Abstract
Sulfated glycosaminoglycans (GAGs), including heparan sulfate and chondroitin sulfate, are synthesized on the so-called common GAG-protein linkage region (GlcUAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser) of core proteins, which is formed by the stepwise addition of monosaccharide residues by the respective specific glycosyltransferases. Glucuronyltransferase-I (GlcAT-I) is the key enzyme that completes the synthesis of this linkage region, which is a prerequisite for the conversion of core proteins to functional proteoglycans bearing GAGs. The Xyl and Gal residues in the linkage region can be modified by phosphorylation and sulfation, respectively, although the biological significance of these modifications remains to be clarified. Here we present evidence that these modifications can significantly influence the catalytic activity of GlcAT-I. Enzyme assays showed that the synthetic substrates, Gal-Gal-Xyl(2-O-phosphate)-O-Ser and Gal-Gal(6-O-sulfate)-Xyl(2-O-phosphate)-O-Ser, served as better substrates than the unmodified compound, whereas Gal(6-O-sulfate)-Gal-Xyl(2-O-phosphate)-O-Ser exhibited no acceptor activity. The crystal structure of the catalytic domain of GlcAT-I with UDP and Gal-Gal(6-O-sulfate)-Xyl(2-O-phosphate)-O-Ser bound revealed that the Xyl(2-O-phosphate)-O-Ser is disordered and the 6-O-sulfate forms interactions with Gln318 from the second GlcAT-I monomer in the dimeric enzyme. The results indicate the possible involvement of these modifications in the processing and maturation of the growing linkage region oligosaccharide required for the assembly of GAG chains.
Proteoglycans (PGs),2 which bear various sulfated glycosaminoglycan (GAG) side chains (1), are distributed in the extracellular matrix of every tissue and at the surface of every cell type (2, 3). These GAGs include chondroitin sulfate (CS), dermatan sulfate (DS), heparan sulfate (HS), and heparin (Hep). Accumulating evidence suggests that PGs function in development and pathophysiological processes. HS-PGs play critical roles in signaling pathways through specific molecular interactions with protein ligands (4, 5), such as Wingless and Hedgehog as well as fibroblast growth factors in Drosophila (5, 6) and likely in the corresponding mammalian systems as well. CS/DS-PGs have been implicated in the signaling of heparin-binding growth factors (7-11) and in the neural network formation in developing mammalian brains (12-17).
The expression of these functions of PGs must be strictly regulated by biosynthesis especially of the GAG chains, which are classified into galactosaminoglycans (CS and DS) and glucosaminoglycans (HS and Hep) (1). Both types of GAG chains are synthesized on the common GAG-protein linkage region of core proteins, GlcUAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser (18), which is formed by the stepwise addition of monosaccharide residues by the respective specific glycosyltransferases, Xyl transferase, Gal transferases I and II, and GlcUA transferase (GlcAT-I) (19). The repeating disaccharide region [(-4GlcUAβ1-4GlcNAcα1-)n] of HS/Hep is synthesized on the linkage region by the HS co-polymerase complex of EXT1 and EXT2 (20-24). In contrast, the repeating disaccharide region [(-4GlcUAβ1-3GalNAcβ1-)n] of CS/DS is synthesized on the linkage region by the enzyme complexes of bifunctional chondroitin synthases 1 and 2, and chondroitin polymerizing factor (25-27). However, the regulatory mechanism of the enzymatic transfer of the first GlcNAc and GalNAc, by which the selective assembly of CS/DS and HS/Hep on the common linkage region tetrasaccharide is controlled, remains a long-standing enigma.
It is conceivable that the biosynthesis of the linkage region tetrasaccharide is strictly regulated, because it is located at the critical determining point not only for the biosynthetic selective chain assembly of HS/Hep and CS/DS, but also for converting glycoproteins into PGs. A number of PG precursor proteins often lack GAG side chains, and hence are called part-time PGs (3, 19). However, the biosynthetic mechanism, by which a given protein containing the GAG attachment consensus amino acid sequence becomes a PG, remains obscure. It is essential to clarify the substrate specificities and regulatory mechanisms of the glycosyltransferases involved in the synthesis of the linkage region tetrasaccharide sequence. In this respect, it should be noted that the linkage region can be modified by phosphorylation and sulfation (19).
We have carried out a series of structural studies of the GAG-protein linkage region, based on the working hypothesis (29) that possible structural differences in the linkage region among the various GAG chains may exist and determine the type and/or character of the GAG species to be synthesized. These and other studies led to the identification of novel modified structures such as GlcUAβ/IdoUAα1-3Gal(4-O-sulfate) β1-3Gal(±6-O-sulfate)β1-4Xyl, GlcUAβ1-3Gal(±6-O-sulfate)β1-3Gal(±6-O-sulfate)β1-4Xyl, and GlcUAβ1-3Galβ1-3Galβ1-4Xyl(2-O-phosphate) for CS/DS chains (29-34). Remarkably, sulfated Gal residues have been demonstrated in the linkage region of CS and DS, but not in HS or Hep, whereas Xyl-2-O-phosphate has been found in both HS/Hep and CS, indicating that the sulfate groups on the Gal residues may be involved in the selective chain assembly of CS/DS, and that the modifying groups may be key elements that control the glycosyltransferases involved in the synthesis of the linkage region thereby regulating the maturation of part-time PGs (19).
We have cloned and characterized the substrate specificity of GlcAT-I (35, 36). A crystal structure of the ternary complex of the enzyme with the donor substrate product UDP and the acceptor substrate analogue, Galβ1-3Galβ1-4Xyl, revealed the key amino acid residues required for the recognition of the donor and acceptor substrates (37). Here we investigated whether the enzyme could recognize the modifying groups on the linkage region trisaccharide by conducting enzymatic analyses and x-ray crystallography to clarify the possible involvement of the 2-O-phosphate group on the Xyl residue and the 6-O-sulfate groups on the Gal residues.
EXPERIMENTAL PROCEDURES
Materials—UDP-[U-14C]GlcUA (285.2 mCi/mmol) and unlabeled UDP-GlcUA were purchased from PerkinElmer Life Sciences and Sigma (St. Louis, MO), respectively. Calf intestine alkaline phosphatase and bovine liver β-glucuronidase (EC 3.2.1.31) were purchased from Roche Molecular Biochemicals (Tokyo, Japan) and Sigma, respectively. The linkage trisaccharide serines, Galβ1-3Galβ1-4Xylβ1-O-Ser, Galβ1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser, Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser, and Gal(6-O-sulfate)β1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser were chemically synthesized (38, 39). Galβ1-3Gal(6-O-sulfate)β1-4Xylβ1-O-Ser and Gal(6-O-sulfate)β1-3Galβ1-4Xylβ1-O-Ser were prepared by digestion of Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser and Gal(6-O-sulfate)β1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser with alkaline phosphatase, respectively.
Expression of the Soluble Form of GlcAT-I and Enzyme Assay—The construction of a soluble form of GlcAT-I fused with the cleavable insulin signal sequence and the protein A IgG-binding domain was carried out as described (35, 36), except for the replacement of the expression vector pSVL with pEF-BOS (40). Each expression plasmid (10 μg) was transfected into COS-1 cells on 100-mm plates using Lipofectamine (Invitrogen) according to the instructions provided by the manufacturer. Two days after transfection, 2 ml of the culture medium was collected and incubated with 10 μl of IgG-Sepharose (Amersham Biosciences) for 1 h at 4 °C. The beads recovered by centrifugation were washed with and then re-suspended in each assay buffer, and tested for glucuronyltransferase activities using linkage region trisaccharide serines (1 nmol each) as acceptor substrates. The assay mixture contained 10 μl of the re-suspended beads, 1 nmol of a linkage trisaccharide serine, 143 μm UDP-[14C]GlcUA (1.66 × 105 dpm), 50 mm MES buffer, pH 6.5, and 2 mm MnCl2 in a total volume of 30 μl. Reaction mixtures were incubated at 37 °C for 10 min, and then radiolabeled products were separated from UDP-[14C]GlcUA by gel filtration using a Superdex Peptide column (Amersham Biosciences) equilibrated with 0.25 m NH4HCO3, 7% 1-propanol. The recovered labeled products were quantified by liquid scintillation counting.
Characterization of the Reaction Products—Isolation of the products from the GlcAT reaction using various linkage trisaccharide serines was carried out by gel filtration on a Superdex Peptide column as described above. The radioactive peak containing the product was pooled and evaporated dry. The isolated product was digested with 100 mIU of β-glucuronidase in a total volume of 50 μl of 0.1 m sodium acetate buffer, pH 4.5, for 4 h at 37 °C or with 10 units of alkaline phosphatase in a total volume of 60 μl of 0.07 m glycine/NaOH buffer, pH 9.9, containing 0.5 m MgCl2 for 4 h at 37 °C. The digest was analyzed using the same Superdex Peptide column as that noted above.
Crystallization of Enzyme and Data Collection—The catalytic domain of GlcAT-1 (Thr76-Val335) was expressed with an amino-terminal His tag, MGSSHHHHHHSSGLVPRGSHM, and purified as previously described (37). Crystals of the catalytic domain were obtained using the vapor diffusion hanging drop method. Protein at 15 mg/ml in 25 mm Hepes buffer, pH 7.5, 50 mm NaCl, 10 mm MnCl2, and 10 mm UDP-GlcUA was mixed with an equal volume of reservoir solution, consisting of 0.1 m MES buffer, pH 6.0, and 21% monomethylether polyethylene glycol 2000, for a total drop volume of 16 μl. Crystals were harvested and transferred in four steps into a cryoprotection solution containing 23% monomethylether polyethylene glycol 2000, 10 mm UDP, 10 mm MnCl2, 0.1 m MES buffer, pH 6.2, and 10% ethylene glycol. Finally the crystals were transferred into the cryoprotection solution with 20 mm Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser added, and allowed to soak overnight. The crystals were flash-frozen in a stream of nitrogen gas at -170 °C for data collection. Diffraction data were collected on a RAXIS4 image plate system using a RUH3R rotating anode x-ray source. Data were processed using DENZO and SCALPACK (41) (Table 1). CNS (42) was used for refinement of the model to the data and O (43) was used for model building and manual refinement.
TABLE 1.
Data collection and statistics
| Data statistics | |
| Source | RUH3R rotating anode |
| Wavelength (Å) | 1.542 |
| Space group | P21 |
| Unit cell | a = 57.05, b = 48.39, c = 103.71 |
| α = γ = 90.0°; β = 92.40° | |
| No. of crystals | 1 |
| Resolution (Å) | 1.9 |
| No. of observations | 164,388 |
| No. of unique reflections | 44,763 |
| Redundancy | 3.7 |
| % Completeness (last shell) | 99.6 (97.1) |
| Rsym (last shell)a | 0.06 (0.35) |
|
I/σI (last shell)
|
16.2 (6.45)
|
| Refinement statistics | |
| Resolution (Å) | 50-1.9 |
| Rcryst/Rfreeb (%) | 21.0/23.4 |
| No. of waters | 281 |
| Mean B value (Å2) | 31.0 |
| Root mean square deviation from ideal value | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 1.3 |
| Dihedral angle (°) | 23.6 |
|
Improper angle (°)
|
0.90
|
| Ramachandran statistics | |
| Residues in | |
| Most favored regions (%) | 87.9 |
| Additionally allowed regions (%) | 11.6 |
| Generously allowed regions (%) | 0.5 |
| Disallowed regions (%) | 0.0 |
Rsym = (∑|I - <I>|)/∑(I).
Rcryst = ∑||Fo| - |Fc||/∑|Fo| calculated from working data set. Rfree is calculated from 5% of data randomly chosen not to be included in refinement.
The final model contains residues 75-140 and 152-335 of molecule A, and residues 75-140 and 155-335 of molecule B. Each molecule contains one UDP, one Mn2+, and one substrate analogue. Interpretable electron density existed only for the Galβ1-3Gal(6-O-sulfate) portion of each acceptor substrate analogue.
RESULTS
Catalytic Activities of the Recombinant GlcAT-I toward the Linkage Region Trisaccharide Serines—To examine whether GlcAT-I could recognize the phosphate and/or sulfate on the GAG-protein linkage region trisaccharide, a soluble form of GlcAT-I was generated by replacing the first 43 amino acids of GlcAT-I with a cleavable insulin signal sequence and a protein A IgG-binding domain as described (35, 36), and then the soluble GlcAT-I was expressed in COS-1 cells as a recombinant enzyme fused with the protein A IgG-binding domain. The fused enzyme expressed in the medium was absorbed on IgG-Sepharose beads to eliminate endogenous glucuronyltransferase, and then the purified enzyme-bound beads were used as an enzyme source after the purity of the enzyme was confirmed by Western blotting as previously reported (36). The bound fusion protein was assayed for GlcAT-I activity using various linkage region trisaccharide serines as acceptor substrates. As shown in Table 2, the phosphorylated trisaccharide serine, Gal-Gal-Xyl(2-O-phosphate)-Ser, and the sulfated trisaccharide serine, Gal-Gal(6-O-sulfate)-Xyl-Ser, served as better acceptors than the non-modified counterpart. The trisaccharide with both of these modifications also served as a better substrate than the non-modified counterpart. In strong contrast, unlike the 6-O-sulfate group on the Gal(I) residue, a 6-O-sulfate on the non-reducing terminal Gal(II) residue abolished the acceptor activity completely as demonstrated using the phosphorylated and sulfated trisaccharide serine, Gal(6-O-sulfate)-Gal-Xyl(2-O-phosphate)-Ser.
TABLE 2.
Acceptor specificity of a soluble form of GlcAT-I produced by transfected COS-1 cells
Glucuronyltransferase assays were carried out using a recombinant soluble form of GlcAT-I and various synthetic trisaccharide serines as acceptor substrates as detailed under “Experimental Procedures.” The values represent the averages of two independent experiments.
| Acceptor | GlcAT-I activity | Relative activity |
|---|---|---|
| nmol/ml | % | |
| medium/h | ||
| Galβ1-3Galβ1-4Xylβ1-O-Ser | 0.49 | 100 |
| Galβ1-3Galβ1-4Xyl(2P)β1-O-Sera | 1.6 | 330 |
| Galβ1-3Gal(6S)β1-4Xylβ1-O-Ser | 1.1 | 220 |
| Galβ1-3Gal(6S)β1-4Xyl(2P)β1-O-Ser | 1.4 | 290 |
| Gal(6S)β1-3Galβ1-4Xyl(2P)β1-O-Ser | NDb | 0 |
| Gal(6S)β1-3Galβ1-4Xylβ1-O-Ser | ND | 0 |
2P, 4S, and 6S represent 2-O-phosphate, 4-O-sulfate, and 6-O-sulfate, respectively.
ND, not detected (<0.1 pmol/ml medium/h).
To investigate the catalytic efficiency of GlcAT-I for the three compounds serving as acceptors, the kinetic parameters of GlcAT-I for these substrates were determined and are shown in Table 3. When compared with the non-modified substrate, the phosphorylated compound, Gal-Gal-Xyl(2-O-phosphate)-Ser, gave a 1.8-fold smaller Km and a 2.3-fold larger Vmax, resulting in a 4.2-fold higher Vmax/Km value. The sulfated compound, Gal-Gal(6-O-sulfate)-Xyl-Ser, showed a similar Km and a 2.8-fold larger Vmax, yielding a 2.6-fold higher Vmax/Km value. The phosphorylated and sulfated compound, Gal(6-O-sulfate)-Xyl(2-O-phosphate)-Ser, gave a slightly smaller (76%) Km and a 3.3-fold larger Vmax, resulting in a 4.2-fold higher Vmax/Km value. Thus, the catalytic efficiency expressed by Vmax/Km for the three modified compounds was significantly (2.6- to 4.3-fold) higher than that for the unmodified counterpart, suggesting that the 2-O-phosphorylation of Xyl and 6-O-sulfation of Gal(I) stimulate the completion of the linkage region tetrasaccharide and may be involved in the regulation of GAG biosynthesis.
TABLE 3.
Kinetic properties of a soluble form of GlcAT-I produced by transfected COS-1 cells
Kinetic parameters of GlcAT-I for various acceptor substrates were determined using a recombinant soluble form of the enzyme as detailed under “Experimental Procedures.” Km and Vmax values were calculated from Lineweaver-Burk plots (see supplemental Fig. S1).
| Acceptor | Km | Vmax | Vmax/Km |
|---|---|---|---|
| μm | nmol/ml | ||
| medium/h | |||
| Galβ1-3Galβ1-4Xylβ1-O-Ser | 46 | 1.2 | 0.03 |
| Galβ1-3Galβ1-4Xyl(2P)β1-O-Sera | 25 | 2.8 | 0.11 |
| Galβ1-3Gal(6S)β1-4Xylβ1-O-Ser | 49 | 3.3 | 0.07 |
| Galβ1-3Gal(6S)β1-4Xyl(2P)β1-O-Ser | 35 | 3.9 | 0.11 |
2P, 4S, and 6S represent 2-O-phosphate, 4-O-sulfate, and 6-O-sulfate, respectively.
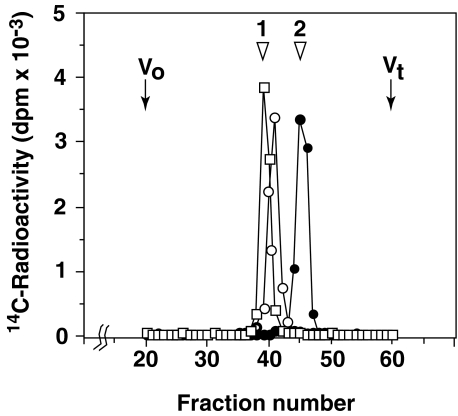
Identification of GlcAT-I Reaction Products—To confirm the β-configuration of the GlcUA residue incorporated into the phosphorylated acceptor substrates, we tested the sensitivity of the GlcAT-I reaction products, obtained with the optimal acceptor Galβ1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser, to the action of β-glucuronidase or alkaline phosphatase. As shown in Fig. 1, the [14C]GlcUA-labeled products were completely digested with β-glucuronidase, quantitatively yielding a 14C-labeled peak at the position of free [14C]GlcUA. In addition, the labeled products were completely digested by alkaline phosphatase, quantitatively yielding a 14C-labeled peak at the position of the unmodified tetrasaccharide serine GlcUAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser. The phosphorylated tetrasaccharide serine was eluted behind the unmodified tetrasaccharide serine presumably because of the interaction of the phosphate with the gel. These results together confirmed that GlcUA was indeed transferred to the phosphorylated acceptor through a β-linkage.
FIGURE 1.
Characterization of the glucuronyltransferase reaction products using β-glucuronidase and alkaline phosphatase. The linkage region trisaccharide serine Galβ1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser was used as an acceptor substrate for the GlcAT-I reaction. The 14C-labeled reaction products recovered from a Superdex Peptide column were subjected to digestion with β-glucuronidase or alkaline phosphatase as described under “Experimental Procedures.” The β-glucuronidase digest (filled circles), the alkaline phosphatase digest (open squares), or the undigested sample (open circles) was applied to a column of Superdex Peptide (1.0 × 30 cm), and the respective effluent fractions (0.4 ml each) were analyzed for radioactivity. Arrowheads 1 and 2 indicate the elution position of the authentic linkage region tetrasaccharide serine GlcUAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser and free GlcUA, respectively.
X-ray Crystallographic Analysis of the Molecular Interaction of GlcAT-I and Various Acceptor Substrates—To help interpret the different activities of the different acceptors, the crystal structure of the catalytic domain of GlcAT-I was determined in the presence of Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser plus UDP to compare it to that of the existing structure of GlcAT-I with Galβ1-3Galβ1-4Xyl plus UDP (37).
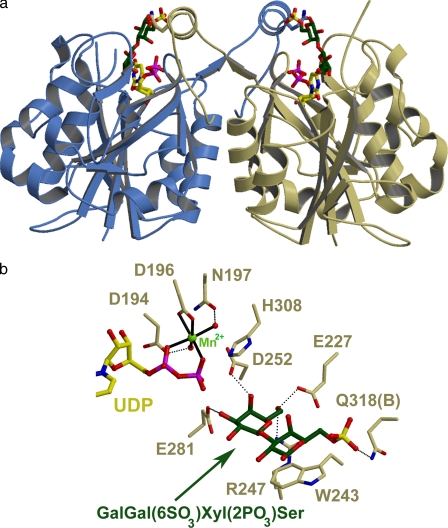
GlcAT-I crystallizes as a dimer whereby the COOH-terminal tail of each monomer contributes to the active site of the other (Fig. 2, a and b) (37). Recent biochemical data on GlcAT-I (44) supports that this homodimer is the physiological complex as do the crystal structure of GlcAT-P (45) and the molecular modeling of GlcAT-1 (44).
FIGURE 2.
Crystal structure of GlcAT-I in a complex with UDP, Mn2+, and Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser. a, ribbon diagram of the proposed GlcAT-I dimer. The monomers are colored blue and khaki, the UDP molecules are colored yellow, and Gal-Gal(6-O-sulfate) is colored green. Residue Gln318 from each monomer is displayed interacting with 6-O-sulfate of the acceptor substrate in the active site of the other monomer. b, active site of GlcAT-I. The UDP molecule is colored yellow, the Mn2+ ion, light green, and the acceptor analogue, dark green. Bonds representing coordination to the metal ions are shown with solid black lines, whereas hydrogen bond interactions are shown with dotted lines. Residue Gln318 (B) is from the other monomer of GlcAT-I in the proposed dimer (48, 49).
The substrate Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser bound in the same orientation and position to GlcAT-I as did the Galβ1-3Galβ1-4Xyl molecule (Fig. 3) from the previous study (37). Like in the previous study, interpretable density only exists for the Gal-Gal(6-O-sulfate) portion of the substrate. There did appear to be some weak density for the 4Xyl(2-O-phosphate), but it was not possible to determine the exact orientation and position. However, it was clear from the density that this portion of the acceptor extended away from the enzyme into the solvent and appeared to form no specific interactions with the protein molecule. Interestingly when the acceptor substrate is not phosphorylated or sulfated, only one substrate binds the dimer (37), but in this study active sites of both monomers were occupied when Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser was bound (Fig. 2b). The nonsulfated Gal-Gal-Xyl(2-O-phosphate)-Ser was not tested in the co-crystal due to the limited amount of the material, which would be expected to give no interpretable density for the phosphorylated xylose based on the results from crystallography of the Gal-Gal(6-O-sulfate)-Xyl(2-O-phosphate)-Ser.
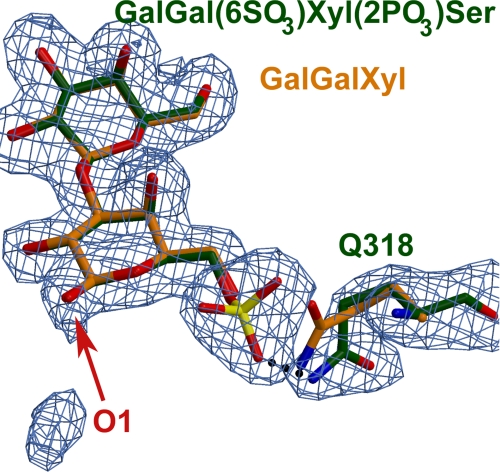
FIGURE 3.
Superposition of the two structures of GlcAT-I with the substrate analogue Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1- O-Ser (orange) and Galβ1-3Galβ1-4Xyl (green). In both structures, the position of the Xyl residue cannot be determined due to a lack of clear electron density. This figure shows the difference in conformation of Gln318(B) in the two structures. The hydrogen bond between NE2 of Gln318(B) and the sulfate oxygen is represented with a black dotted line. The (Fo - Fc) Fourier difference annealed omit map of the Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser acceptor and Gln318(B) is shown contoured at 3 σ and colored blue (48, 49).
The acceptor substrate with the terminal Gal(II) is buried in the catalytic pocket with the acceptor O-3 hydroxyl group 5.1 Å from the β-phosphate group of the UDP molecule (Fig. 2b). The hydrogen bonding interactions between GlcAT-I and Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser were conserved with respect to the terminal Gal(II). Asp252 forms a hydrogen bond with the O-4 hydroxyl group, Glu227 and Arg247 interact with the O-6 hydroxyl group, and an oxygen atom from the carboxyl group of the proposed catalytic base, Glu281 (37), is 2.7 Å from the acceptor O-3 hydroxyl group of the terminal Gal(II) (Fig. 2b).
In the GlcAT-I/Galβ1-3Galβ1-4Xyl structure, the only possible hydrogen bond interaction between Gal(I) of the acceptor substrate and the enzyme was likely formed by Gln318 from the second monomer of the dimer and the O-6 hydroxyl group of Gal(I) (37). Intriguingly, placement of a sulfate on the O-6 oxygen atom, as was seen in the GlcAT-I/Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser structure, creates a conformational change in the side chain of Gln318 from the second monomer (Fig. 3). In this orientation, atom NE2 of Gln318 is located 3.0 Å from O-4 of the sulfate moiety (Figs. 2, a and b, 3). This represented the only distinguishable change in the interactions between the two acceptor molecules and the GlcAT-I molecule.
Recent mutational data on GlcAT-I obtained using a Gal-Gal(6-O-sulfate)-O-methoxyphenyl analogue suggest that Gln318 does not play a significant role in the binding of substrates with Gal(I) sulfated at the C-6 carbon (46, 47). Rather, Lys317 is suggested to increase binding. In the crystal structure, however, the side chain for Lys317 is disordered and does not form an ordered hydrogen bond with the substrate, although it is near in proximity. Three possibilities that could account for this discrepancy are: (a) effects due to a different analogue (methoxyphenyl instead of Xyl(2-O-phosphate)β1-O-Ser) near the Lys317 position, (b) buffer effects of the crystallization condition do not favor interactions between Lys317 and the 6-O-sulfate, or (c) Lys317 may be involved in the formation of the proper conformation of the enzyme, thereby being indirectly involved in the expression of the catalytic activity.
In contrast, the compound Gal(6-O-sulfate)β1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser was not a substrate for GlcAT-I (Table 2). It is clear from the structural data that placement of a sulfate group on the O-6 hydroxyl group of the terminal Gal(II) residue would create steric conflict with conserved residues Glu227 and Arg247, which interact with the O-6 hydroxyl group on this Gal(II) residue (Fig. 2b). It is likely that such conflict would either inhibit binding or re-orient the positioning of the molecule such that catalysis could not proceed. This could explain the lack of activity with this acceptor (Table 2).
DISCUSSION
In this study, enzyme assays demonstrated that the synthetic compounds, Galβ1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser, Galβ1-3Gal(6-O-sulfate)β1-4Xylβ1-O-Ser, and Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser, served as better substrates for the truncated form of the recombinant human GlcAT-I than the unmodified trisaccharide serine, suggesting that both the 6-O-sulfate group on Gal(I) and the 2-O-phosphate group on Xyl enhance the acceptor activity. However, the increase in the acceptor activity of these analogues cannot be clearly explained by the current structural data due to the lack of strong density between the phosphate and the enzyme. It should be noted that a sulfation of the Gal and a phosphorylation of the Xyl have not been simultaneously found on the same chain and are mutually exclusive (19).
Interestingly, x-ray crystallography revealed that the 6-O-sulfate group on the Gal(I) residue of the phosphorylated and sulfated compound, buried in the substrate-binding pocket of one monomer of a dimeric enzyme, was likely recognized by the specific residue Gln318 of the other monomer. The sulfate at the O-6 hydroxyl group of the Gal(I) residue can be accommodated by the enzyme through a rearrangement of Gln318 of the second monomer. Therefore, it is conceivable that the increase in activity with Galβ1-3Gal(6-O-sulfate)β1-4Xyl(2-O-phosphate)β1-O-Ser and also with Galβ1-3Gal(6-O-sulfate)β1-4Xylβ1-O-Ser over Galβ1-3Galβ1-4Xylβ1-O-Ser was partly due to the hydrogen bond between Gln318 and the sulfate on the acceptor. This speculation relies on the assumption that the dimer in the crystal is the physiological dimer such that Gln318 of one monomer can interact with the substrate bound to the active site of the other monomer, which is consistent with the results from the kinetic studies presented here.
The present findings suggest that the 2-O-phosphorylation of the Xyl residue and the 6-O-sulfation of the Gal(I) residue can take place before the formation of the tetrasaccharide by the glucuronyl transfer reaction, and are consistent with previous finding by Moses et al. (50) using pulse-chase experiments that the linkage region was transiently almost fully phosphorylated on Xyl of the Gal-Gal-Xyl trisaccharide formed on the core protein of decorin, which is followed by addition of the first GlcUA and then dephosphorylation. If the modifications indeed take place before the GlcUA transfer reaction, the modifying kinase should exist in the early Golgi or endoplasmic reticulum. Such a putative Xyl kinase remains to be identified. A partially purified bovine serum chondroitin 6-O-sulfotransferase and the recombinant human chondroitin 6-O-sulfotransferase have been shown to catalyze 6-O-sulfation of Gal(I).3
Syndecan-1 is a hybrid-type PG bearing both HS and CS chains (51). Simultaneous chemical analysis of the linkage regions of both the HS and CS chains of syndecan-1 demonstrated two kinds of linkage region tetrasaccharide structures, GlcUA-Gal-Gal-Xyl and GlcUA-Gal-Gal-Xyl(2-O-phosphate) for the HS chains, whereas an additional sulfated tetrasaccharide structure, GlcUA-Gal(4-O-sulfate)-Gal-Xyl, was demonstrated for the CS chains (52), providing conclusive evidence for the previously proposed hypothesis (19, 29) that 4-O-sulfation of Gal is peculiar to CS chains in contrast to phosphorylation of Xyl, which is common to both HS and CS chains. However, there does not appear to be sufficient space for a sulfate of the C-4 position of the Gal(II) saccharide in the active site of GlcAT-I. In fact, recombinant GlcAT-I shows no significant activity toward a Gal(4-O-sulfate)-Gal-O-methoxyphenyl analogue (44). 4-O-Sulfation of this group may occur after the addition of GlcUA. If the sulfate group is involved in the assembly of CS/DS chains, it may be recognized by the chondroitin synthase complexes (25-27), and may enhance the activity of the chondroitin synthase complexes. In addition, the sulfate group may inhibit the addition of GlcNAc onto the linker region by the EXT1/EXT2 complex (23), EXTL2 (53), or EXTL3 (54) that commit the linker to HS synthesis. It is not conclusive from studying the crystal structure of EXTL2 with UDP and GlcUA-Gal(II) bound if substitution at the 4-O position of Gal(II) could be accommodated without altering the position of the acceptor OH on the GlcUA saccharide (55).
In view of the results from the present study and those reported by Moses et al. (50), it is reasonable to assume that phosphorylation takes place before the transfer of GlcUA to the trisaccharide. However, dephosphorylation seems to occur immediately after the formation of the tetrasaccharide sequence, at least in the rat fibroblast system (50). However, phosphorylated Xyl in the linkage region has also been identified in mature proteoglycans as well (19, 29). Thus, dephosphorylation may not always be required for chain elongation. The phosphorylation of the Xyl residue and sulfation of the Gal residues may be required for biosynthetic maturation of the linkage region tetrasaccharide sequence, which may be a prerequisite for the initiation and efficient elongation of the repeating disaccharide region of GAG chains and/or biosynthetic selective assembly of CS and HS chains.
Thrombomodulin, which bears a mature CS chain, is known as β-thrombomodulin and functions as an anticoagulant on the endothelial cell surface. Some thrombomodulin proteins are part time PGs and some are devoid of a CS chain, and are known as α-thrombomodulin, which has a truncated un-modified tetrasaccharide linkage region, GlcUA-Gal-Gal-Xyl (56). It is also known that human urinary thrombomodulin bears an unusually modified truncated tetrasaccharide linkage region, GlcUA(3-O-sulfate)β1-3Galβ1-3(± sialic acidα2-6)Galβ1-4Xyl (28). These results altogether appear to indicate the importance of the appropriate ordered modification of the linkage region for proteins in the maturation of PGs. Unless it is modified or inappropriately modified, a part time PG core protein ends up as a glycoprotein with a truncated linkage region oligosaccharide. Hence, a better understanding of the control mechanism of the biosynthetic modification of the linkage region is required for clarifying the mechanisms underlying the assembly of GAG chains on the putative GAG attachment sites and selective assembly of CS/DS and HS/Hep on the common linkage region tetrasaccharide.
6-O-Sulfated Gal(II) has been demonstrated as Gal(6-O-sulfate)-Gal(6-O-sulfate)-Xyl in the linkage region of the polymer CS chains from shark cartilage (31). However, in the present study the compound Gal(6-O-sulfate)β1-3Galβ1-4Xyl(2-O-phosphate)β1-O-Ser did not serve as an acceptor (Table 2). These results together suggest that 6-O-sulfation of Gal(II) takes place after elongation of the repeating disaccharides on the linkage region tetrasaccharide. It would be interesting to investigate the possibility that the 6-O- and 4-O-sulfate groups (29, 31, 32) on Gal(II) and/or the 6-O-sulfate group (31) on Gal(I) function as a marker for intracellular transport of CS chains to the Golgi compartment for biosynthetic processing or elongation and maturation, thus discriminating against HS chain formation. Alternatively, the possibility cannot be excluded that GlcAT-I of shark cartilage may bind acceptor sugars end-labeled by sulfate on Gal(II) and/or disulfated on both Gal(I) and Gal(II).
The atomic coordinates and structure factors (code 3CU0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by a National Institutes of Health grant from the Intramural Research Program (NIEHS). This work was supported in part by the Scientific Research Promotion Fund from the Japan Private School Promotion Foundation, Grants-in-aid for Scientific Research-B 19390025 (to H. K.) and 18390030 (to K. S.), and for Scientific Research on Priority Areas 14082207 (to K. S.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Core Research for Evolutional Science and Technology (CREST) program of the Japan Science and Technology (JST) Agency (to H. K. and K. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PG, proteoglycan; CS, chondroitin sulfate; DS, dermatan sulfate; GAG, glycosaminoglycan; HS, heparan sulfate; Hep, heparin; GlcUA, D-glucuronic acid; GalNAc, N-acetyl-d-galactosamine; IdoUA, l-iduronic acid; GlcAT, glucuronyltransferase; MES, 2-(N-morpholino)ethanesulfonic acid.
H. Kitagawa, K. Tsutsumi, A. Ikegami, F. Goto, T. Ogawa, and K. Sugahara, unpublished data.
References
- 1.Rodén, L. (1980) in The Biochemistry of Glycoproteins and Proteoglycans (Lennarz, W. J., ed) pp. 267-371, Plenum Publishing, New York
- 2.Poole, A. R. (1986) Biochem. J. 236 1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fransson, L. Å. (1987) Trends Biochem. Sci. 12 406-411 [Google Scholar]
- 4.Salmivirta, M., Lidholt, K., and Lindahl, U. (1996) FASEB J. 10 1270-1279 [DOI] [PubMed] [Google Scholar]
- 5.Perrimon, N., and Bernfield, M. (2000) Nature 404 725-728 [DOI] [PubMed] [Google Scholar]
- 6.Lander, A. D., and Selleck, S. B. (2000) J. Cell Biol. 148 227-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deepa, S. S., Umehara, Y., Higashiyama, S., Itoh, N., and Sugahara, K. (2002) J. Biol. Chem. 277 43707-43716 [DOI] [PubMed] [Google Scholar]
- 8.Deepa, S. S., Yamada, S., Zako, M., Goldberger, O., and Sugahara, K. (2004) J. Biol. Chem. 279 37368-37376 [DOI] [PubMed] [Google Scholar]
- 9.Bao, X., Mikami, T., Yamada, S., Faissner, F., Muramatsu, T., and Sugahara, K. (2005) J. Biol. Chem. 280 9180-9191 [DOI] [PubMed] [Google Scholar]
- 10.Bao, X., Muramatsu, T., and Sugahara, K. (2005) J. Biol. Chem. 280 35318-35328 [DOI] [PubMed] [Google Scholar]
- 11.Li, F., Shetty, A. K., and Sugahara, K. (2007) J. Biol. Chem. 282 2956-2966 [DOI] [PubMed] [Google Scholar]
- 12.Lander, A. D. (1993) Curr. Opin. Neurobiol. 3 716-723 [DOI] [PubMed] [Google Scholar]
- 13.Oohira, A., Matsui, F., Tokita, Y., Yamauchi, S., and Aono, S. (2000) Arch. Biochem. Biophys. 374 24-34 [DOI] [PubMed] [Google Scholar]
- 14.Sugahara, K., and Yamada, S. (2000) Trends Glycosci. Glycotechnol. 12 321-349 [Google Scholar]
- 15.Sugahara, K., Mikami, T., Uyama, T., Mizuguchi, S., Nomura, K., and Kitagawa, H. (2003) Curr. Opin. Struct. Biol. 13 612-620 [DOI] [PubMed] [Google Scholar]
- 16.Morgenstern, D. A., Asher, R. A., and Fawcett, J. W. (2002) Prog. Brain Res. 137 313-332 [DOI] [PubMed] [Google Scholar]
- 17.Sugahara, K., and Mikami, T. (2007) Curr. Opin. Struct. Biol. 17 536-545 [DOI] [PubMed] [Google Scholar]
- 18.Lindahl, U., and Rodén, L. (1972) in Glycoprotein (Gottschalk, A., ed) pp. 491-517, Elsevier, Amsterdam
- 19.Sugahara, K., and Kitagawa, H. (2000) Curr. Opin. Struct. Biol. 10 518-527 [DOI] [PubMed] [Google Scholar]
- 20.McCormick, C., Leduc, Y., Martindale, D., Mattison, K., Esford, L. E., Dyer, A. P., and Tufaro, F. (1998) Nat. Genet. 19 158-161 [DOI] [PubMed] [Google Scholar]
- 21.Lind, T., Tufaro, F., McCormick, C., Lindahl, U., and Lidholt, K. (1998) J. Biol. Chem. 273 26265-26268 [DOI] [PubMed] [Google Scholar]
- 22.Senay, C., Lind, T., Muguruma, K., Tone, Y., Kitagawa, H., Sugahara, K., Lidholt, K., Lindahl, U., and Kusche-Gullberg, M. (2000) EMBO Rep. 1 282-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, B.-T., Kitagawa, H., Tanaka, J., Tamura, J., and Sugahara, K. (2003) J. Biol. Chem. 278 41618-41623 [DOI] [PubMed] [Google Scholar]
- 24.Busse, M., and Kusche-Gullberg, M. (2003) J. Biol. Chem. 278 41333-41337 [DOI] [PubMed] [Google Scholar]
- 25.Kitagawa, H., Uyama, T., and Sugahara, K. (2001) J. Biol. Chem. 276 38721-38726 [DOI] [PubMed] [Google Scholar]
- 26.Kitagawa, H., Izumikawa, T., Uyama, T., and Sugahara, K. (2003) J. Biol. Chem. 278 23666-23671 [DOI] [PubMed] [Google Scholar]
- 27.Izumikawa, T., Uyama, T., Okuura, Y., Sugahara, K., and Kitagawa, H. (2007) Biochem. J. 403 545-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakabayashi, H., Natsuka, S., Mega, T., Otsuki, N., Isaji, M., Naotsuka, M., Koyama, S., Kanamori, T., Sakai, K., and Hase, S. (1999) J. Biol. Chem. 274 5436-5442 [DOI] [PubMed] [Google Scholar]
- 29.Sugahara, K., Yamashina, I., De Waard, P., Van Halbeek, H., and Vliegenthart, J. F. G. (1988) J. Biol. Chem. 263 10168-10174 [PubMed] [Google Scholar]
- 30.Sugahara, K., Ohi, Y., Harada, T., de Waard, P., and Vliegenthart, J. F. (1992) J. Biol. Chem. 267 6027-6035 [PubMed] [Google Scholar]
- 31.de Waard, P., Vliegenthart, J. F., Harada, T., and Sugahara, K. (1992) J. Biol. Chem. 267 6036-6044 [PubMed] [Google Scholar]
- 32.Sugahara, K., Ohkita, Y., Shibata, Y., Yoshida, K., and Ikegami, A. (1995) J. Biol. Chem. 270 7204-7212 [DOI] [PubMed] [Google Scholar]
- 33.Lauder, R. M., Huckerby, T. N., and Nieduzynski, I. A. (2000) Biochem. J. 347 339-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Beer, T., Inui, A., Tsuda, H., Sugahara, K., and Vliegenthart, J. F. G. (1996) Eur. J. Biochem. 240 789-797 [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa, H., Tone, Y., Tamura, J., Neumann, K., Ogawa, T., Oka, S., Kawasaki, T., and Sugahara, K. (1998) J. Biol. Chem. 273 6615-6618 [DOI] [PubMed] [Google Scholar]
- 36.Tone, Y., Kitagawa, H., Imiya, K., Oka, S., Kawasaki, T., and Sugahara, K. (1999) FEBS Lett. 459 415-420 [DOI] [PubMed] [Google Scholar]
- 37.Pedersen, L. C., Tsuchida, K., Kitagawa, H., Sugahara, K., Darden, T. A., and Negishi, M. (2000) J. Biol. Chem. 275 34580-34585 [DOI] [PubMed] [Google Scholar]
- 38.Tamura, J., and Nishihara, J. (1999) Bioorg. Med. Chem. Lett. 9 1911-1914 [DOI] [PubMed] [Google Scholar]
- 39.Tamura, J., and Nishihara, J. (2001) J. Org. Chem. 66 3074-3083 [DOI] [PubMed] [Google Scholar]
- 40.Mizushima, S., and Nagata, S. (1990) Nucleic Acids Res. 18 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 42.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., and Gros, P. (1998) Acta Crystallogr. Sect. D 54 905-921 [DOI] [PubMed] [Google Scholar]
- 43.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 44.Ouzzine, M., Gulberti, S., Netter, P., Magdalou, J., and Fournel-Gigleux, S. (2000) J. Biol. Chem. 275 28254-28260 [DOI] [PubMed] [Google Scholar]
- 45.Kakuda, S., Shiba, T., Ishiguro, M., Tagawa, H., Oka, S., Kajihara, Y., Kawasaki, T., Wakatsuki, S., and Kato, R. (2004) J. Biol. Chem. 279 22693-22703 [DOI] [PubMed] [Google Scholar]
- 46.Gulberti, S., Lattard, V., Fondeur, M., Jacquinet, J. C., Mulliert, G., Netter, P., Magdalou, J., Ouzzine, M., and Fournel-Gigleux, S. (2005) J. Biol. Chem. 280 1417-1425 [DOI] [PubMed] [Google Scholar]
- 47.Fondeur-Gelinotte, M., Lattard, V., Gulberti, S., Oriol, R., Mulliert, G., Coughtrie, M. W. H., Magdalou, J., Netter, P., Ouzzine, M., and Fournel-Gigleux, S. (2007) Glycobiology 17 857-867 [DOI] [PubMed] [Google Scholar]
- 48.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24 940-950 [Google Scholar]
- 49.Merritt, E. A., and Bacon, D. J. (1997) Methods Enzymol. 277 505-524 [DOI] [PubMed] [Google Scholar]
- 50.Moses, J., Oldberg, A., and Fransson L. A. (1999) Eur. J. Biochem. 260 879-884 [DOI] [PubMed] [Google Scholar]
- 51.Kokenyesi, R., and Bernfield, M. (1994) J. Biol. Chem. 269 12304-12309 [PubMed] [Google Scholar]
- 52.Ueno, M., Yamada, S., Zako, M., Bernfield, M., and Sugahara, K. (2001) J. Biol. Chem. 276 29134-29140 [DOI] [PubMed] [Google Scholar]
- 53.Kitagawa, H., Shimakawa, H., and Sugahara, K. (1999) J. Biol. Chem. 274 13933-13937 [DOI] [PubMed] [Google Scholar]
- 54.Kim, B.-T., Kitagawa, H., Tamura, J., Saito, T., Kusche-Gullberg, M., Lindahl, U., and Sugahara, K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7176-7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedersen, L. C., Dong, J., Taniguchi, F., Kitagawa, H., Krahn, J., Pedersen, L. G., Sugahara, K., and Negishi, M. (2003) J. Biol. Chem. 278 14420-14428 [DOI] [PubMed] [Google Scholar]
- 56.Nadanaka, S., Kitagawa, H., and Sugahara, K. (1998) J. Biol. Chem. 273 33728-33734 [DOI] [PubMed] [Google Scholar]