Abstract
The human immunodeficiency virus type 1 (HIV-1) capsid protein plays a critical role in virus core particle assembly and is an important target for novel therapeutic strategies. In a previous study, we characterized the binding affinity of a hydrocarbon stapled helical peptide, NYAD-1, for the capsid protein (Kd ∼ 1 μm) and demonstrated its ability to penetrate the cell membrane (Zhang, H., Zhao, Q., Bhattacharya, S., Waheed, A. A., Tong, X., Hong, A., Heck, S., Goger, M., Cowburn, D., Freed, E. O., and Debnath, A. K. (2008) J. Mol. Biol. 378, 565–580). In cell-based assays, NYAD-1 colocalized with the Gag polyprotein during traffic to the plasma membrane and disrupted the formation of mature and immature virus particles in vitro systems. Here, we complement the cellular and biochemical data with structural characterization of the interactions between the capsid and a soluble peptide analogue, NYAD-13. Solution NMR methods were used to determine a high resolution structure of the complex between the inhibitor and a monomeric form of the C-terminal domain of the capsid protein (mCA-CTD). The intermolecular interactions are mediated by the packing of hydrophobic side chains at the buried interface and unperturbed by the presence of the olefinic chain on the solvent-exposed surface of the peptide. The results of the structural analysis provide valuable insight into the determinants for high affinity and selective inhibitors for HIV-1 particle assembly.
Worldwide 30 million people are infected with human immunodeficiency virus type 1 (HIV-1),2 and it has claimed more lives than some of the deadliest epidemics in human history. HIV-1 belongs to the retroviral family, and significant progress made in understanding its life cycle has fueled the development of diverse therapeutic strategies. Important targets for intervention include inhibiting the fusion of the virus at the surface of CD4+T cells, reverse transcription of the viral RNA, and processing of the gag polyprotein by the HIV-1 protease (1). However, none of these treatment strategies has proven to be fully effective against the rapid emergence of drug-resistant variants of the virus.
As we gain further insight into the structural biology of the virus particle itself and the mechanism of its assembly from the gag polyprotein, the latter has emerged as an important new target for drug development (2). The newly synthesized gag protein migrates to the inner surface of the cell membrane, where it buds into an immature particle that encapsulates the genomic material and important viral proteins. Subsequent proteolytic cleavage of the gag protein into the matrix protein, capsid, and nucleocapsid protein lead to repacking of the core and the formation of the mature virus particle. The interactions involving the capsid protein play a crucial role in the formation of the virus core particle with the correct morphology and are an important target for disrupting the assembly step.
The dimeric capsid protein consists of N- and C-terminal domains that are flexibly linked in each monomer, and dimerization is mediated by the C-terminal domain. In mature particles, the capsid protein assembles into a fullerene-like structure with densely packed hexamers of the capsid protein. The formation of the hexamer is mediated by the N-terminal domain of the capsid, and the hexameric rings are bridged by the C-terminal dimerization domain (3, 4). The first target for disrupting the CA assembly was the identification of small molecules that could bind to the N-terminal domain of the capsid protein (5, 6). Subsequently, a short peptide (CAI) bound to the C-terminal dimerization domain of the capsid protein was shown to disrupt the formation of the budding virus particle in vitro (7, 8). The possible clinical application of these compounds is limited by weak binding affinity, toxicity and, in the case of CAI, low membrane permeability.
The lack of insight into the mechanism of inhibition by these small molecules was remedied by the recently published cryo-electron microscopy image reconstruction of the capsid assembly (4). We now know that both CAP-1 and CAI bind onto a surface that could potentially disrupt the interactions between the N- and C-terminal domains of two different CA molecules packed in the hexameric rings of the lattice (4, 6, 9).
In a previous study starting with the sequence of CAI, we presented the structural basis for a redesigned peptide NYAD-1 that is cell-permeable and has a 10-fold higher affinity for CA-CTD (Kd ∼ 1 μm) (10). In the x-ray structure of CAI in complex with CA-CTD, the peptide forms an amphipathic helix with the hydrophobic surface buried at the complex interface (8). The location of charged residues Asp-4 and Glu-8 on the solvent-exposed surface of the peptide was ideal for chemical modification by hydrocarbon stapling, a well known technique for stabilizing the secondary structure of helical peptides by cross-linking the i and i + 4 positions with an olefinic chain via β-alanine analogues (13). The olefinic chain is also known to enhance cell permeability of peptides and has been used quite successfully for designing an activator from the BID (Bcl-2 interacting protein) BH3 domain for BAX (Bcl-2 associated X protein) (11) and reactivation of p53 tumor suppressor pathway by a stapled p53 peptide (12). Residues Asp-4 and Glu-8 from CAI were replaced with a non-standard amino acid (S)-2-(2′-pentenyl) alanine whose side chains were then cross-linked with an olefinic bond (13) (Scheme 1). As anticipated, the redesigned peptide NYAD-1 formed a stable helix in solution and was cell-permeable. Fluorescent tagging showed the peptide colocalized with the gag protein in the cell and specifically disrupted the formation of mature HIV-1 particles in a dose-dependent manner (IC50 ∼ 4–15 μm).
SCHEME 1.
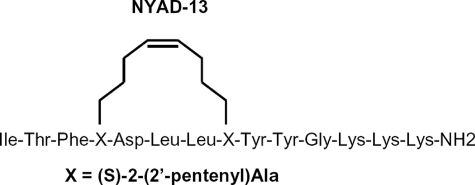
Schematic representation of the NYAD-13 peptide structure. The non-standard amino acid, (S)-2-(2′-pentenyl) alanine, is indicated by X in the amino acid sequence of the peptide.
The binding site of the peptide was mapped from the back-bone chemical shift differences and shown to correspond closely to the site identified in the x-ray structure of CAI in complex with CA-CTD (7, 8). The rationally designed model predicts the presence of the bulky linker on the distal site of the peptide, not altering the binding site of CA-CTD, but perturbations in the peptide structure might still be apparent.
In this report, we extend the previously published biochemical data to the complete structural characterization of the complex formed between a soluble homologue NYAD-13 and the monomeric form of the C-terminal domain of the capsid protein. The peptide NYAD-13 was derived from NYAD-1 by replacing the proline at the C-terminal with three lysines. The rationale for the substitution was two-fold; although the C-terminal end of CAI is not engaged in intermolecular interactions, the positive charges would increase the solubility of the peptide NYAD-13 and facilitate structural studies. The principal objective of determining a high resolution NMR structure of the complex was to probe the role of the olefinic chain in the bound peptide structure and its impact on the binding mode. It is our goal to employ the analysis of the NMR ensemble to obtain a more dynamic view of the CA-CTD binding site and identify the determinants of binding affinity of the HIV-1 capsid domain. The structural insights are expected to help us in optimizing the peptide-based inhibitors with respect to both affinity and specific antiviral activity.
EXPERIMENTAL PROCEDURES
Peptide Synthesis—The NYAD-13 peptide was synthesized by replacing the fourth (i) and eight (i + 4) residues of the linear peptide CAI ITFEDLLDYYGKKK by the olefin-bearing unnatural amino acid (S)-2-(2′-pentenyl) alanines stapled by the olefin metathesis using the Grubbs catalyst (13). The helical conformation of the free peptide was confirmed (data not shown) from circular dichroism (10). In the NMR assignment and structure, the peptide is numbered 1–14.
NMR Samples—Uniformly 15N/13C-enriched protein samples of mCA-CTD were produced by expressing the pET14b plasmid encoding the gene in Escherichia coli BL21 (DE3) cells cultured in M9-minimal medium containing 15NH4Cl (Cambridge Isotope Laboratories) and [13C6]glucose as sole nitrogen and carbon source, respectively. Recombinant proteins were isolated from bacteria as described previously (10), and the integrity of the samples was confirmed by mass spectrometry. NMR samples were prepared from U-15N,13C-labeled C-CA sample complexed with unlabeled NYAD-13 peptide dissolved in 100 mm NH4Ac buffer, 95% H2O, 5% D2O, and 2 mm dithiothreitol at pH 7.0. The molecular concentrations were estimated from the UV absorbance at 280 nm with extinction coefficients of 2,980 m–1 cm–1 (peptide) and 3,105 M–1 cm–1 (protein), respectively. Resonance assignments were made on a sample containing 400 μl of 1.9 mm U-15N,13C-labeled mCA-CTD complexed with 1.8 mm unlabeled NYAD-13 in the acetate buffer. NOE data were collected on two samples containing 600 μm U-15N,13C-labeled mCA-CTD complexed with 900 μm unlabeled peptide in 90% H2O, 10% D2O, and 100% D2O. In the NMR assignment and structure, the CA protein is numbered from the natural sequence, 147–231 GI: 2801 504.
NMR Resonance Assignments—NMR data were acquired at 25 °C on Bruker AVANCE spectrometers equipped with Z axis gradient triple axis CryoProbes. A standard suite of backbone (HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO) and side-chain experiments (HCCCONH, HCCH-TOCS, HCCH-COSY) was acquired at 500 MHz for chemical shift assignments (14, 15). Distance restraints were obtained from 13C-edited three-dimensional NOESY-HSQC (100-ms mixing time) optimized for aliphatic and aromatic resonances and acquired on both H2O and D2O samples. Information for the amide region was obtained from a 15N-edited three-dimensional NOESY-HSQC acquired at 800 MHz.
The peptide NYAD-13 was assigned using two-dimensional 15N,13C f1, f2-filtered NOESY acquired at 900 MHz (16). Inter-molecular NOEs were obtained from two-dimensional 15N,13C f2-filtered NOESY at 900 MHz and three-dimensional 15N,13C f1-filtered 13C-edited NOESY-HSQC experiment recorded with 100-ms mixing time at 800 MHz on both H2O and D2O samples (17).
Structure Calculations—NMR data were processed in Topspin 1.3 from Bruker Biospin and analyzed using CARA1.5 (18). The structure of the peptide-bound complex was calculated in two stages using the programs CYANA 2.1 (19, 20) for the automatic assignment of the NOE peak lists, and a final water refinement was done in ARIA 2.2 (21). The upper limits of the distance restraints were calculated from the NOESY cross-peak intensities using the calibration routine of CYANA, and the dihedral angles were obtained from TALOS (22). The upper limit for the intermolecular distances were initially set to 4.5 and 6.0 Å but subsequently scaled to minimize the target function without increasing restraint violations. During the CYANA runs, the peptide was calculated without the olefinic linker, and a lysine was used instead of the non-standard amino acid (S)-2-(2′-pentenyl) alanine. The ensemble of the 20 best structures had a backbone r.m.s.d. of 0.36 ± 0.06 Å (residues 128–218, 3–10) and an average target function of 0.78 ± 0.06. There were no distance violations greater than 0.2 Å and no dihedral violations greater than 5°. The CYANA-generated distance and angle restraints were converted into the CNS format in CCPN (23). The structure for the non-standard amino acid linker was generated and energy-minimized in PRODRG2 (24). The new definitions of the bond lengths and angles were introduced in topollhdg5.3.pro and parallhdg5.3.pro files for incorporation during the CNS annealing run and are available in the supplemental material. A total of 1,000 structures were calculated, and the 200 lowest energy structures were subjected to water refinement and further analysis by PROCHECK_NMR (25). The structural statistics of the 20 best structures are reported in Table S1.
RESULTS AND DISCUSSION
The wild type C-terminal domain of the HIV-1 capsid protein forms a symmetric dimer with a Kd ∼ 18 μm (26). The dimer interface is stabilized by hydrophobic interactions involving the side chains of Trp-184 and Met-185 and salt bridges (26). Dimerization of the capsid domain is completely abolished by mutating either one of these sites (Trp-184 and Met-185) to alanine, and the defective variants inhibited assembly of mature particles and infectivity (27). Despite the loss of functionality, the recently published NMR structure of mCA-CTD (28) suggests that the monomer structure is not perturbed significantly when compared with the dimer. The remarkable stability of the monomeric form has been attributed to hydrophobic packing and a network of hydrogen bonds formed by conserved residues belonging to the MHR motif (26). It has been previously reported that the dimer in solution exists as multiple conformations that interchange on the slower time scale (7). The functional relevance of the flexible dimer interface can be rationalized as a structural requirement in the packing of the hexameric rings mediated by the CA-CTD (4, 26). In solution, binding of the peptide NYAD-13 to CA-CTD does not quench the conformational flexibility of the dimer entirely,3 and using the analogy of the CAI complex structure, we can speculate that the peptide destabilizes rather than dissociates the dimer (7). The absence of any change in line width between the free and the peptide-bound forms of CA-CTD further supported this conclusion. To meet the immediate goal of this study and circumvent the problem of differential line-broadening from conformational exchange, we decided to work with the monomeric form of the mutant capsid protein.
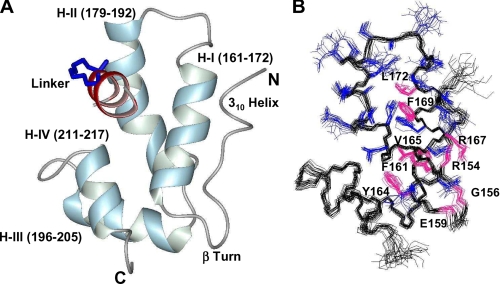
Structure of NYAD-13 in Complex with mCA-CTD—The monomeric form of CA-CTD as shown in Fig. 1 consists of a four helix bundle (161–172, 179–192, 196–205, 211–217) stabilized by the hydrophobic core of the protein. The N-terminal region has a short 3–10 helix (150–152) and a strand (153–158) connected by a type I β turn (156–159) to helix I. The extreme C-terminal residues 219–231 form an unstructured floppy tail. The absence of residual structure at the C terminus was confirmed by backbone amide-proton relaxation experiments.
FIGURE 1.
A, the helical representation of a single structure of mCA-CTD (148–221) and NYAD-13 (2–11). The secondary structure consists of an N-terminal 310 helix, a type 1 β-turn, and a four-helix bundle. B, the side chains (blue) from residues in helix I and helix II are represented in the ensemble of NMR structures. For clarity, the peptide has been removed from the structure. Conserved residues from the MHR motif are colored in magenta. The structural representations were generated in MOLMOL 2.1 (30).
Helix I, which is also part of the highly conserved MHR motif, plays a crucial role in stabilizing the helical scaffold of the protein. Important hydrophobic contacts are manifested in a large number of long range NOEs involving the aromatic rings of Phe-161, Tyr-164, and Tyr-169 and the methyl groups of Val-165 and Leu-172 (Fig. 1B). The tight packing of side chains in the buried core of the protein is reinforced by the absence of Tyr-164 ring flips on the fast NMR time scale. In the peptide complexed structure of mCA-CTD, helix 2 is very well defined with a kink starting at Thr-188, and this is in contrast to the partial loss in structure reported in previous studies (28, 29). Binding of the peptide thus appears to stabilize the secondary structure of helix 2.
In the x-ray structure of CA-CTD (PDB code 1AUM), conserved residues from the MHR motif, Arg-167, Glu-159, Gly-156, and Arg-154, participate in a network of hydrogen bonds vital to the stability of the capsid fold (26). Although we did not see evidence of these hydrogen bonds in all our structures, we did observe the following donor-acceptor pairs (GluHN-159–GlyCO-156, ArgHH21-167–ArgCO-154) within hydrogen-bonding distance in at least 40% of the ensemble.
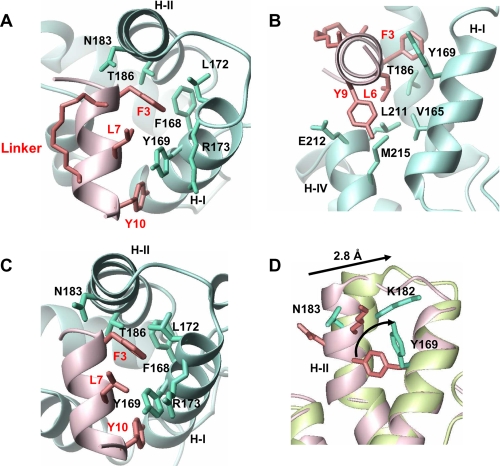
The peptide binding site on the protein is a shallow hydrophobic pocket lined by residues from all four helices. The bound peptide (residues 3–11) adopts a helical conformation where the hydrophobic side chains make extensive intermolecular contacts with specific direct experimental confirmation from NOEs. The long axis of the peptide is perpendicular to both helix I and helix II with the N terminus buried against helix II and stabilized by the capping interactions involving Thr-2 and Asn-183 (8). The importance of Asn-183 at this position is supported by observing several NOEs between Asn-183 side-chain protons and Thr-2 and Phe-3 resonances. The peptide forms an amphipathic helix that buries several hydrophobic side chains of Phe-3, Leu-6, Tyr-9, and Tyr-10 at the interface of the complex. The most important residue appears to be the aromatic ring of Phe-3, which inserts itself partly between helix I and helix II without disrupting the core packing of the protein. Phe-3 makes a large number of important contacts with the side chains of Tyr-169, Leu-172, Arg-173, and Thr-186 (Fig. 2A). The aromatic ring of Phe-3 appears to partially fill a hydrophobic cavity and is a strong candidate for substitution that could potentially disrupt the core structure. Leu-6 anchors the lower surface of the peptide through interactions with Val-165 and Leu-211 (Fig. 2B). The partly solvent-exposed side chain of Tyr-9 on the lower surface of the peptide interacts with Leu-211 and Glu-212. The ring of Tyr-10 packs against the side chains of Val-165 and Tyr-169 and is important for anchoring the C-terminal end of the helical peptide. The unstructured C-terminal end of the peptide (11GKKK14) is completely exposed to the solvent and does not participate in intermolecular interactions.
FIGURE 2.
Structural details of intermolecular contacts with ribbon representation of the protein (blue) and peptide (pink) backbone. A, the top view of the binding surface displays the interactions between the side chains of Phe-3 and Tyr-10 from the peptide and helix I and II of mCA-CTD. B, the side view of the complex displays the interactions that anchor Leu-6 and Tyr-9 from the peptide using Leu-211 and Met-215 from helix IV. C, the top view of the x-ray structure of CAI in complex with CA-CTD (2BUO). D, superposition of the backbone Cα atoms of CA-CTD (pink) and mCA-CTD (green) based on alignment generated from residues in helix I, helix III, and helix IV (r.m.s.d. = 0.8 Å). When helix II is included, the r.m.s.d. increases to 1.3 Å. Residues that are important for binding the target peptide and rearranged through the helix movement are indicated in the figure. The PDB code for CA-CTD structure used in the alignment is 1A8O. The figures were generated in MOLMOL 2.1 (30).
The olefinic linker connecting positions 4 and 8 on the solvent-accessible surface of the peptide did not yield any intermolecular NOEs. In the final ensemble of structures, the Asn-183 side chain was found to be closest in distance to the linker, but this interaction was not observed in the NOE data. There are no previous published structures of a stapled peptide using this linker, so the chemical structure of the hydrocarbon ring was confirmed by the intra-ring NOEs. The eight-membered carbon component is sufficiently long that no kinking is needed. In the 20 lowest target structures, the Cα-Cα distances for Lya-4—Lya-8 ranged from 6 to 8 Å. The range of flexibility of the induced helix and of the ring is likely restricted by the trans-olefinic bond, as indicated by observed NOEs to the side chain of Asp-5, and this will be the subject of additional future investigation.
In summary, the interactions between NYAD-13 and mCA-CTD are mediated by hydrophobic contacts, and the presence of the olefinic chain on the exposed surface of the peptide does not perturb these interactions. The principal effect of the olefinic chain is to impose constraints on the backbone of NYAD-13, and the flexibly linked peptide side chains can easily adapt to the binding surface of mCA-CTD.
Structural Comparison with CA-CTD and CAI+CA-CTD Complex—The overall similarities between the three-dimensional structures of mCA-CTD in complex with NYAD-13 and CA-CTD (PDB code 2BUO) in the presence and absence of CAI are remarkably high (Fig. 2). The most significant differences between the three structures are observed in the relative position of helix II, which plays a critical role in dimer formation (26). The backbone Cα atom r.m.s.d. of all four helices when compared with CA-CTD in the absence of CAI is 0.8 Å, whereas the superposition of helix I (161–172), helix III (196–205), and helix IV (211–217) is better than 0.4 Å. When complexed to NYAD-13, the C terminus of helix II in mCA-CTD is displaced by 2.8 Å when compared with the free protein (Fig. 2D). This repositioning of helices I and II results in a much larger (6 Å) movement in the wild type CA-CTD complexed with CAI (8). This movement of helix II is also shown to be coupled to a decrease in the buried surface area at the dimer interface and predicted to destabilize the structure of the dimer (8).
In the complex of mCA-CTD with NYAD-13, the movement of helix II appears to be restricted and may simply reflect the need to preserve the packing between the two long helices in the absence of the stabilizing effect of the dimerization contacts. As such, the rotation of helix II does not trigger a major conformational change in the hydrophobic core of the protein, but the flipping of side chains essentially remolds the topology of the binding surface. In support of an induced fit mechanism, the coupled rearrangement of several side chains creates new cavities and provides access to buried residues. The importance of these changes is highlighted in Fig. 2D. The aromatic ring of Tyr-169 flips by 90°, which facilitates the docking of Phe-3 into a newly created cavity lined by Leu-172 and Phe-168. The lateral movement of helix II also removes Lys-182 from the N terminus of the peptide, and this position is now occupied by Asn-183 important for capping interactions.
The similarities in the packing of side chains at the intermolecular surface of the NYAD-13 and CAI complexes suggest a common mechanism of binding that relies on the recognition of hydrophobic side chains. The packing of the aromatic rings of Phe-3, Tyr-9, and Tyr-10 play a crucial role in anchoring the peptide to the protein. The relative positions and average orientation of the rings is preserved in the two structures. We do see a difference in the stereochemistry of the methyl groups of the two leucines. The side chain of Leu-6 is completely buried at the complex interface and thus favors a single rotamer in NYAD-13 but is flipped when compared with CAI. This is in contrast to Leu-7, which is not constrained by interactions on the solvent-exposed surface of the peptide. In conclusion, the relative flexibility of the helix I/II interface and the floppiness of side chains presented by helix I at the binding surface play a critical role in eliminating steric clashes and maximizing the buried apolar surface area in the complex.
The total buried surface (1,346 Å2) in the NYAD-13-bound complex of mCA-CTD is distributed equally between mCA-CTD (635 Å2, 10% of SA surface) and the peptide (712 Å2, 40% of SA). These numbers are remarkably close to those of CA-CTD bound to CAI (8). Therefore, the 10-fold higher affinity of NYAD-13 (Kd ∼ 1 μm) for mCA-CTD when compared with CAI for mCA-CTD (Kd ∼ 15 μm) can simply be attributed to a loss of unfavorable entropic contributions as the stapled peptide is already folded into a helical conformation. The issue of in vivo effectiveness of the related NYAD-1 likely reflects the difference of cell permeability and of stability rather than of structural interaction.
In summary, the hydrocarbon stapling renders the biologically active peptide inhibitor NYAD-1 cell-permeable and enhances the binding affinity for CA-CTD (10). In this study, the biochemical data are complemented with a high resolution NMR structure of mCA-CTD in complex with NYAD-13 and provides understanding of the structural role of the linker. Confirming the absence of interactions between the linker and mCA-CTD was therefore the first step in refining the structure of NYAD-13 as a legitimate drug candidate for HIV-1. Future efforts will focus on improving the peptide solubility and binding affinity without sacrificing some of its favorable drug like properties.
A strategic goal relevant to future design efforts is to understand the molecular basis for specificity of such peptide inhibitors against retroviruses in general. In the previous study, we have shown the peptide analogue NYAD-1 to be active only in inhibiting mature and immature HIV particle assembly and not against EIAV particles (10). In the sequence alignment of p24 domains (supplemental material), the highly conserved residues belong to the MHR motif, but elsewhere, there are few similarities. In the EIAV sequence, the two substitutions (Y169L and N183K) in the hydrophobic binding surface could dictate the ability to bind NYAD-1 with sufficient affinity. The conclusions drawn from the sequence analysis are highly speculative and do not include the effect of EIAV particle morphology. Structural details of the capsid formation in the presence and absence of inhibitors are crucial for addressing some of these issues.
Supplementary Material
Acknowledgments
We acknowledge the expertise of Dr. Kaushik Dutta on implementing structure calculations in ARIA 2.2. The 900 MHz system was purchased with funds from National Institutes of Health Grant P41 GM-66354, the Keck Foundation, and the Member Institutions of the New York Structural Biology Center (NYSBC).
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM 47021 and P41 GM-66354. This work was also supported by intramural funding from the New York Blood Center. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (code 2K1C) have been deposited in theProtein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures and two supplemental tables.
Footnotes
The abbreviations used are: HIV-1, human immunodeficiency virus type 1; CA, capsid; CA-CTD, wild type C-terminal domain of capsid; mCA-CTD, monomeric and mutant form (W184A/M185A) of C-terminal domain of HIV-1 capsid protein; EIAV, equine infectious anemia virus; HSQC, heteronuclear single-quantum coherence; Lya, lysine-analog residue; MHR, major homology region; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; PDB, Protein Data Bank; r.m.s.d., root mean square deviation.
S. Bhattacharya, H. Zhang, A. K. Debnath, and D. Cowburn, unpublished results.
References
- 1.De Clercq, E. (2007) Nat. Rev. Drug Discov. 6 1001–1018 [DOI] [PubMed] [Google Scholar]
- 2.Li, F., and Wild, C. (2005) Curr. Opin. Investig. Drugs 6 148–154 [PubMed] [Google Scholar]
- 3.Li, S., Hill, C. P., Sundquist, W. I., and Finch, J. T. (2000) Nature 407 409–413 [DOI] [PubMed] [Google Scholar]
- 4.Ganser-Pornillos, B. K., Cheng, A., and Yeager, M. (2007) Cell 131 70–79 [DOI] [PubMed] [Google Scholar]
- 5.Tang, C., Loeliger, E., Kinde, I., Kyere, S., Mayo, K., Barklis, E., Sun, Y., Huang, M., and Summers, M. F. (2003) J. Mol. Biol. 327 1013–1020 [DOI] [PubMed] [Google Scholar]
- 6.Kelly, B. N., Kyere, S., Kinde, I., Tang, C., Howard, B. R., Robinson, H., Sundquist, W. I., Summers, M. F., and Hill, C. P. (2007) J. Mol. Biol. 373 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sticht, J., Humbert, M., Findlow, S., Bodem, J., Muller, B., Dietrich, U., Werner, J., and Krausslich, H. G. (2005) Nat. Struct. Mol. Biol. 12 671–677 [DOI] [PubMed] [Google Scholar]
- 8.Ternois, F., Sticht, J., Duquerroy, S., Krausslich, H. G., and Rey, F. A. (2005) Nat. Struct. Mol. Biol. 12 678–682 [DOI] [PubMed] [Google Scholar]
- 9.Sundquist, W. I., and Hill, C. P. (2007) Cell 131 17–19 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, H., Zhao, Q., Bhattacharya, S., Waheed, A. A., Tong, X., Hong, A., Heck, S., Goger, M., Cowburn, D., Freed, E. O., and Debnath, A. K. (2008) J. Mol. Biol. 378 565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walensky, L. D., Pitter, K., Morash, J., Oh, K. J., Barbuto, S., Fisher, J., Smith, E., Verdine, G. L., and Korsmeyer, S. J. (2006) Mol. Cell 24 199–210 [DOI] [PubMed] [Google Scholar]
- 12.Bernal, F., Tyler, A. F., Korsmeyer, S. J., Walensky, L. D., and Verdine, G. L. (2007) J. Am. Chem. Soc. 129 2456–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafmeister, C. E., Po, J., and Verdine, G. L. (2000) J. Am. Chem. Soc. 122 5891–5892 [Google Scholar]
- 14.Sattler, M., Schleucher, J., and Greisinger, C. (1999) Prog. NMR Spectrosc. 34 93–158 [Google Scholar]
- 15.Cavanagh, J., Fairbrother, W.J., III, Palmer, A. G., and Skelton, N. J. (1996) Protein NMR Spectroscopy: Principles and Practice, Academic Press, San Diego
- 16.Ikura, M., and Bax, A. (1992) J. Am. Chem. Soc. 114 2433–2440 [Google Scholar]
- 17.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenbalt, J., Konrat, R., and Kay, L. E. (1997) J. Am. Chem. Soc. 119 6711–6721 [Google Scholar]
- 18.Keller, R. (2004) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment. Ph.D. thesis, Eidgenössische Technische Hochschule (ETH) Zurich, Switzerland.
- 19.Herrmann, T., Guntert, P., and Wuthrich, K. (2002) J. Mol. Biol. 319 209–227 [DOI] [PubMed] [Google Scholar]
- 20.Güntert, P., Mumenthaler, C., and Wüthrich, K. (1997) J. Mol. Biol. 273 283–298 [DOI] [PubMed] [Google Scholar]
- 21.Nilges, M., Macias, M. J., O'Donoghue, S. I., and Oschkinat, H. (1997) J. Mol. Biol. 269 408–422 [DOI] [PubMed] [Google Scholar]
- 22.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289–302 [DOI] [PubMed] [Google Scholar]
- 23.Fogh, R., Ionides, J., Ulrich, E., Boucher, W., Vranken, W., Linge, J. P., Habeck, M., Rieping, W., Bhat, T. N., Westbrook, J., Henrick, K., Gilliland, G., Berman, H., Thornton, J., Nilges, M., Markley, J., and Laue, E. (2002) Nat. Struct. Biol. 9 416–418 [DOI] [PubMed] [Google Scholar]
- 24.Schuttelkopf, A. W., and van Aalten, D. M. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1355–1363 [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R. A., Rullman, J. A. C., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477–486 [DOI] [PubMed] [Google Scholar]
- 26.Gamble, T. R., Yoo, S., Vajdos, F. F., von Schwedler, U. K., Worthylake, D. K., Wang, H., McCutcheon, J. P., Sundquist, W. I., and Hill, C. P. (1997) Science 278 849–853 [DOI] [PubMed] [Google Scholar]
- 27.von Schwedler, U. K., Stray, K. M., Garrus, J. E., and Sundquist, W. I. (2003) J. Virol. 77 5439–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong, H. C., Shin, R., and Krishna, N. R. (2008) Biochemistry 47 2289–2297 [DOI] [PubMed] [Google Scholar]
- 29.Alcaraz, L. A., del Alamo, M., Barrera, F. N., Mateu, M. G., and Neira, J. L. (2007) Biophys. J. 93 1264–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koradi, R., Billeter, M., and Wuthrich, K. (1996) J. Mol. Graphics 14 51–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.