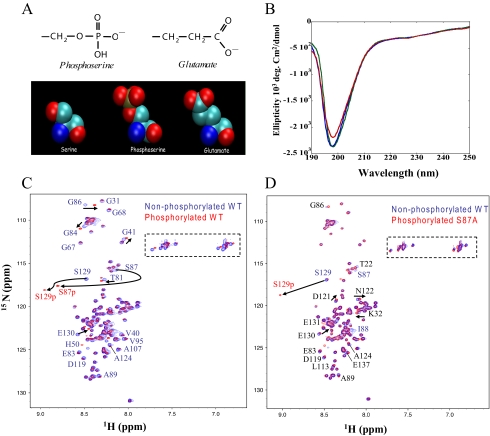
Generation of Ser(P)-129 and the Phosphomimics S129E/D—The structural and electrostatic similarities between Glu/Asp and phospho-Ser suggest that this type of substitution represents a reasonable approach to mimic constitutive phosphorylation at a specific Ser residue (Fig. 1A). The phosphate moiety at pH 7 exists predominately in the fully deprotonated state, resulting in a net charge of -2, in contrast to the single negative charge contributed by Glu/Asp. To determine whether a substitution of Ser-129 by an acidic residue is sufficient to mimic the effect of phosphorylation, we prepared mutants in which Ser-129 was replaced by Glu (S129E) or Asp (S129D). To provide mechanistic insight into the in vivo studies performed with the phosphorylation mimics, the S129A variant was also prepared. The S129E, S129D, and S129A mutations were introduced within the human cDNA sequence of SNCA by two-step PCR mutagenesis, and the mutant proteins were expressed in Escherichia coli and purified to greater than 95% homogeneity, as verified by SDS-PAGE and mass spectrometry.
FIGURE 1.
α-Syn is disordered independent of phosphorylation. A, the atomic structures of the side chains of Glu and phospho-Ser demonstrate the structural and electrostatic similarities between the two moieties. B, CD spectra of WT (blue line), S129A (green line), and S129E (red line) α-syn (10 μm). C, comparison of two-dimensional 1H,15N HSQC spectra of unphosphorylated WT (blue) and phosphorylated WT (red) α-syn. D, comparison of two-dimensional 1H,15N HSQC spectra of unphosphorylated S87A (blue) and phosphorylated S87A (red) α-syn. Resonance assignments are indicated with residue numbers. A dashed rectangle marks Gln and Asn side-chain resonances.
To determine whether mutation of Ser-129 into a charged residue is sufficient to reproduce the effect of phosphorylation on the structural and aggregation properties of α-syn in vitro, we prepared the Ser(P)-129 α-syn using CK1. We found that CK1 phosphorylates α-syn more efficiently than casein kinase II (supplemental Fig. 2). To block CK1-mediated phosphorylation at Ser-87, the S87A mutant of α-syn was expressed, purified, and phosphorylated in vitro to generate S87A/Ser(P)-129 α-syn (supplemental Fig. 3). To verify that phosphorylation by CK1 occurs at Ser-129, we made use of the anti-phospho-Ser-129 antibody (supplemental Fig. 4). Ser(P)-129 can be detected as early as the first 3 h of incubation (data not shown) and increases with time and increasing CK1 concentration.
α-Syn Is Disordered Independent of Phosphorylation—To elucidate the consequences of phosphorylation on the structure and dynamics of monomeric α-syn, we performed a series of CD measurements and high resolution NMR studies. NMR resonances in 1H,15N HSQC spectra of WT and S87A α-syn, unphosphorylated and phosphorylated, were recorded at 15 °C. For all proteins, the resonances were sharp and showed only a limited dispersion of chemical shifts, reflecting a high degree of backbone mobility (Fig. 1). Chemical shifts in WT and S87A α-syn in the unphosphorylated form were highly similar except in direct vicinity of the mutation site (supplemental Fig. 4). Upon phosphorylation of WT α-syn by CK1, the resonances of Ser-87 and Ser-129 were strongly attenuated at the position seen in the HSQC of the unphosphorylated protein. At the same time, new signals appeared in the region in which resonances of phosphorylated amino acids are usually found (Figs. 1, C and D, and supplemental Fig. 1). Three-dimensional NMR spectra assigned the new signals to Ser-87 and Ser-129. For S87A α-syn, phosphorylation at Ser-87 was blocked such that only the resonance of Ser-129 was attenuated at its original position and appeared at its phosphorylated position (Fig. 1D). Other chemical shift changes induced by phosphorylation were generally small. Detailed analysis, however, showed that the chemical shifts of residues down to residue 90 were influenced by phosphorylation of Ser-129 in S87A α-syn. In addition, weak chemical shift changes were observed for the 60 N-terminal residues (Fig. 2C) in S87A α-syn, in agreement with recent studies (23).
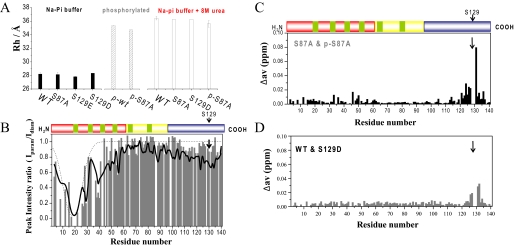
FIGURE 2.
Phosphorylation disrupts long-range interactions in monomeric α-syn. A, hydrodynamic radii of various α-syn mutants in phosphate buffer at 15 °C with and without 8 m urea. B, comparison of paramagnetic broadening of amide protons between unphosphorylated WT α-syn (black) and phosphorylated S87A α-syn (gray). In both cases the paramagnetic MTSL spin label was attached to residue 18, which was mutated from Ala to Cys. HSQC spectra in the presence (paramagnetic) and absence (diamagnetic) of spin label were recorded at 15 °C, and the intensity ratio of the resonance peaks was determined. Dashed lines indicate paramagnetic effects expected for a random coil polypeptide. C and D, normalized weighted average 1H,15N chemical shift differences between unphosphorylated and phosphorylated S87A α-syn (C) and between WT and S129D α-syn (D).
3J(HN,Hα) scalar couplings are highly sensitive to the dihedral angles of the protein backbone. We measured 3J(HN,Hα) couplings in WT α-syn and phosphorylated S87A α-syn. 3J(HN,Hα) couplings were mostly close to their random coil values and were very similar for unphosphorylated WT and phosphorylated Ser-87α-syn (data not shown), indicating that phosphorylation has no apparent effect on the secondary structure of α-syn. Far UV-CD spectra of unphosphorylated WT, S129E, S129D, S129A, and S87A α-syn and phosphorylated WT and S87A α-syn were virtually identical and consistent with a predominantly random coil structure (Figs. 1B and 3).
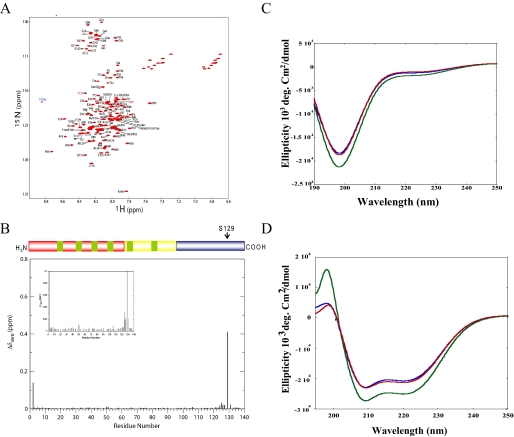
FIGURE 3.
Phosphorylation at Ser-129 does not alter the structure of micelle-bound α-syn. A, comparison of two dimensional 1H,15N HSQC spectra of unphosphorylated (black) and phosphorylated (red) S87A α-syn bound to detergent micelles. B, normalized weighted average chemical shift differences between phosphorylated and unphosphorylated S87A α-syn in its micelle-bound state. C, CD spectra of WT (blue line), S87A (green line), and S87A/Ser(P)-129 (red line) α-syn (10 μm). D, circular dichroism spectroscopy of WT (blue line), S87A (green line), and S87A/Ser(P)-129 (red line) α-syn in the presence of POPG vesicles. α-syn, POPG mass ratio is 1:20.
Phosphorylation Expands the Ensemble of Conformations Populated by α-Syn—Pulse field gradient NMR experiments allow accurate determination of the diffusion coefficient of a molecule. From the diffusion coefficient, a hydrodynamic radius Rh can be calculated that provides an estimation of the overall dimensions of a biomolecule. For WT α-syn, we determined a hydrodynamic radius of 28.2 Å (Fig. 2A). Upon the addition of 8 m urea, it increased to 36.4 Å. For S87A α-syn, the corresponding values were 28.1 and 36.2 Å, respectively. If α-syn were a true random coil, a hydrodynamic radius of 36.9 Å would be expected. When WT α-syn was phosphorylated, its hydrodynamic radius increased by 7.1 to 35.3 Å (Fig. 2A). Similarly, phosphorylation of S87A α-syn at Ser-129 increased Rh by 6.6 to 34.7 Å (Fig. 2A). The addition of 8 m urea to phosphorylated S87A α-syn only slightly further increased the Rh value to 35.6 Å, indicating that phosphorylation extends the ensemble of conformations populated by α-syn close to its fully random coil-like dimensions.
Phosphorylation Disrupts Transient Intramolecular Long-range Interactions—To probe the effect of phosphorylation on transient long-range interactions, we measured paramagnetic relaxation enhancement of NMR resonances. The interactions between a specifically attached paramagnetic nitroxide radical and nearby (less than ∼25 Å) protons cause broadening of their NMR signals because of an increase in transverse relaxation rate (24). This effect has an r-6 dependence on the electron-proton distance and, thus, allows the measurement of long-range distances between the spin label and the affected amide protons in proteins (25). Because the primary sequence of α-syn lacks cysteine, Ala-18 was mutated into a cysteine to provide an attachment point for the nitroxide radical MTSL. Neither the mere introduction of the mutation nor the addition of the MTSL radical modified the hydrodynamic radius or altered the time course of aggregation for α-syn (17). Fig. 2B compares the paramagnetic relaxation enhancement profile observed for unphosphorylated WT and phosphorylated S87A α-syn. In unphosphorylated WT α-syn, the profile of intensity ratios showed a broad paramagnetic effect extending to residue 60 and long-range interactions with C-terminal residues 115-140, in agreement with previous measurements (17). In phosphorylated S87A α-syn, the paramagnetic broadening was largely restricted to the vicinity of the spin label; signal intensities ratios for residues 45-60 and for C-terminal residues 120-135 approached values close to one.
To probe the influence of phosphorylation at Ser-129 on backbone dynamics, we measured 15N spin relaxation times in WT and S87A α-syn. For most of the backbone amides of unphosphorylated WT and S87A α-syn, steady-state heteronuclear 1H,15N NOEs were below 0.3 (supplemental Fig. 7) (15). Residues 68-80 and 90-97 showed 1H,15N NOE values slightly below average, as previously noted (15). Phosphorylation at Ser-129 did not induce strong changes in the NOE profile, indicating that pico-to-nanosecond motions of the backbone of α-syn were largely unaffected. Similarly, 15NR1ρ relaxation rates that are sensitive to motions of the backbone occurring on the micro- to millisecond time scale were largely unaffected by phosphorylation (data not shown).
S129E/D Do Not Reproduce the Structural Consequences of Phosphorylation—To evaluate if mutation into a charged residue can mimic the effect of phosphorylation on the conformation of α-syn, we characterized monomeric S129D and S129E α-syn by NMR spectroscopy. The 1H,15N HSQC spectra of S129D/E α-syn was highly similar to that observed for the WT protein. Chemical shift changes were restricted to the direct vicinity of the mutation site, and 3J(HN,Hα) were largely unchanged (Fig. 2D and supplemental Fig. 6). Using pulse field gradient NMR experiments, we determined hydrodynamic radii of 28.3 and 27.8 Å for S129D and S129E α-syn (Fig. 2A), respectively, values very similar to WT protein (Rh = 28.2 Å) but significantly smaller than that observed for phosphorylated α-syn (Rh = 35.3 Å). Thus, mutation of Ser-129 into aspartate or glutamate does not disrupt the long-range interactions that stabilize WT α-syn.
Phosphorylation at Ser-129 Does Not Alter the Structure of Micelle-bound α-Syn—To investigate potential effects of Ser-129 phosphorylation on the structure of membrane-bound α-syn, NMR 1H,15N HSQC spectra of SDS micelle-bound Ser-129-phosphorylated S87A α-syn were compared with equivalent spectra of unphosphorylated protein (Fig. 3, A and B). The conformation of the micelle-bound protein is thought to effectively mimic that of the membrane-bound form of the protein (26). Significant chemical shift changes were observed only near the site of phosphorylation, indicating that Ser-129 phosphorylation does not lead to large scale structural changes of this conformation of the protein (Fig. 3B). In the SDS micelle-bound state, no long range interactions occur between the C-terminal and the lipid binding N-terminal domain (26-28), and thus, no long range effects of Ser-129 phosphorylation are expected. There is some evidence, however, that membrane binding subtly influences the conformation of the C-terminal tail of α-syn (28, 29), and a potential perturbation of such an effect by Ser-129 phosphorylation cannot be excluded based on these two-dimensional NMR spectra alone.
α-Syn is known to undergo a random coil to α-helical transformation upon binding to phospholipids. To determine the effect of phosphorylation at Ser-129 on α-syn-lipid interactions, we examined the secondary structure of the purified monophosphorylated S87A (S87A/Ser(P)-129) in the absence (Fig. 3C) and presence (Fig. 3D) of phospholipids by CD. In all cases we observed that phosphorylation does not perturb the overall conformation of S87A (Fig. 3C) and its ability to adopt α-helical conformations upon binding to synthetic vesicles (Fig. 3D). Detailed studies are currently under way to accurately quantify the effect of phosphorylation on membrane binding by multiple methods.
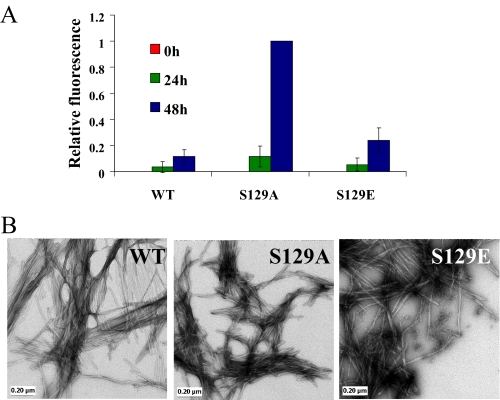
S129E Exhibits Similar in Vitro Aggregation Properties as the WT—To determine the effect of the phospho-mimicking mutations on the aggregation properties of α-syn, we compared the aggregation propensity of S129E and S129A to that of the WT protein as a function of time using the ThT binding assay (Fig. 4A) and TEM (Fig. 4B). In a few experiments, the S129E variant formed slightly more fibrils than WT protein (Fig. 4B), but these differences are not statistically significant, and in other experiments there were no differences in amyloid formation between the two proteins (WT and S129E). In contrast, we consistently observed that the S129A fibrillates more rapidly and forms significantly more fibrils than both the WT and the phosphorylation mimic, S129E. Both S129A and S129E α-syn formed classical amyloid-like fibrils with morphological features similar to that of WT α-syn (Fig. 4B).
FIGURE 4.
S129E exhibits similar in vitro aggregation properties as the WT protein, but S129A forms significantly more fibrils than WT. A, ThT fluorescence was monitored at indicated time points from samples of 100 μm solutions of WT, S129A, and S129E α-syn incubated at 37 °C. The error bars represent the S.D. of four independent experiments. B, TEM images of WT, S129A, and S129E α-syn after 48 h of aggregation (scale bar, 0.20 μm).
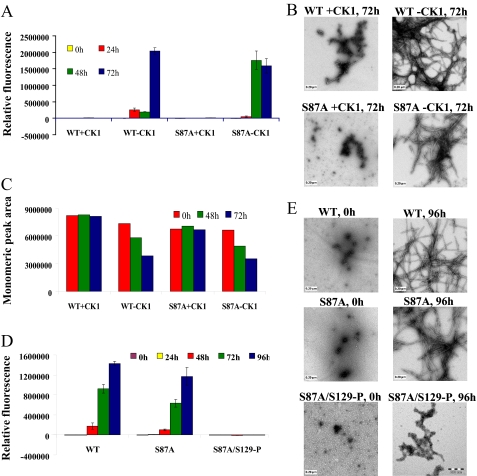
CK1-mediated Phosphorylation Inhibits α-Syn Aggregation—To investigate the effect of in vitro phosphorylation on α-syn fibril formation, WT α-syn was co-incubated with CK1 in the appropriate reaction buffer at 30 °C for 24 h, after which the samples were transferred to 37 °C, and fibril formation was monitored by ThT and TEM and compared with control samples that were subjected to the same experimental conditions in the absence of the kinase. We consistently observed that CK1-mediated phosphorylation significantly inhibits α-syn fibril formation, even after 48 h of incubation under agitating conditions as discerned from the ThT (Fig. 5A) and TEM studies (Fig. 5B). To verify the ThT and TEM results, we also determined the amount of soluble α-syn species at each time point using SEC by monitoring the area of the peak corresponding to the monomer. Fig. 5C shows that the amount of monomeric α-syn in samples subjected to CK1 phosphorylation remains unchanged even after 72 h of incubation under fibril formation conditions, whereas the amount of monomeric unphosphorylated α-syn decreased with time, consistent with the TEM and ThT results. In all samples, we did not observe any accumulation of stable soluble oligomers that can be separated by SEC, although we cannot rule out the possibility of oligomer dissociation in the column.
FIGURE 5.
CK1-mediated phosphorylation at Ser-129 inhibits the fibrillation of WT and S87Aα-syn. A, ThT fluorescence measurements of CK1-phosphorylated WT and S87A α-syn and their unphosphorylated controls (100 μm); results are the average of three readings ± S.D. B, TEM images of phosphorylated and unphosphorylated S87A α-syn after 72 h of aggregation at 37 °C (scale bar, 0.20 μm). C, graph representing the area of monomeric peak after 0, 48, and 72 h of aggregation at 37 °C. D, ThT fluorescence measurement of WT, S87A, and S87A/Ser(P)-129 α-syn as a function of time (average of three readings ± S.D.). E, TEM images of WT, S87A, and S87A/Ser(P)-129 α-syn at 0 h and after 96 h of aggregation at 37 °C (scale bar, 0.20 μm).
Phosphorylation at Ser-129 Is Sufficient to Inhibit the Aggregation of α-Syn—Previous studies showed that CK1 phosphorylates α-syn at Ser-87 and Ser-129 (30, 31). To determine the relative contribution of phosphorylation at Ser-129 to the CK1-induced inhibition of α-syn fibril formation, we examined the effect of CK1-mediated phosphorylation on the fibrillation of S87A mutant of α-syn. Prephosphorylation of S87A with CK1 results in significant inhibition of α-syn fibrillation relative to the unphosphorylated forms of the protein. After 72 h of incubation under agitation conditions, S87A formed significant amount of amyloid fibrils, whereas the prephosphorylated form of S87A (S87A/Ser(P)-129) showed predominantly soluble α-syn species (Fig. 5, A and B). Consistent with the TEM and ThT results, the amount of monomeric prephosphorylated S87A (S87A/Ser(P)-129) remained unchanged during the course of the aggregation experiment (72 h), whereas the amount of monomeric unphosphorylated S87A decreased significantly with time (Fig. 5C).
To further demonstrate that inhibition of α-syn fibrillogenesis is due to phosphorylation at Ser-129, we purified the phosphorylated form of S87A (S87A/Ser(P)-129) (supplemental Fig. 3) and compared its aggregation properties to that of S87A and WT α-syn. When purified WT, S87A and S87A/Ser(P)-129 α-syn were subjected to aggregation conditions at 37 °C with continuous agitation, and S87A/Ser(P)-129 did not form any fibrils and remained in a predominantly monomeric state even after 96 h of incubation, whereas the unphosphorylated S87A and WT α-syn exhibited high levels of fibrillation as demonstrated by the increased ThT fluorescence (Fig. 5D) and the significant decrease in levels of monomeric species with time (SEC) (Fig. 5C) and detection of fibrillar structures by TEM (Fig. 5E). However, longer incubations (>10 days) of S87A/Ser(P)-129 resulted in the formation of ThT-fluorescent aggregates that are morphologically distinct from those formed by unphosphorylated S87A and WT α-syn (supplemental Fig. 8).
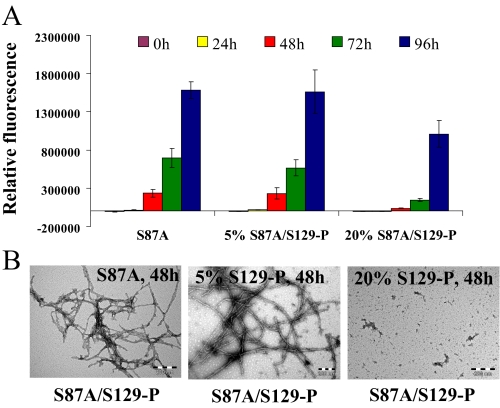
S87A/Ser(P)-129 Inhibits the Fibrillation of S87A in a Dose-dependent Manner—Given that only ∼5% of α-syn is thought to be phosphorylated at Ser(P)-129 under physiological conditions (32), we sought to determine whether the two forms (Ser-129 and Ser(P)-129) of S87A α-syn interact and the consequences of such interaction on the aggregation of α-syn. In the presence of 5% of S87A/Ser(P)-129 (mimicking physiological conditions), we did not observe any significant differences in the fibrillation of S87A. However, in the presence of 20% S87A/Ser(P)-129, we observed significant inhibition of S87A α-syn fibrillation as discerned by TEM, ThT, and SEC (Fig. 6 and supplemental Fig. 9). The aggregation of S87A containing 20% of S87A/Ser(P)-129 α-syn was reduced by ∼85% after 48 h, 80% after 72 h, and 40% after 96 h of aggregation, whereas S87A containing 5% of S87A/Ser(P)-129 α-syn aggregated to similar level as S87A at all time points. These findings suggest that inhibition of S87A aggregation by S87A/Ser(P)-129 is concentration-dependent.
FIGURE 6.
S87A/Ser(P)-129 interacts with S87A and inhibits its aggregation in a dose-dependent manner. S87A α-syn (70 μm) was mixed with the appropriate volume of 70 μm S87A/Ser(P)-129 to generate samples containing 5 and 20% of the latter protein. A, ThT fluorescence measurement of the 70 μm S87A, S87A containing 5% S87A/Ser(P)-129 α-syn and S87A containing 20% S87A/Ser(P)-129 α-syn (average of 3 readings ± S.D.). B, TEM images of S87A and S87A containing 5 or 20% S87A/Ser(P)-129 after 48 h of aggregation at 37 °C (scale bar, 0.20 μm).
DISCUSSION
Phosphorylation of α-syn at Ser-129 has been implicated in the pathogenesis of PD and related synucleinopathies. However, the exact mechanisms by which phosphorylation modifies the physiological and pathogenic properties of α-syn in vivo remain unknown. To understand the structural basis underlying the effect of phosphorylation on the oligomerization and fibrillation properties of α-syn, we probed the effect of phosphorylation at Ser-129 on the conformational and dynamic properties of monomeric α-syn.
Phosphorylation at Ser-129 Disrupts Transient Long-range Interactions in Monomeric α-Syn—Previous studies demonstrated that monomeric α-syn assumes conformations that are stabilized by long-range interactions and act to inhibit oligomerization and aggregation (17, 33). Polyamine binding and temperature increase, conditions that induce aggregation in vitro, release this inherent tertiary structure (17). NMR spectroscopy also showed that high concentrations of chemical denaturants disrupt the autoinhibitory conformations of monomeric α-syn, leading to an extended conformation (Fig. 2A). Here we investigated the effect of phosphorylation on the stabilizing interactions of α-syn. Hydrodynamic radii determined by NMR spectroscopy demonstrated that phosphorylation of α-syn at Ser-129 strongly increased the hydrodynamic radius of α-syn (Fig. 2A). In addition, paramagnetic relaxation enhancement of amide protons and chemical shifts showed that long-range interactions were disrupted upon phosphorylation at Ser-129 (Fig. 2). Thus, phosphorylation strongly perturbs the ensemble of conformations populated by monomeric α-syn in solution. These perturbations may influence both its pathogenic behavior as well as any physiological function of α-syn.
Ser-129 is located outside the region of α-syn that is known to modulate its interaction with membranes. Therefore, phosphorylation at these residues is not expected to significantly disrupt α-syn membrane interactions. Phosphorylation at Ser-129 did not prevent the binding of α-syn to liposomes nor did it interfere with the formation of helical structure upon binding, as indicated by the CD data (Fig. 3). Furthermore, the precise conformation of the protein in its micelle-bound state is also essentially unaltered by Ser-129 phosphorylation, with NMR chemical shift changes restricted to the immediate vicinity of the phosphorylation site within the acidic C-terminal tail and not extending into the helical lipid binding domain. This indicates that any influence of Ser-129 phosphorylation on the normal function of α-syn, which is believed to be associated with its membrane-bound conformation, is likely to be mediated by the effects of the modification on protein-protein interactions involving the C-terminal tail.
S129E/D Do Not Reproduce the Structural Consequences of Phosphorylation at Ser-129—Although phosphorylation-mimicking mutations (S→ E and S→ D) are commonly used to probe the structural and functional consequences of protein phosphorylation, a direct comparison between the phosphorylation mimics and the corresponding phosphorylated form of the protein is rarely investigated. Our in vitro studies demonstrate that the phosphomimics S129E and S129D do not reproduce the effect of phosphorylation on the structural and aggregation properties of α-syn in vitro. Mutation of Ser-129 into Asp or Glu did not lead to an extended conformation of monomeric α-syn (Fig. 2A and 7). Only the conformation and dynamics of residues close to the site of mutation were influenced, indicating that mutation of Ser or Thr residues to Asp or Glu cannot fully mimic the effect of phosphorylation on the structure and dynamics of α-syn and potentially other intrinsically disordered proteins. Together, these findings highlight the critical importance of the increased negative charge and/or bulkiness of the phosphate group relative to the carboxylate of Glu and Asp in mediating the structural and functional consequences of phosphorylation. Several studies have reported that protein-protein interactions mediated by phosphoserines are abolished when serine is replaced by glutamate. In some of these studies it was shown that the bulkiness, steric hindrance (34), or conformational changes (35) induced by the phosphate group, rather than its negative charge, is responsible for the observed effects of phosphorylation. On this basis, one can speculate that phosphorylation of α-syn may constitute an important and specific regulator of α-syn aggregation and interactions with other proteins. These observations have important implications for use of phosphomimics to elucidate the role of phosphorylation in modulating biological processes in vivo and underscore the importance of exercising caution when interpreting results obtained using phospho-mimicking mutants.
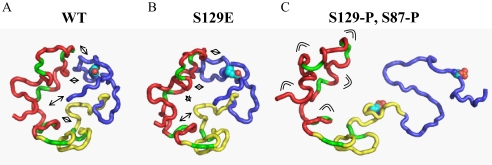
FIGURE 7.
Schematic illustration of the disruption of long-range interactions in α-syn upon phosphorylation at Ser-129. A, representative member of the native state ensemble of conformations populated by α-syn (17). The basic N-terminal region, the highly amyloidogenic NAC region, and the acidic C-terminal domain are shown in red, yellow, and blue, respectively. The side chain of Ser-129 is shown. Long-range interactions are indicated by arrows. B, phosphomimics do not mimic the structural consequences of phosphorylation. Same as B, but the side chain at position 129 was replaced by a Glu. C, upon phosphorylation at Ser-129, long-range interactions are disrupted, and the ensemble of conformations is extended. The higher flexibility of the N-terminal domain is indicated.
Mechanistic and Therapeutic Implications for PD—A detailed molecular understanding of the functional consequences of phosphorylation may reveal new insight into the normal biology of α-syn and the mechanisms by which it contributes to neurodegeneration in PD and related disorders. The first attempt to address the effect of phosphorylation on α-syn aggregation and toxicity was carried out by Feany and co-worker (7) in a Drosophila model of PD overexpressing WT, S129D, or S129A. Interestingly, the aggregation properties of the phosphomimics (S129D and S129E) and WT α-syn in vivo (Drosophila) bears striking resemblance to what we observed in vitro, with WT and S129E exhibiting the same number of inclusion, whereas S129A forms more (4×) aggregates than WT and S129E (Fig. 4A). The S129D mutant showed enhanced toxicity and reduced levels of α-syn inclusions, whereas expression of the S129A mutant suppressed dopaminergic cell loss and increased the amount of α-syn aggregates formed relative to the WT or S129D mutant. One possible explanation for these observations is that Ser(P)-129 may influence the formation and accumulation of toxic oligomeric species of α-syn rather than of mature fibrils, which may in fact be protective. However, our in vitro studies are not entirely consistent with such a hypothesis, since we observe that Ser-129 phosphorylation inhibits both mature fibril formation and also the conversion of monomeric α-syn to oligomeric species.
In addition to modulating the oligomerization and fibrillogenesis of α-syn, phosphorylation may be involved in regulating its physiological properties by modulating its interactions with other neuronal proteins and/or synaptic vesicles and phospholipids. For example, α-syn interacts with the microtubule associated protein tau and stimulates tau phosphorylation (36) and fibrillogenesis (37) both in vitro and in vivo. The tau binding site was mapped to the C-terminal region (amino acids 87-140) of α-syn. Phosphorylation at Ser-129, but not Tyr-125 was also reported to reduce the rate of α-syn transport (38). Together, these findings suggest that reversible phosphorylation within the C-terminal region (Tyr-125 or Ser-129) may be involved in regulating the association/dissociation with tau and other neuronal proteins (e.g. tau, synphilin (39), phospholipase D (31, 40), 14-3-3 (41), metals (3), and lipids. Phosphorylation within the C-terminal region (Ser-129 or Tyr-125) or the incorporation of phosphorylation mimicking mutations at these residues also reportedly reduces membrane binding and blocks α-syn-mediate inhibition of phospholipase D2 (30, 31, 40), an enzyme involved in the hydrolysis of phosphatidylcholine and vesicular trafficking. Although our results demonstrate that Ser(P)-129 α-syn is capable of binding to membranes in an apparently unperturbed conformation (compared with the WT protein), we have not measured the effects of phosphorylation on membrane affinity, which may indeed be reduced.
Does Phosphorylation of α-Syn Occur before or after LB Formation?—Our findings that monophosphorylated α-syn is capable of forming ThT positive fibril-like aggregates, albeit after longer incubations, could explain the presence of Ser(P)-129 α-syn within LBs. The morphology of the fibrils formed by Ser(P)-129 is distinct from those formed by the unphosphorylated protein (supplemental Fig. 8); however, the pathogenic consequences of this difference in structure or morphology has not been investigated. Further studies are required to determine the molecular and cellular determinants that influence the aggregation of phosphorylated α-syn and whether phosphorylation precedes aggregation or occurs after fibrillation and LB formation. Recent studies have shown that Ser(P)-129 α-syn accumulations colocalize with several kinases known to phosphorylate α-syn at this residue, including CK2 (42) and GRK5 (43) in LBs. These findings combined with the fact that the C-terminal region containing Ser-129 remains exposed in the aggregated forms (protofibrils and fibrils) of α-syn support the hypothesis that phosphorylation of α-syn could also occur within LBs and is not a prerequisite for α-syn fibrillation and LB formation in PD.
Together, our findings suggest that phosphorylation may constitute an important regulator of α-syn aggregation and toxicity. Thus, the identification of the kinases and phosphatases involved in regulating α-syn phosphorylation could result in new viable targets for small molecule-based therapeutic strategies for PD and related synucleinopathies. Several kinases have emerged as viable therapeutic targets for neurodegenerative diseases, including Alzheimer disease (44), and are likely to be the focus of intense efforts because of the wealth of knowledge and expertise on the development of kinase inhibitors.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AG019391 (from NIA). This work was also supported by the Swiss Federal Institute of Technology Lausanne (to H. A. L., K. E. P., and A. W. S.), Swiss National Science Foundation Grant 310000-110027 (to H. A. L.), by the Max Planck society (to M. Z. and C. O. F.), a Deutsche Forschungsgemeinschaft Heisenberg scholarship to M. Z. (ZW 71/2-1 and 3-1), Agencia de Promocion Cientifica y Tecnologica, Fundación Antorchas (to C. O. F.), Alexander von Humboldt Foundation (to C. O. F.), and the Irma T. Hirschl Foundation, and a gift from Herbert and Ann Siegel (to D. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-9.
Footnotes
The abbreviations used are: α-syn, α-synuclein; PD, Parkinson disease; SEC, size exclusion chromatography; ThT, thioflavin T; TEM, transmission electron microscopy; MTSL, 1-oxy-2, 2, 5, 5-tetramethyl-d-pyrroline-3-methyl)-methanethiosulfonate; CK, casein kinase; POPG, 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt); MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; HSQC, heteronuclear single quantum coherence; NOE, nuclear Overhauser effect; LB, Lewy body; WT, wild type; HPLC, high performance liquid chromatography; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; PBS, phosphate-buffered saline.
References
- 1.Trojanowski, J. Q., and Lee, V. M. (2003) Ann. N. Y. Acad. Sci. 991107 -110 [DOI] [PubMed] [Google Scholar]
- 2.Abeliovich, A., Schmitz, Y., Farinas, I., Choi-Lundberg, D., Ho, W. H., Castillo, P. E., Shinsky, N., Verdugo, J. M., Armanini, M., Ryan, A., Hynes, M., Phillips, H., Sulzer, D., and Rosenthal, A. (2000) Neuron 25239 -252 [DOI] [PubMed] [Google Scholar]
- 3.Cooper, A. A., Gitler, A. D., Cashikar, A., Haynes, C. M., Hill, K. J., Bhullar, B., Liu, K., Xu, K., Strathearn, K. E., Liu, F., Cao, S., Caldwell, K. A., Caldwell, G. A., Marsischky, G., Kolodner, R. D., Labaer, J., Rochet, J. C., Bonini, N. M., and Lindquist, S. (2006) Science 313324 -328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Outeiro, T. F., and Lindquist, S. (2003) Science 3021772 -1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahle, P. J., Neumann, M., Ozmen, L., and Haass, C. (2000) Ann. N. Y. Acad. Sci. 920 33-41 [DOI] [PubMed] [Google Scholar]
- 6.George, J. M., Jin, H., Woods, W. S., and Clayton, D. F. (1995) Neuron 15 361-372 [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., and Feany, M. B. (2005) Nat. Neurosci. 8657 -663 [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara, H., Hasegawa, M., Dohmae, N., Kawashima, A., Masliah, E., Goldberg, M. S., Shen, J., Takio, K., and Iwatsubo, T. (2002) Nat. Cell Biol. 4160 -164 [DOI] [PubMed] [Google Scholar]
- 9.Anderson, J. P., Walker, D. E., Goldstein, J. M., de Laat, R., Banducci, K., Caccavello, R. J., Barbour, R., Huang, J., Kling, K., Lee, M., Diep, L., Keim, P. S., Shen, X., Chataway, T., Schlossmacher, M. G., Seubert, P., Schenk, D., Sinha, S., Gai, W. P., and Chilcote, T. J. (2006) J. Biol. Chem. 28129739 -29752 [DOI] [PubMed] [Google Scholar]
- 10.Kahle, P. J., Neumann, M., Ozmen, L., Muller, V., Jacobsen, H., Spooren, W., Fuss, B., Mallon, B., Macklin, W. B., Fujiwara, H., Hasegawa, M., Iwatsubo, T., Kretzschmar, H. A., and Haass, C. (2002) EMBO Rep. 3583 -588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, M., Kanuka, H., Fujiwara, H., Koyama, A., Hasegawa, M., Miura, M., and Iwatsubo, T. (2003) Neurosci. Lett. 336155 -158 [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, M., Fujiwara, H., Nonaka, T., Wakabayashi, K., Takahashi, H., Lee, V. M., Trojanowski, J. Q., Mann, D., and Iwatsubo, T. (2002) J. Biol. Chem. 27749071 -49076 [DOI] [PubMed] [Google Scholar]
- 13.Smith, W. W., Margolis, R. L., Li, X., Troncoso, J. C., Lee, M. K., Dawson, V. L., Dawson, T. M., Iwatsubo, T., and Ross, C. A. (2005) J. Neurosci. 255544 -5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessler, J. C., Rochet, J. C., and Lansbury, P. T., Jr. (2003) Biochemistry 42 672-678 [DOI] [PubMed] [Google Scholar]
- 15.Bussell, R., Jr., and Eliezer, D. (2001) J. Biol. Chem. 27645996 -46003 [DOI] [PubMed] [Google Scholar]
- 16.Hoyer, W., Antony, T., Cherny, D., Heim, G., Jovin, T. M., and Subramaniam, V. (2002) J. Mol. Biol. 322383 -393 [DOI] [PubMed] [Google Scholar]
- 17.Bertoncini, C. W., Jung, Y. S., Fernandez, C. O., Hoyer, W., Griesinger, C., Jovin, T. M., and Zweckstetter, M. (2005) Proc. Natl. Acad. Sci. U. S. A. 1021430 -1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lashuel, H. A., Petre, B. M., Wall, J., Simon, M., Nowak, R. J., Walz, T., and Lansbury, P. T., Jr. (2002) J. Mol. Biol. 3221089 -1102 [DOI] [PubMed] [Google Scholar]
- 19.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 20.Johnson, B. A., and Blevins R. A. (1994) J. Biomol. NMR 4603 -614 [DOI] [PubMed] [Google Scholar]
- 21.Wilkins, D. K., Grimshaw, S. B., Receveur, V., Dobson, C. M., Jones, J. A., and Smith, L. J. (1999) Biochemistry 3816424 -16431 [DOI] [PubMed] [Google Scholar]
- 22.Jones, J. A., Wilkins, D. K., Smith, L. J., and Dobson C. M. (1997) J. Biomol. NMR 10 199-203 [Google Scholar]
- 23.Sasakawa, H., Sakata, E., Yamaguchi, Y., Masuda, M., Mori, T., Kurimoto, E., Iguchi, T., Hisanaga, S. I., Iwatsubo, T., Hasegawa, M., and Kato, K. (2007) Biochem. Biophys. Res. Commun. 363795 -799 [DOI] [PubMed] [Google Scholar]
- 24.Gillespie, J. R., and Shortle, D. (1997) J. Mol. Biol. 268158 -169 [DOI] [PubMed] [Google Scholar]
- 25.Kosen, P. A., Scheek, R. M., Naderi, H., Basus, V. J., Manogaran, S., Schmidt, P. G., Oppenheimer, N. J., and Kuntz, I. D. (1986) Biochemistry 252356 -2364 [DOI] [PubMed] [Google Scholar]
- 26.Eliezer, D., Kutluay, E., Bussell, R., Jr., and Browne, G. (2001) J. Mol. Biol. 3071061 -1073 [DOI] [PubMed] [Google Scholar]
- 27.Bussell, R., Jr., and Eliezer, D. (2003) J. Mol. Biol. 329763 -778 [DOI] [PubMed] [Google Scholar]
- 28.Ulmer, T. S., Bax, A., Cole, N. B., and Nussbaum, R. L. (2005) J. Biol. Chem. 2809595 -9603 [DOI] [PubMed] [Google Scholar]
- 29.Tamamizu-Kato, S., Kosaraju, M. G., Kato, H., Raussens, V., Ruysschaert, J. M., and Narayanaswami, V. (2006) Biochemistry 4510947 -10956 [DOI] [PubMed] [Google Scholar]
- 30.Okochi, M., Walter, J., Koyama, A., Nakajo, S., Baba, M., Iwatsubo, T., Meijer, L., Kahle, P. J., and Haass, C. (2000) J. Biol. Chem. 275390 -397 [DOI] [PubMed] [Google Scholar]
- 31.Pronin, A. N., Morris, A. J., Surguchov, A., and Benovic, J. L. (2000) J. Biol. Chem. 27526515 -26522 [DOI] [PubMed] [Google Scholar]
- 32.Hirai, Y., Fujita, S. C., Iwatsubo, T., and Hasegawa, M. (2004) FEBS Lett. 572227 -232 [DOI] [PubMed] [Google Scholar]
- 33.Dedmon, M. M., Lindorff-Larsen, K., Christodoulou, J., Vendruscolo, M., and Dobson, C. M. (2005) J. Am. Chem. Soc. 127476 -477 [DOI] [PubMed] [Google Scholar]
- 34.Corbit, K. C., Trakul, N., Eves, E. M., Diaz, B., Marshall, M., and Rosner, M. R. (2003) J. Biol. Chem. 27813061 -13068 [DOI] [PubMed] [Google Scholar]
- 35.Tabor, S., and Richardson, C. C. (1985) Proc. Natl. Acad. Sci. U. S. A. 821074 -1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen, P. H., Hager, H., Nielsen, M. S., Hojrup, P., Gliemann, J., and Jakes, R. (1999) J. Biol. Chem. 27425481 -25489 [DOI] [PubMed] [Google Scholar]
- 37.Frasier, M., Walzer, M., McCarthy, L., Magnuson, D., Lee, J. M., Haas, C., Kahle, P., and Wolozin, B. (2005) Exp. Neurol. 192274 -287 [DOI] [PubMed] [Google Scholar]
- 38.Saha, A. R., Hill, J., Utton, M. A., Asuni, A. A., Ackerley, S., Grierson, A. J., Miller, C. C., Davies, A. M., Buchman, V. L., Anderton, B. H., and Hanger, D. P. (2004) J. Cell Sci. 1171017 -1024 [DOI] [PubMed] [Google Scholar]
- 39.Lee, G., Tanaka, M., Park, K., Lee, S. S., Kim, Y. M., Junn, E., Lee, S. H., and Mouradian, M. M. (2004) J. Biol. Chem. 2796834 -6839 [DOI] [PubMed] [Google Scholar]
- 40.Payton, J. E., Perrin, R. J., Woods, W. S., and George, J. M. (2004) J. Mol. Biol. 3371001 -1009 [DOI] [PubMed] [Google Scholar]
- 41.Ostrerova, N., Petrucelli, L., Farrer, M., Mehta, N., Choi, P., Hardy, J., and Wolozin, B. (1999) J. Neurosci. 195782 -5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu, M. Y., Kim, D. W., Arima, K., Mouradian, M. M., Kim, S. U., and Lee, G. (2007) J. Neurol. Sci. 266 9-12 [DOI] [PubMed] [Google Scholar]
- 43.Arawaka, S., Wada, M., Goto, S., Karube, H., Sakamoto, M., Ren, C. H., Koyama, S., Nagasawa, H., Kimura, H., Kawanami, T., Kurita, K., Tajima, K., Daimon, M., Baba, M., Kido, T., Saino, S., Goto, K., Asao, H., Kitanaka, C., Takashita, E., Hongo, S., Nakamura, T., Kayama, T., Suzuki, Y., Kobayashi, K., Katagiri, T., Kurokawa, K., Kurimura, M., Toyoshima, I., Niizato, K., Tsuchiya, K., Iwatsubo, T., Muramatsu, M., Matsumine, H., and Kato, T. (2006) J. Neurosci. 269227 -9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klafki, H. W., Staufenbiel, M., Kornhuber, J., and Wiltfang, J. (2006) Brain 1292840 -2855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.