Abstract
Histone deacetylases (HDACs) catalyze the removal of acetyl groups from histones and contribute to transcriptional repression. In addition, the HDAC inhibitors induce apoptosis in cancer cells through alterations in histone acetylation and activation of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) apoptotic pathway. Lysophosphatidic acid (LPA) is a growth factor that promotes survival of cancer cells through activation of G protein-coupled receptors. Here we show that HDAC inhibitors can induce apoptosis through activation of the TRAIL apoptotic pathway, and LPA prevented HDAC inhibitor-induced apoptosis and increased TRAIL receptor DR4 (death receptor 4) protein expression. This was associated with increased HDAC1 recruitment to the DR4 promoter following LPA treatment and a reduction in HDAC inhibitor-induced histone acetylation in the DR4 promoter. In addition, LPA induces HDAC enzyme activity in a dose- and time-dependent manner, and this is associated with HDAC1 activation and increased binding of HDAC1 to HDAC2. Reducing the expression of HDAC1 significantly lowered LPA-induced HDAC activity and increased histone acetylation. LPA induction of HDAC activity was blocked by the LPA receptor antagonist, Ki16425, or by inhibiting receptor activation with pertussis toxin. Reducing the expression of the LPA receptor LPA1 also blocked LPA-induced HDAC activation. In addition, LPA reduced histone acetyltransferase enzymatic activity. Finally, LPA attenuated the ability of the HDAC inhibitor to reduce HDAC activity. Thus, LPA enhances survival of cancer cells by increasing HDAC activity and reducing histone acetylation.
Transcription in eukaryotic cells is influenced by the chromatin structure within which DNA is tightly packaged (1). The nucleosome is the basic unit of chromatin and consists of 146 bp of DNA wrapped around a histone octamer. The histone tail domains are subjected to post-transcriptional modifications, such as acetylation, phosphorylation, methylation, and ubiquitination (2, 3). Compared with methylation and phosphorylation, the acetylation of core histones is probably the best understood type of modification (4, 5). Acetylation of histone tails correlates with transcriptional activity in many genes, allowing DNA to unfold and providing access for transcription factors to bind to their targeted promoters. The turn-over of acetylated histones is regulated by the opposing activities of histone acetyltransferases (HATs)2 and histone deacetylases (HDACs), where HATs generally allow transcription and HDACs repress transcription (4, 5). In cancer, deregulation of HAT or HDAC activity often occurs (6-8). Based on sequence similarities, HDACs are divided into three functional classes as follows: class I (HDAC1, -2, -3, and -8), class II (HDAC4-7, -9, and -10), and class III (HDAC11) (9). The class I enzyme, HDAC1, belongs to a family of highly conserved enzymes and was the first protein shown to have histone deacetylating activity in mammals (10). HDAC1 is a nuclear protein and can heterodimerize with the closely related deacetylase HDAC2 (11, 12). Both enzymes are found in three major multiprotein complexes, named Sin3, NuRD, and Co-REST (13, 14). HDAC1 can repress gene transcription either directly or as part of these multiprotein complexes when recruited by a variety of transcriptional regulators, including SP1/SP3, nuclear receptors, the pocket proteins pRB, p107, and p130, and the tumor suppressor p53 (15, 16).
HDAC inhibitors (HDIs) preferentially induce apoptosis in cancer cells through activation of both the death receptor and the mitochondrial apoptotic signaling pathways (17). In acute myeloid leukemia cells, HDIs induce the expression of death receptor (DR) 4, DR5, and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL); these changes all contribute to HDI-induced apoptosis (18). Furthermore, HDIs sensitize cancer cells to TRAIL-induced apoptosis, as a synergistic apoptotic response is seen when the cells are treated with a combination of an HDAC inhibitor and TRAIL (19, 20). In chronic lymphocytic leukemia (CLL), HDIs sensitize the leukemia cells to TRAIL-induced apoptosis through activation of DR4 (21). However, it has been unclear as to which HDAC is responsible for this effect, as the inhibitors used affect the catalytic activity of most class I and class II deacetylases (17, 22). More recently, it has been shown in CLL cells that inhibition of class I but not class II HDACs sensitizes the cells to TRAIL-induced apoptosis (23). However, little is known about the individual roles of mammalian deacetylases in transcriptional control or the relevant target genes for HDIs.
Lysophosphatidic acid (LPA; monoacylglycerol 3-phosphate) is a naturally occurring soluble glycerophospholipid that was initially identified as an intermediate in a de novo lipid biosynthesis pathway (24, 25). Effects of LPA include stimulation of cellular proliferation, platelet aggregation, smooth muscle cell contraction, and tumor cell invasion (26). In B cells, LPA acts as a growth factor increasing cell proliferation, intracellular calcium, and immunoglobulin formation (27). LPA binds to G protein-coupled receptors (GPCR) as follows: LPA1 (Edg-2), LPA2 (Edg-4), LPA3 (Edg-7), LPA4 (GPR23), and LPA5 (GPR92) (27, 28). The LPA1 receptor is widely expressed, whereas LPA2 and LPA3 have more restricted expression patterns. The LPA4 and LPA5 receptors belong to the G2Y family of GPCRs, structurally distinct from the LPA1-3 receptors of the Edg family (28, 29). Expression of LPA receptors is aberrant in cancer cells from multiple different lineages (28). LPA1 expression is increased in CLL and colon cancer cells, whereas LPA2 and LPA3 are elevated in ovarian cancer cells suggesting that LPA receptors are involved in tumorigenesis (30, 31, 34). In addition, elevated levels of LPA have been found in the ascites of ovarian cancer patients and in the bone marrow of multiple myeloma patients. This elevated level has been suggested to contribute to tumor invasiveness, proliferation, and resistance to apoptosis (24, 32). LPA protects ovarian cancer cells against chemotherapeutic drug-induced apoptosis and prevents apoptosis in serum-deprived renal proximal tubular cells, fibroblasts, and macrophages (30, 33). In addition, LPA effectively blocks chemotherapeutic drug-induced apoptosis in CLL cells (34). The mechanism of this survival response to LPA is not well understood.
Here we demonstrate that LPA prevents HDI-induced apoptosis through reduced DR4 expression. LPA also activates HDAC1 enzymatic activity and reduces HAT enzymatic activity. Preventing LPA from binding to its receptors with a selective antagonist or inhibiting LPA1 expression blocked LPA-induced HDAC enzymatic activity and inhibited the protective effects of LPA against HDI-induced apoptosis. Together, our results revealed for the first time a novel mechanism for regulation of histone acetylation by LPA that increases cell survival and antagonizes the effects of HDIs.
EXPERIMENTAL PROCEDURES
Materials—LPA (18:1) was purchased from Sigma and dissolved in phosphate-buffered saline containing 0.1% fatty acid-free bovine serum albumin. Anti-HDAC1 and -HDAC2 antibodies were purchased from Affinity BioReagents; anti-acetyl-H3, anti-acetyl-H4, anti-histone H3, and anti-p300 for ChIP experiments were purchased from Upstate Biotechnology; anti-histone H3 and H4 were from Cell Signaling Technology; and soluble super-TRAIL was purchased from Alexis. Monoclonal antibodies against β-actin, DR4, and HDAC1 were purchased from Santa Cruz Biotechnology. Anti-LPA1 antibody was purchased from Exalpha Biologicals Inc. Anti-DR5 antibody was purchased from MEDICORP (Toronto, Canada). Trichostatin (TSA) and valproic acid (VPA) were purchased from Sigma and were dissolved in dimethyl sulfoxide. GW9662 was purchased from Cayman.
Patients and Lymphocyte Isolation—Peripheral blood samples were obtained from CLL patients or normal age-matched individuals following informed consent, in agreement with the Research Ethics Board at the University of Manitoba. The cells were isolated as described by Hu et al. (34). Briefly, mononuclear cells were isolated from the buffy coat of CLL patients or normal individuals using Ficoll-Paque density gradient centrifugation. The cell isolates were washed with ice-cold phosphate-buffered saline, pH 7.4, and a second wash with ice-cold 10 mm Tris, 140 mm NH4Cl to lyse residual erythrocytes. When the peripheral blood lymphocyte counts were <30 × 109/liter, contaminating monocytes were removed from the isolates using CD33 antibodies coupled to magnetic beads, and T cells were removed by sheep red cell resetting. The remaining cells were purified B cells.
Cell Culture and Treatment Conditions—Lymphocytes were cultured at 37 °C in a humidified, 5% CO2 incubator at an initial density of 5 × 106 cells/ml in serum-free RPMI 1640 culture medium. BJAB, I-83, and CaOv3 cells were also grown in RPMI 1640 medium supplemented with 100 units of penicillin, 100 μg of streptomycin, and 10% fetal bovine serum. The cells were always incubated in serum-free conditions overnight prior to treatment. Human embryonic kidney cell line 293 (HEK293) was maintained in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen).
HDAC Activity Assay—HDAC activity was determined using the fluorometric HDAC activity assay kit (BioVision Inc.). Nuclear extracts (10 μg) from the cells treated with LPA (10 μm) were assayed for HDAC activity. The HDAC reaction was initiated by adding t-butoxycarbonyl-Lys(AC)-amido-4-methylcoumarin substrate (Boc-Lys(AC)AMC), and incubation was performed at 37 °C for 30 min with the nuclear extracts. After 30 min, the lysine developer was added, and the mixture was incubated for another 30 min at 37 °C. Fluorescence was measured using a Spectra Max M5 fluorescent plate reader (Molecular Devices), with excitation at 360 nm and emission at 460 nm. No enzyme control or inhibitor controls were included, and all reactions were set up in duplicate. Results represent three independent experiments.
HAT Activity Assay—HAT activity was determined using the colorimetric HAT activity assay kit (BioVision Inc.). Nuclear extracts (50 μg) from the cells treated with LPA (10 μm) were assayed for HAT activity. The reaction was initiated by adding peptide substrate to the nuclear lysate, containing active HATs, and incubated at 37 °C for 1-3 h. Enzyme activity was measured using a Spectra Max M5 plate reader (Molecular Devices). The cell lysate was boiled for 15 min and taken as negative control. Results represent three independent experiments.
Immunoprecipitation—Cells were treated with LPA and lysed in lysis buffer (50 mm HEPES, pH 7.25, 150 mm NaCl, 50 μm ZnCl3, 50 μm NaF, 2 mm EDTA, 1 mm sodium vanadate, 1% Nonidet P-40, and 2 mm phenylmethylsulfonyl fluoride). 1 mg/ml protein from each pre-cleared cell lysate was incubated with 4 μg of anti-HDAC1 or anti-HDAC2 antibody overnight at 4 °C, and then 50 μl of protein A-Sepharose beads was added and incubated for an additional 2 h. The immunoprecipitation complex was washed four times with lysis buffer and boiled in loading buffer for 10 min, and immunoblotting was performed as described below. Anti-HDAC1 or anti-HDAC2 was used as the primary antibody. For the HDAC activity assay after the immunoprecipitation of HDAC1 or HDAC2, the beads were washed two times and assayed directly for HDAC activity as described above.
Immunoblots—The cells were lysed in Nonidet P-40 lysis buffer (50 mm HEPES, pH 7.25, 150 mm NaCl, 50 μm ZnCl2, 50 μm NaF, 2 mm EDTA, 1 mm sodium vanadate, 1.0% Nonidet P-40, 2 mm phenylmethylsulfonyl fluoride). The cell debris was removed by centrifugation at 13,000 × g for 5 min at 4 °C, and the protein concentration was determined by the Bio-Rad assay. After SDS-PAGE, the proteins were transferred to nitro-cellulose membranes. The membranes were blocked with 5% milk in Tris/Tween 20 solution for 1 h, followed by overnight incubation with primary antibody at 4 °C, and finally incubated for 1 h with secondary antibody conjugated with horseradish peroxidase (Bio-Rad). Detection was by ECL-Western Lightning Chemiluminescence reagent (Amersham Biosciences). The blots were stripped in 10% SDS buffer and re-probed with β-actin to normalize for protein loading in each lane.
ChIP—The ChIP assay was performed as described by Shetty et al. (35) with some modifications. I-83 or primary CLL cells were fixed to cross-link protein bound to DNA by adding form-aldehyde at a final concentration of 1% for 10 min at room temperature. The cell lysate was sonicated at 30% output on ice four times with 15-s intervals and pre-cleared with protein A-Sepharose 4B (Amersham Biosciences). The lysate was precipitated with the primary antibodies to acetylated histone 3 (3 μg) or HDAC1 (4 μg) or p300 (4 μg) overnight at 4 °C. Immunoprecipitates were tested to ensure that the targeted proteins were completely immunoprecipitated (data not shown). The DNA was purified using the QIA-AMP DNA purification system (Qiagen). PCR products were detected, using primers specific for DR4 (forward primer, 5′-TCTCCCGTGGTTTAAGGACTTCA-3′ and reverse primer, 5′-TCAAGCGATTTTCCTGCCTCA-3′), for 30 cycles with cycling conditions as follows: 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. For each treatment point, 1% of the total DNA was assayed for equal loading (input). Input was conducted for each condition tested.
Transfection of HEK293 Cells with siRNA and LPA1 Vector—HEK293 cells were plated in 6-well plates and grown to 70% confluence. siRNA (40 nm) against HDAC1 or LPA1 (Smart Pool, Dharmacon) was used. Transfections were performed using Lipofectamine2000 as described in the manufacturer's instructions. After 48 h, the cells were treated with 10 μm LPA for an additional 2 h. The nuclear extracts were then assayed for HDAC activity as described above. Scrambled siRNA was used as control. For the LPA1 overexpression experiment, the cells were grown to ∼60% confluence and transfected with 5 μg of cDNA for human LPA1 in pcDNA3 vector containing a FLAG tag. Transfections were performed using Lipofectamine as described in the manufacturer's instructions. After 48 h, the cells were treated with 10 μm LPA for an additional 2 h.
Assessment of Cell Surface Receptor Expression—Cells were resuspended in blocking buffer (10% normal goat serum in PBS), incubated for 30 min on ice to block nonspecific binding, and then incubated with anti-TRAIL-R1/DR4 fluorescein isothiocyanate-conjugated antibody (Alexis) or an isotopematched control antibody (Pharmingen) for 1 h on ice. The samples were washed twice by centrifugation at 1200 rpm for 5 min with ice-cold PBS and resuspended in PBS and analyzed immediately by flow cytometry.
Cell Death Detection and Membrane Permeability Assay—The cells were resuspended in 100 μl of medium and stained with a mixture of acridine orange (100 μg/ml) and ethidium bromide (100 μg/ml) in phosphate-buffered saline. A cell suspension was then applied onto a microscope slide and covered with a coverslip, and the cells were viewed using a fluorescence microscope with a fluorescein filter set to quantitate condensed DNA in apoptotic cells. Random fields were scored, and the percentage of apoptotic cells was determined from cells containing condensed DNA to cells with normal DNA. At least 200 cells were counted for each condition tested.
Caspase-8 Activity Assay—Cells were collected and lysed as described above in immunoblots. 50 μg of cell lysate was taken and diluted in 2× reaction buffer (BioVision) and labeled with IETD-p-nitroanilide substrate (BioVision) at 200 μm final concentration. Sample was incubated at 37 °C for 1-2 h and read by plate reader (Molecular Devices), at 400 or 405 nm.
Annexin V/7-AAD Staining—Cells (5 × 106) were collected in 5-ml tubes, centrifuged, resuspended in 1× binding buffer (10 mm HEPES/NaOH, pH 7.4, 140 mm NaCl, 2.5 mm CaCl2), and supplemented with annexin V-fluorescein isothiocyanate (Pharmingen) and 2.5 μg of 7-AAD (Molecular Probes, Eugene, OR). After 15 min of incubation at room temperature in the dark, an excess of 1× binding buffer was added to a final volume of 500 μl. The cells were then analyzed for fluorescence-activated cell sorter with a Caliber flow cytometer.
Statistical Analysis—All experiments were repeated at least three times, and each experiment was performed at least in duplicate. The data were expressed as means ± S.E. Statistical analysis was performed by using Student's t test. The criterion for statistical significance was p < 0.05.
RESULTS
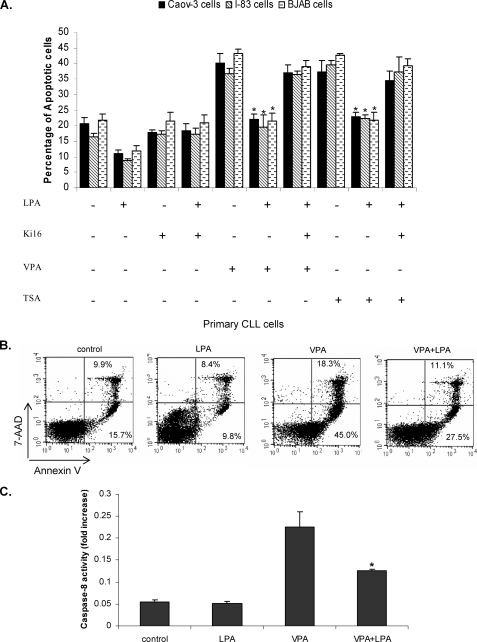
LPA Blocks HDI-induced Apoptosis—LPA protects multiple types of cancer cells from spontaneous or drug-induced apoptosis (34). We determined whether LPA could protect tumor cells from HDI-induced apoptosis. I-83 (CLL-like cell line), BJAB (Burkitt's lymphoma cell line), and CaOv-3 (ovarian cancer cell line) cells were treated with 75 nm TSA or 1 mm VPA, alone or in combination with 10 μm LPA, and apoptosis was measured after 48 h by membrane permeability assay (Fig. 1A). In untreated cells, the levels of apoptosis was ∼20%, and LPA reduced this to 9-12% (Fig. 1A). In cells treated with TSA or VPA alone, the number of apoptotic cells ranged from 36 to 43%, and the addition of LPA decreased apoptosis to 19-23%. At the same time, cells were also treated with Ki16425. Ki16425 selectively inhibits LPA receptor-mediated actions, especially through LPA1 and LPA3. This blocked LPA protection against HDI-induced apoptosis (Fig. 1A). In primary CLL cells, LPA reduced VPA-induced apoptosis from 45 to 27.5% (Fig. 1B). Previous studies have indicated that caspase-8 is activated by HDI (18). To confirm that HDI-induced apoptosis was blocked by LPA, we examined the effect of VPA alone or in combination with LPA on caspase-8 activation as described under “Experimental Procedures.” HDI have been shown previously to activate caspase-8 (18), and we showed that in untreated or LPA-treated cells caspase-8 activity was detected at a basal level (Fig. 1C), whereas in VPA-treated cells the caspase-8 level was increased ∼5-fold in I-83 cells. When VPA was combined with LPA, activity of caspase-8 was dramatically reduced. This implies that HDI-induced apoptosis was blocked by LPA (Fig. 1C).
FIGURE 1.
LPA blocks HDAC inhibitor-induced apoptosis. A, I-83, BJAB, and CaOv-3 cells were pretreated with 1 μm Ki16425, 1 mm VPA, or 75 nm TSA, following 10 μm LPA treatment in serum-free medium. Apoptosis was determined by membrane permeability assay as described under “Experimental Procedures.” Data represent a trend in four independent experiments. B, primary CLL cells were treated with LPA and VPA, as in A, and apoptosis was measured by flow cytometry following annexin V and 7-AAD staining. Cells that were 7-AAD-negative and annexin V-positive were undergoing apoptosis. The percentage of cells in each quadrant is indicated in the quadrant. C, I-83 cells were treated with VPA alone or in combination with LPA. The 50-μg cell lysate was used for caspase-8 activation assay as described under “Experimental Procedures.” Data represent a trend in four independent experiments.
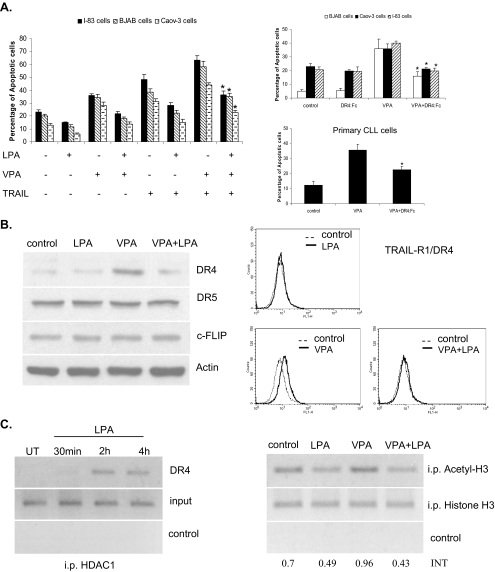
LPA Blocked HDI- and TRAIL-induced Apoptosis—Several studies have suggested that HDIs up-regulate TRAIL DR expression in tumor cells (18, 19, 21). The up-regulation of the TRAIL death receptors can lead to activation of the TRAIL apoptotic pathway (18, 19, 37). In addition, we have shown that VPA activates caspase-8 activity, which is a death receptor-activated caspase. To determine whether HDIs activate the TRAIL apoptotic pathway, we incubated BJAB, I-83, and CaOv-3 cells with 1 mm VPA for 48 h in the presence or absence of 1 ng/ml TRAIL. Combined treatment with VPA and TRAIL increased the apoptotic response over either agent alone, and apoptosis was decreased from 44-63 to 23-36% by the addition of LPA (Fig. 2A). In addition, the above cell lines and primary CLL cells were treated with 1 mm VPA alone or in combination with 100 ng/ml DR4:Fc protein (sequesters TRAIL away from its endogenous receptors) for 24 h. HDI-induced apoptosis was determined by membrane permeability assay, as described under “Experimental Procedures.” VPA induced 36-40% apoptosis in all tested cells, but in the presence of DR4:Fc, VPA-induced apoptosis was reduced to 16-21% (Fig. 2A).
FIGURE 2.
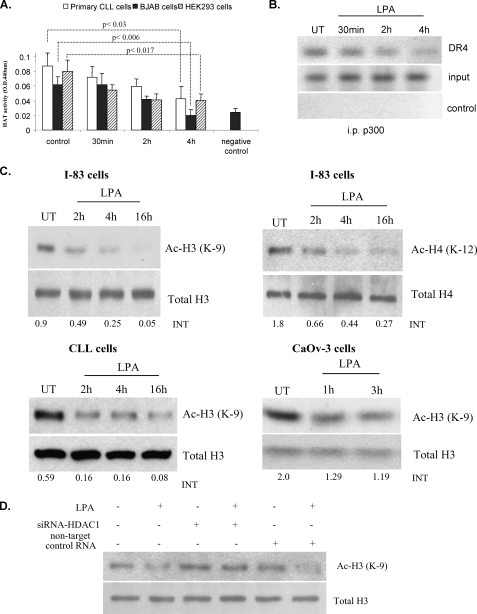
LPA prevents the HDIs and TRAIL-induced apoptosis via the death receptor suppression through HDAC1 binding to the promoter of DR4. A, BJAB, I-83, and CaOv-3 cells were treated with VPA (1 mm) and TRAIL (1 ng/ml) individually or in combination with LPA, as indicated, for 48 h. The percentages of apoptotic cells were determined by membrane permeability assay. Apoptotic cells were scored, with a total of 200 cells being counted for each condition. Standard error was determined on the basis of three independent experiments, and an asterisk represents significant difference (a p value of <0.005) between VPA + TRAIL-treated cells in the presence of LPA compared with the absence of LPA. BJAB, I-83, CaOv-3, and primary CLL cells were treated with VPA (1 mm). The cells were also incubated with DR4:Fc protein (100 ng/ml) as indicated. The amount of apoptosis was determined by membrane permeability assay. Error bars represent the standard errors for three independent experiments, and an asterisk represents a significant difference (p value of <0.005) between VPA-treated cells and VPA + DR4:Fc-treated cells. B, I-83 cells were treated with 1 mm VPA alone or in combination with LPA for 36 h. The protein levels of DR4, DR5, and c-FLIP were examined by Western blotting. The same treated cells were used to determine the cell surface expression of DR4 by flow cytometry. C, cells were treated with LPA (10 μm) over a time course and analyzed using the ChIP assay with anti-HDAC1 antibody. DNA was analyzed by PCR using primers specific for the DR4 promoter. Figure is representative of three separate experiments. Samples with no antibody added were used as a negative control, and total DNA was used as the input control, and the same ChIP experiments were carried out using acetyl-H3 and H3 antibodies, when the cells were treated with VPA alone or in combination with LPA.
HDI-induced DR4 Protein Level Was Decreased by LPA—We have previously demonstrated that fludarabine and chlorambucil treatment increased DR4 and DR5 expression in CLL cells, and this effect contributed to their cytotoxicity (37). Furthermore, DR4 is preferentially activated by HDIs in CLL cells (21). We investigated whether HDI-induced death receptor expression is altered by LPA treatment. I-83 cells were treated with 1 mm VPA in the presence or absence of 10 μm LPA, and DR4 and DR5 protein levels were determined by immunoblot analysis. The expression of DR4 protein was significantly increased by VPA, and this was prevented by the addition of LPA (Fig. 2B). However, DR5 and c-FLIP protein levels were not changed with the same treatment (Fig. 2B). It is known that DRs have to translocate to the cell surface in response to treatment with TRAIL or other drugs (38, 39); therefore, we determined the expression of DR4 receptor on the cell surface by flow cytometric analysis using specific monoclonal antibody against DR4. The expression of DR4 on the cell surface was significantly higher in cells treated with HDI than untreated control, and this was prevented by pretreatment with LPA (Fig. 2B).
Because it has been shown that HDAC1 can be recruited to the promoter region of the gene, thereby silencing the gene activation (12, 35), we determined whether the regulation of DR4 gene expression by LPA involved the recruitment of HDAC1 to the DR4 promoter. Chromatin was isolated from untreated and LPA-treated I-83 cells, and a ChIP assay was performed. HDAC1 or acetyl histone H3 or histone H3 was immunoprecipitated, and purified DNA was analyzed by semiquantitative PCR using primers specific for the DR4 promoter. In LPA treated I-83 cells, HDAC1 was present in a time-dependent manner on the promoter region of DR4 gene (Fig. 2C). Similar results were found in primary CLL cells (data not shown). Furthermore, the binding of acetyl histone H3 to the promoter of DR4 gene was increased by VPA, and this was reduced when the cells were incubated with VPA and LPA (Fig. 2C). These results suggest that HDAC1 is as an important regulator in the LPA-mediated repression of DR4 gene expression.
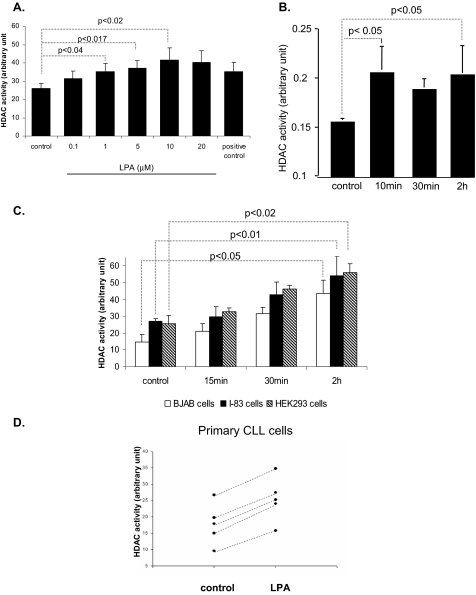
LPA Induces HDAC Enzyme Activity—LPA and HDACs play a global role in the regulation of gene transcription, cell growth, survival, and proliferation. Their aberrant expression or activity contributes to cancer progression (9, 17, 32). To determine whether LPA affects HDAC activation, total nuclear HDAC activity was analyzed after treatment with different concentrations of LPA. The results indicated that LPA induced HDAC activity in a dose-dependent manner (Fig. 3A). Although as little as 1 μm of LPA could induce HDAC activity (from 25 to 35 arbitrary units (AU)) in the CLL-like I-83 cells, 10 μm LPA increased HDAC activity to a greater extent (25-45 AU) (Fig. 3A). Therefore, we used 10 μm LPA throughout this study. The nuclear extract of HeLa cells, as a source of HDAC activity, was taken for positive control. LPA-induced HDAC activity was only detected in the nuclear extract in the I-83 cells, and no activity was detected in the cytoplasmic fraction (data not shown). The time course analysis for the ability of LPA to activate HDAC enzyme activity was also carried out in a variety of different cancer cell lines. The results revealed that HDAC activity increased as early as 10 min after exposure to LPA in the ovarian cancer cell line, CaOv3 (Fig. 3B), whereas an increase was only detected after 30 min of treatment with LPA in I-83, BJAB, and HEK293 cells (Fig. 3C). The highest HDAC activity was determined after 2 h of LPA treatment in all cell lines. Because LPA induces HDAC activity in I-83, primary CLL cells were analyzed for HDAC activity after 2 h of treatment with LPA. Five of eight CLL patient samples showed an increased HDAC activity after LPA treatment (Fig. 3D).
FIGURE 3.
LPA increased HDAC enzyme activity. A, I-83 cells were treated with LPA in various concentrations. 10 μg of nuclear extract was assayed for HDAC enzyme activity as described under “Experimental Procedures.” CaOv-3 (B), BJAB, I-83, and HEK293 (C), and primary CLL cells (D) were treated with LPA at a concentration 10 μm for as long as 2-h time course in serum-free medium. 10 μg of nuclear extract was assayed for HDAC enzyme activity as described under “Experimental Procedures.” Error bars represent the standard deviations for three independent experiments.
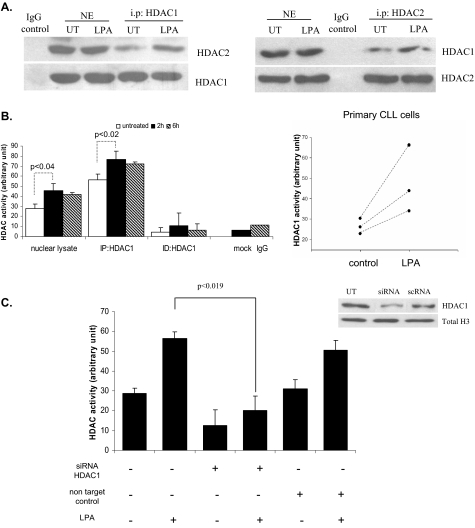
HDAC enzymes are activated in protein complexes. HDAC1 and HDAC2 proteins bind to each other and are recruited to complexes such as the mSin3, Mi2-NURD, and Co-REST complexes. These complexes consist of proteins necessary for modulating the deacetylase activity (9, 13, 14). We therefore performed co-immunoprecipitation experiments to determine whether formation of the HDAC1/2 complex is induced by LPA. HDAC1 was precipitated from nuclear extracts from I-83 cells untreated or treated with LPA. Subsequent immunoblot analysis with the HDAC2 antibody showed an increased association of HDAC2 to HDAC1 after LPA treatment (Fig. 4A). Similarly, increased binding of HDAC1 to HDAC2 was observed after LPA treatment when HDAC2 was immunoprecipitated, and immunoblotting for HDAC1 was performed (Fig. 4A). These data imply that LPA increased the complex formation of HDAC1/2 proteins. Furthermore, to analyze enzyme activity in the HDAC1 complex, the HDAC1 was immunoprecipitated and assayed for HDAC activity as described under “Experimental Procedures.” As shown in Fig. 4B, HDAC1 immunoprecipitation showed increasing HDAC activity (56-77 AU) over a 6-h time course in I-83 cells and a similar increase in total HDAC activity (25-46 AU) (Fig. 3, A and C). However, the HDAC1 immunodepleted complex failed to show HDAC activity. Isotope matching control antibody was used as a negative control. Similar to the above observation, primary CLL cells showed significantly increased HDAC1 activity after LPA treatment, as compared with untreated control cells (Fig. 4B). To confirm the importance of HDAC1 for the increase in HDAC activity observed with LPA, siRNA directed against HDAC1 was transfected into HEK293 cells. Because of the relatively low efficiency with introducing siRNA into suspension cells, we chose HEK293 cells for transfection studies. The transfected cells showed a dramatic reduction (from 60-20 AU) in HDAC activity (Fig. 4C). No increase in HDAC activity was observed when these cells were treated with LPA, suggesting a role for the HDAC1 complex for LPA-induced HDAC activity. HDAC activity was unaffected in cells transfected with nontargeting control siRNA. To confirm siRNA transfection efficiency, HDAC1 protein expression was detected by immunoblot analysis using an HDAC1 antibody in cells untreated, transfected with siRNA, or transfected with nontargeting control siRNA (scRNA). HDAC1 expression was significantly reduced only following siRNA against HDAC1 transfection (Fig. 4C). Total histone H3 represents loading control for the blot (Fig. 4C). The nuclear HDAC1 protein levels did not change following LPA indicating that LPA directly induces HDAC enzymatic activity, and HDAC1 expression was not detected in the pellet (data not shown). Taken together, these results imply that the HDAC1 complex may be the target for LPA-induced HDAC activity.
FIGURE 4.
HDAC1 plays a role in LPA-induced HDAC activity. A, I-83 cells were treated for 2 h by LPA in serum-free medium. Nuclear lysates were co-immunoprecipitated (i.p.) with antibodies to HDAC1 or HDAC2 or a negative control (IgG) followed by immunoblotting for the presence of endogenous HDAC2 or HDAC1. The lower band that is detected by the same antibody used for immunoprecipitation confirmed equal loading. B, HDAC1 immunoprecipitated (IP) complex or immunodepleted (ID) lysate was assayed for HDAC activity as described above. Isotope matching rabbit IgG antibody or immunoprecipitation without antibody were negative controls. The same experiment was carried out in primary CLL cells from three different patients, and the amount of HDAC1 activity was determined after 2 h of LPA treatment. C, HEK293 cells were transfected with siRNA-HDAC1 or siRNA nontarget control. Cells were untreated (UT) or treated with LPA for an additional 2 h, and the nuclear extracts (NE) were assayed for HDAC activity. Western blotting indicates that HDAC1 expression is knocked down by siRNA to HDAC1. Total H3 represents the loading control, and intensity of the blot was determined by densitometry.
LPA Decreases Histone Acetylation—Acetylation is one of the important post-transcriptional modifications of histones (4-8). Acetylation and deacetylation are opposing actions by the HATs and HDACs (5, 8). In previous figures we showed that LPA induced HDAC enzyme activity in primary CLL cells and multiple cancer cell lines. We then investigated whether LPA can reduce cellular HAT enzyme activity in BJAB, HEK293, and primary CLL cells. Compared with untreated control cells, there was a reduction of HAT activity in cells treated with LPA over a 4-h time course. The most dramatic decrease of HAT enzymatic activity (from 0.1-0.06 to 0.04-0.02 OD) occurred at 4 h following LPA treatment in all cell types tested (Fig. 5A). In addition, we determined whether LPA treatment can reduce the recruitment of p300 to the DR4 promoter by ChIP experiment. In LPA-treated cells, reduced binding of p300 to the promoter region of DR4 gene was observed over a 4-h time course (Fig. 5B). This reduction in binding of p300 was consistent with the result showing increased recruitment of HDAC1 to the DR4 promoter (Fig. 2C). Interestingly, it was demonstrated that HDI TSA induced acetylation of histone H3 at lysine 9 (43). In the case of prostate cancer, recent studies have demonstrated that global changes in histone acetylation occur, in particular acetylation of histone H4K12 and H3K9 (36). Therefore, as LPA reduced HAT activation and increased HDAC activation, we speculated that LPA would decrease histone acetylation. We evaluated acetylation of histone H3 at lysine 9 and H4 at lysine 12 sites. As expected, decreased acetylation of histone H3 and H4 were detected by immunoblotting following LPA treatment in I-83 cells (Fig. 5C). Also, treatment of the CLL cells with LPA caused a decrease in the expression of acetylated histone H3, as detected by immunoblotting. The decrease in histone H3 acetylation was also found as early as an hour after LPA treatment in CaOv3 cells (Fig. 5C). In addition, immunoblot analysis showed that LPA treatment failed to decrease acetyl histone H3 in the presence of siRNA against HDAC1 in HEK293 cells (Fig. 5D). Taken together, these results suggest that LPA represses HAT activity and activating the HDAC1 enzyme complex decreases histone acetylation.
FIGURE 5.
LPA reduces acetylation of histones H3 and H4. A, BJAB, HEK293, and primary CLL cells were treated with LPA at a concentration 10 μm for a 4-h time course in serum-free medium. Nuclear extract was assayed for HAT enzyme activity as described under “Experimental Procedures.” Error bars represent the standard deviations for three independent experiments. B, cells treated with LPA (10 μm) over a time course and analyzed using the ChIP assay with anti-p300 antibody. DNA was analyzed by PCR using primers specific for the DR4 promoter. Samples boiled for 15 min were used as a negative control, and total DNA was used as the input control. UT, untreated; i.p., immunoprecipitated. C, acetylation of histones H3 (Ac-H3) and H4 (Ac-H4) was analyzed by immunoblotting the nuclear extract of LPA-treated or -untreated I-83 cells. Total H3 and H4 were taken as loading controls. The same experiments were carried out with primary CLL and CaOv-3 cells. Intensity (INT) of the blot was determined by densitometry program (Bio-Rad). D, HEK293 cells were transfected with siRNA-HDAC1. After 48 h of transfection the cells were treated with LPA for an additional 2 h. Nuclear extracts were analyzed by immunoblotting with antibody against acetyl histone H3. Blots were stripped and reprobed for total H3 as a loading control.
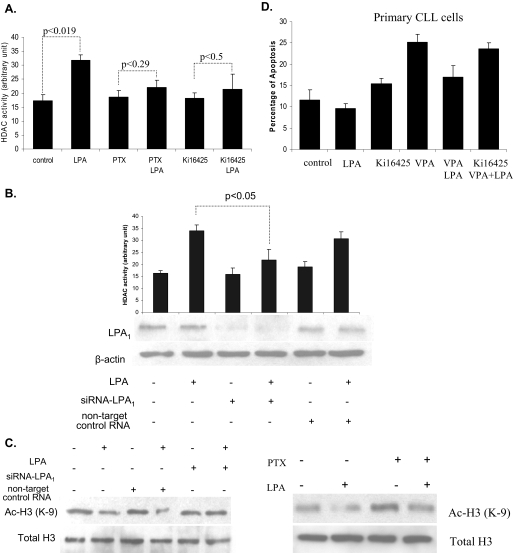
LPA-induced HDAC Activity Is Mediated by LPA1—LPA exerts its biological effects via its specific cognate GPCRs (28, 29). The expression of LPA1 receptors is elevated in colon cancer and CLL cells (31, 34). Therefore, to determine whether LPA receptors are required for LPA-induced HDAC activity, we used the G protein inhibitor, pertussis toxin (PTX). As compared with untreated cells, LPA did not significantly increase HDAC activity in the I-83 cells pretreated with 100 ng/ml PTX (22 AU compared with 18 AU in untreated cells) (Fig. 6A). This implies that LPA-induced HDAC activity is mediated through a Gi-dependent mechanism. Recently, an LPA1-selective antagonist, Ki16425, was developed (Ki values were 0.25 μm for LPA1 and 0.36 μm for LPA3) (40). As shown in Fig. 6A, in I-83 cells 1 μm Ki16425 inhibited the induction of HDAC activity by LPA (from 35 to 21 AU). Because Ki16425 is also a weak antagonist for LPA3, it was possible that LPA3 was involved in LPA-induced HDAC activity. To address this question we used an LPA3-selective agonist, OMPT. HDAC activity was not affected by 1.0 μm OMPT treatment alone, and HDAC activity was not further increased when OMPT was combined with LPA, as compared with LPA alone (data not shown). To further confirm the role of LPA1 in LPA-mediated HDAC enzyme activity, siRNA against LPA1 was transiently transfected in HEK293 cells. LPA treatment failed to induce HDAC activity in the presence of siRNA against LPA1 (Fig. 6B). HDAC activity was unaffected in cells transfected with nontargeting control siRNA. To confirm transfection efficiency, the LPA1 level was assessed by immunoblot analysis in transfected and nontransfected cells, and this confirmed a significant decrease in LPA1 in siRNA-treated cells. β-Actin was used as the loading control for the blot (Fig. 6B). Acetyl histone H3 protein levels, which were down-regulated in LPA-treated cells, were almost completely restored in cells transfected with siRNA against LPA1 (Fig. 6C). Cells transfected with nontargeting control siRNA showed the same as nontransfected cells. Similar results were observed in cells treated with 100 ng/ml PTX, where PTX prevented the reduction in acetyl histone H3 by LPA (Fig. 6C). LPA1 was over-expressed in HEK293 cells, with an empty vector as a control. As expected, significantly increased HDAC1 activity was detected in LPA1 overexpressing cells as compared with cells transfected with the empty vector (data not shown). Finally, to determine whether LPA1 was involved in the protective effect of LPA against HDAC inhibitor-induced apoptosis, I-83, BJAB, and CaOv-3 cells were treated with 1 μm Ki16425 in combination with LPA and VPA or TSA. As shown in Fig. 1A, the addition of Ki16425 prevented the protective effect of LPA against HDAC inhibitor-induced apoptosis (Fig. 1A). Similarly, Ki16425 treatment reversed the protective effect of LPA against HDAC inhibitor-induced apoptosis in primary CLL cells (Fig. 6D). Taken together, these results indicate that the LPA1 receptor plays a role in LPA-induced HDAC activity via Gi proteins and blocks HDAC inhibitor-induced apoptosis.
FIGURE 6.
LPA-induced HDAC activity was mediated via LPA1 receptor. A, I-83 cells were pretreated with 1 μm Ki16425 or 100 ng/ml PTX for at least 1 h before incubating with LPA. HDAC activity was measured in the nuclear extracts. B, HEK293 cells were transfected with siRNA-LPA1 for 48 h before treatment with LPA, and cell lysate was analyzed for HDAC activity. Values represent mean ± S.E. for three separate experiments. Transfection efficiency was assessed by immunoblotting experiments using rabbit polyclonal antibody anti-EDG2 (LPA1). β-Actin represents a loading control. C, protein level of acetyl histone H3 was examined by immunoblotting experiment from the same lysate as described above. I-83 cells were pretreated with 1 μm Ki16425 or 100 ng/ml PTX for at least 1 h before incubating with LPA. The nuclear lysate was analyzed by Western blotting to determine the level of acetyl histone H3 (Ac-H3). D, CLL cells were treated with 1 μm Ki16425, 1 mm VPA, or 75 nm TSA, or alone or in combination with 10 μm LPA in serum-free medium. Apoptosis was determined by membrane permeability assay as described under “Experimental Procedures.” Standard errors were determined from three independent experiments.
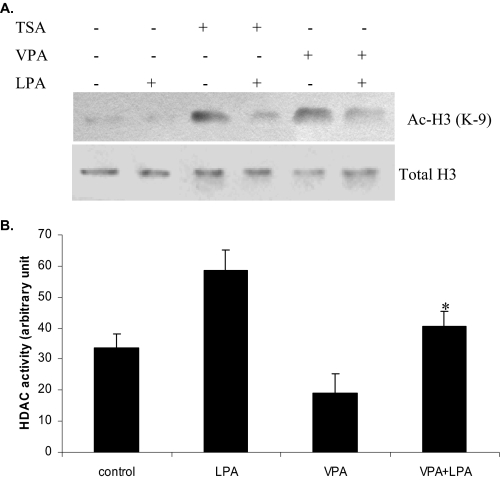
HDI-induced Histone Acetylation Was Attenuated by LPA—HDIs are emerging new class of anticancer agents for the treatment of solid tumors and hematological malignancies (17, 22). We analyzed the ability of LPA to block HDI-induced histone acetylation. To this end, I-83 cells were pretreated with 75 nm TSA and 1 mm VPA, both structurally unrelated HDIs. We found that treatment with these HDIs significantly increased acetyl histone H3 protein levels in the cells (Fig. 7A). However, this increase did not occur when LPA was incubated with the HDIs (Fig. 7A). Total histone H3 was taken as the loading control (Fig. 7A). Also a similar result was observed with CaOv3 cells treated with 0.1-0.5 mm sodium butyrate or 0.1-1.0 mm suberoyl bis-hydroxamic acid alone or in combination with LPA (data not shown). HDAC activity was also assessed in the same cells incubated with VPA alone or in combination with LPA. Treatment with VPA increased the acetylation of histone H3 (Fig. 7A) and reduced HDAC activity (Fig. 7B). However, the addition of LPA to VPA prevented the inhibition of HDAC activity by TSA (Fig. 7B).
FIGURE 7.
LPA reduces HDAC inhibitor-induced histone H3 acetylation and blocks HDAC inhibitor-induced apoptosis. A, I-83 cells were pretreated with 75 nm TSA, 1 mm VPA following 10 μm LPA treatment, and acetyl histone H3 (Ac-H3) was analyzed, as described above. B, cells were treated with LPA and VPA, as in A, and 10 μg of nuclear extract was assayed for HDAC enzyme activity as described under “Experimental Procedures.” Data represent a trend in four independent experiments.
DISCUSSION
Histone acetylation plays an important role in cancer pathogenesis (5-7). HDAC-dependent aberrant transcriptional repression is implicated as the main oncogenic mechanism in specific types of myeloid leukemia, lymphomas, and other types of cancers (15, 22). For example, aberrant recruitment of HDAC activity has been reported in cell lines derived from patients with acute promyelocytic leukemia (41). As a result, HDIs are being evaluated in clinical trials for solid tumors and hematological malignancies (17, 22). However, progress in the field has been hampered by the paucity of knowledge of HDAC biology. In particular, little is known about how HDAC activity is physiologically regulated in normal and neoplastic cells. Environmental and endogenous factors up-regulating cellular HDAC activity have not been described despite the existence of many natural and synthetic inhibitors of HDAC that are empirically being tested in clinical trials on a variety of cancers (4). In this study, we have demonstrated for the first time that HDI-induced apoptosis is effectively blocked by LPA treatment, and LPA blocks HDI-induced DR4 expression. Furthermore, HDAC activity is increased by LPA, and this is mediated through its receptor LPA1. In addition, LPA reduced HAT activation. This is the first demonstrated receptor-mediated HDAC activation reported. This increased HDAC activity and decreased HAT activity induce a reduction in total histone acetylation in several different cancer cell lines and primary CLL cells and increases the recruitment of HDAC1 to the DR4 promoter. Conversely, p300 association with the DR4 promoter is reduced. This represents a novel mechanism for regulating HDAC and HAT activity in cancer cells leading to protection against HDI-induced apoptosis.
HDAC enzymatic activity is regulated by a large protein complex that controls gene transcription (9). Mammalian HDAC1 and HDAC2 proteins are associated with the Sin3, NuRD, and CoREST complexes that include NCo-R and SMR, and these complexes appear to repress gene expression by deacetylating core histones (9, 13, 14). It has been reported that the nuclear receptor peroxisome proliferator-activator γ (PPARγ) binds to the HDAC complexes regulating their function (28). LPA has been shown to bind to PPARγ in monocytes and CV1 cells (42). However, in our studies LPA-induced HDAC activity was not blocked by the PPARγ antagonist GW9662 (data not shown). Serine phosphorylation of HDAC1 has also been implicated in HDAC1 activation and association with protein complexes (41). The kinase responsible for this phosphorylation is unknown, but casein kinase 2 has been implicated (41). However, even in the presence of a casein kinase 2 kinase inhibitor, DMAT, LPA was still capable of inducing HDAC activity (data not shown). Thus the downstream mediators of LPA-induced HDAC activation remain to be determined.
Besides, HDAC complexes, transcription factors regulate HDAC functions through direct association of HDAC proteins. This association represses expression of specific genes because of increased recruitment of HDACs to these genes (14-16). For example, under basal conditions, transcription factor NF-κBis associated with HDAC1 blocking its activation (44). In addition, HDACs induce deacetylation of transcription factors inhibiting their transcriptional activity (9, 14). The tumor suppressor p53 is acetylated on lysine residues that increase its transcriptional activity of pro-apoptotic genes (45). HDIs activate p53, and DR4 induced expression is regulated by p53 (46). NF-κB is also acetylated and associated with increased transcriptional activation (47). LPA-mediated transcriptional repression appears to be mediated by recruitment of HDAC to target genes. We have determined that LPA induces association of HDAC1 with HDAC2, and increased recruitment of HDAC1 to the DR4 promoter correlating with decreased DR4 expression.
LPA reduces HAT activation and p300 recruitment to the DR4 promoter. Because HDAC and HAT enzymatic activity balance each other, the combined effect of increasing HDAC and decreasing HAT enzymatic activity will lead to significant reduction in histone acetylation and alter DR4 expression. However, it has been shown that HDACs can repress as well as activate gene expression (10). Thus, the LPA regulation of and cross-talk between HDAC and HAT enzymatic activity will be the focus for future investigations.
LPA and its receptors have been implicated in cancer progression (26-28). In ovarian cancer, CLL, colon cancer, and multiple myeloma cells, LPA receptors are increased, whereas in the ascites of ovarian cancer patients and bone marrow of multiple myeloma patients, the LPA levels are elevated (30-34). LPA protects ovarian cancer cells from taxol- and cisplatin-induced apoptosis and CLL cells from fludarabine- and chlorambucil-induced apoptosis (33, 34). LPA also protects ovarian, colon, and prostate cancer cell lines from TRAIL-induced apoptosis (48). The mechanism for this protection against apoptosis is mediated by the phosphatidylinositol 3-kinase/AKT pathway, up-regulation of anti-apoptotic proteins such as c-FLIP and Bcl-2, and phosphorylation of BAD. In addition, LPA activation of the Ras/MAPK pathway protects hepatocytes from apoptosis (49). Yang et al. (50) demonstrated that upon activation of MAPK, both co-activator and HDAC complexes are recruited to the repressor complex. Overexpression of LPA2 provides protection from irradiation-induced apoptosis in colon cancer cells, and overexpression of LPA1 protects HEK293 cells from etoposide-induced apoptosis (31, 34). Recently, a direct correlation between G-coupled proteins, Gβγ, and HDACs has been demonstrated (10). In addition, LPA activates gene transcription leading to proliferation in ovarian cancer cells (24).
The HDACs are emerging as important targets in cancer therapy, and the HDIs produce their anti-tumor effect through their ability to modify histone acetylation (9, 14, 15, 22). We and others showed that HDIs potentiate TRAIL-induced apoptosis (18, 19, 23). HDIs also activate the TRAIL death receptor apoptotic signaling pathway leading to HDI cytotoxicity (18, 19, 21). In acute myeloid leukemia cells, HDIs induced the expression of DR4 and DR5 along with TRAIL (18). Using HDIs in combination with TRAIL give a synergistic apoptotic response in many cancer cells. In primary CLL cells, HDIs preferentially activate the DR4 apoptotic pathway sensitizing CLL cells to TRAIL-induced apoptosis (19, 21). This sensitization requires class I HDAC enzymatic activity (23). This agrees with our findings showing LPA activation of HDAC1 (class I HDAC) and increased HDAC1 recruitment to the DR4 gene. In addition LPA effectively blocked HDI-induced apoptosis. This suggests that combining an LPA antagonist with HDIs and/or TRAIL will increase apoptotic response in cancer cells providing a molecular target for cancer treatment.
Acknowledgments
We thank Elizabeth Henson for technical support and advice and Dr. James Davie for suggestions on experimental design. CLL samples from patients were obtained from the Manitoba CLL Tissue Bank. The Manitoba CLL Tissue Bank is part of Cancer-Care Manitoba Translational Research Program.
This work was supported, in whole or in part, by National Institutes of Health Grant CA 102196 (to X. F.). This work was also supported by a translational research grant from the Leukemia and Lymphoma Society and CancerCare Manitoba Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HAT, histone acetyltransferase; CLL, chronic lymphocytic leukemia; LPA, lysophosphatidic acid; HDAC, histone deacetylase; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; GPCR, G protein-coupled receptor; PTX, pertussis toxin; VPA, valproic acid; TSA, trichostatin; 7-AAD, 7-aminoactinomycin D; PBS, phosphate-buffered saline; HDI, HDAC inhibitor; ChIP, chromatin immunoprecipitation; DR, death receptor; siRNA, small interfering RNA; AU, arbitrary unit.
References
- 1.Strahl, B. D., and Allis, C. D. (2000) Nature 403 41-45 [DOI] [PubMed] [Google Scholar]
- 2.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997) Nature 389 251-260 [DOI] [PubMed] [Google Scholar]
- 3.Luger, K., and Richmond, T. J. (1998) Curr. Opin. Genet. Dev. 8 140-146 [DOI] [PubMed] [Google Scholar]
- 4.Gray, S. G., and The, B. T. (2001) Curr. Mol. Med. 1 401-429 [DOI] [PubMed] [Google Scholar]
- 5.Gregory, P. D., Wagner, K., and Horz, W. (2001) Exp. Cell Res. 265 195-202 [DOI] [PubMed] [Google Scholar]
- 6.Mahlknecht, U., and Hoelzer, D. (2000) Mol. Med. 6 623-644 [PMC free article] [PubMed] [Google Scholar]
- 7.Timmermann, S. H., Lehrmann, A., and Harel-Bellan, A. (2001) Cell. Mol. Life Sci. 58 728-736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urnov, F. D., Rebar, E. J., Reik, A., and Pandolfi, P. P. (2002) EMBO Rep. 3 610-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S., and van Kuilenburg, A. B. (2003) Biochem. J. 370 737-749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiegelberg, B. D., and Hamm, H. E. (2005) J. Biol. Chem. 280 41769-41776 [DOI] [PubMed] [Google Scholar]
- 11.Hassig, C. A., Tong, J. K., Fleischer, T. C., Owa, T., Grable, P. G., Ayer, D. E., and Schreiber, S. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3519-3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taplick, J. V., Kurtev, K., Kroboth, M., Posch, T., and Seiser, C. (2001) J. Mol. Biol. 308 27-38 [DOI] [PubMed] [Google Scholar]
- 13.Ahringer, J. (2000) Trends Genet. 16 351-356 [DOI] [PubMed] [Google Scholar]
- 14.Grozinger, C. M., and Schreiber, S. L. (2002) Chem. Biol. 9 3-16 [DOI] [PubMed] [Google Scholar]
- 15.Cress, W. D., and Seto, E. (2000) J. Cell. Physiol. 184 1-16 [DOI] [PubMed] [Google Scholar]
- 16.Ng, H. H., and Bird, A. (2000) Trends Biochem. Sci. 25 121-126 [DOI] [PubMed] [Google Scholar]
- 17.Richon, V. M., and O'Brien, O. P. (2002) Clin. Cancer Res. 8 662-664 [PubMed] [Google Scholar]
- 18.Nebbioso, A., Clarke, N., Voltz, E., Germain, E., Ambrosino, C., Bontempo, P., Alvarez, R., Schiavone, E. M., Ferrara, F., Bresciani, F., Weisz, A., de Lera, A. R., Gronemeyer, H., and Altucci, L. (2005) Nat. Med. 11 77-84 [DOI] [PubMed] [Google Scholar]
- 19.Inoue, S., MacFarlane, M., Harper, N., Wheat, L. M., Dyer, M. J., and Cohen, G. M. (2004) Cell Death Differ. 11 S193-S206 [DOI] [PubMed] [Google Scholar]
- 20.Fulda, S., and Debatin, K. M. (2005) Cancer Biol. Ther. 4 1113-1115 [DOI] [PubMed] [Google Scholar]
- 21.MacFarlane, M., Inoue, S., Kohlhaas, S. L., Majid, A., Harper, N., Fennedy, D. B., Dyer, M. J., and Cohen, G. M. (2005) Cell Death Differ. 12 773-782 [DOI] [PubMed] [Google Scholar]
- 22.Marks, P., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T., and Kelly, W. K. (2001) Nat. Rev. Cancer 1 194-202 [DOI] [PubMed] [Google Scholar]
- 23.Inoue, S., Mai, A., Dyer, M. J., and Cohen, G. M. (2006) Cancer Res. 66 6785-6792 [DOI] [PubMed] [Google Scholar]
- 24.Fang, X., Schummer, M., Mao, M., Yu, S., Tabassam, F. H., Swaby, R., Hasegawa, Y., Tanyi, J. L., LaPushin, R., Eder, A., Jaffe, R., Erickson, J., and Mills, G. B. (2000) Biochim. Biophys. Acta 1582 257-264 [DOI] [PubMed] [Google Scholar]
- 25.Swarthout, J. T., and Wallin, H. W. (2000) Cell. Mol. Life Sci. 57 1978-1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills, G. B., and Moolenaar, W. H. (2003) Nat. Rev. Cancer 3 582-591 [DOI] [PubMed] [Google Scholar]
- 27.Goetzl, E. J., Lee, H., Dolezalova, H., Kalli, K., Conover, C. A., Hu, Y.-L., Azuma, T., Stossel, T. P., Karliner, J. S., and Jaffe, R. B. (2000) Ann. N. Y. Acad. Sci. 905 177-187 [DOI] [PubMed] [Google Scholar]
- 28.Anliker, B., and Chun, J. (2004) J. Biol. Chem. 279 20555-20558 [DOI] [PubMed] [Google Scholar]
- 29.Contos, J. J., Fukushima, N., Weiner, J. A., Kaushal, D., and Chun, J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13384-13389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang, X., Gaudette, D., Furui, T., Mao, M., Estrella, V., Eder, A., Pustilnik, T., Sasagawa, T., Lapushin, R., Yu, S., Jaffe, R. B., Wiener, J. R., Erickson, J. R., and Mills, G. B. (2000) Ann. N. Y. Acad. Sci. 905 188-208 [DOI] [PubMed] [Google Scholar]
- 31.Shida, D., Kitayama, J., Yamaguchi, H., Okaji, Y., Tsuno, N. H., Watanabe, T., Takuwa, Y., and Nagawa, H. (2003) Cancer Res. 63 1706-1711 [PubMed] [Google Scholar]
- 32.Sasagawa, T., Okita, M., Murakami, J., Kato, T., and Watanabe, A. (1999) Lipids 34 17-21 [DOI] [PubMed] [Google Scholar]
- 33.Xu, Y., Fang, X. J., Casey, G., and Mills, G. B. (1995) Biochem. J. 309 933-940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu, X., Haney, N., Kropp, D., Kabore, A. F., Johnston, J. B., and Gibson, S. B. (2005) J. Biol. Chem. 280 9498-9508 [DOI] [PubMed] [Google Scholar]
- 35.Shetty, S., Graham, B. A., Brown, J. G., Hu, X., Vegh-Yarema, N., Harding, G., Paul, J. T., and Gibson, S. B. (2005) Mol. Cell. Biol. 25 5404-5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seligson, D. B., Horvath, S., Shi, T., Yu, H., Tze, S., Grunstein, M., and Kurdistani, S. K. (2005) Nature 435 1262-1266 [DOI] [PubMed] [Google Scholar]
- 37.Johnston, J. B., Kabore, A. F., Strutinsky, J., Hu, X., Paul, J. T., Kropp, D. M., Kuschak, B., Begleiter, A., and Gibson, S. B. (2003) Oncogene 22 8356-8369 [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X. D., Franco, A. V., Nguyen, T., Gray, Ch. P., and Hersey, P. (2000) J. Immunol. 164 3961-3970 [DOI] [PubMed] [Google Scholar]
- 39.Kabore, A. F., Sun, J., Hu, X., McCrea, K., Johnston, J. B., and Gibson, S. B. (2006) Apoptosis 11 1175-1193 [DOI] [PubMed] [Google Scholar]
- 40.Ohta, H., Sato, K., Murata, N., Tomura, H., and Okajima, K. (2003) Mol. Pharmacol. 64 994-1005 [DOI] [PubMed] [Google Scholar]
- 41.Lin, R. J., Nagy, L., Inoue, S., Shao, W., Miller, W. H., Jr., and Evans, R. M. (1998) Nature 391 811-814 [DOI] [PubMed] [Google Scholar]
- 42.Zhang, C., Baker, D. L., Yasuda, S., Makarova, N., Balazs, L., Johnson, L. R., Marathe, G. K., McIntyre, T. M., Prestwich, G. D., Byun, H. S., Bittman, R., and Tigyi, G. (2004) J. Exp. Med. 199 763-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong, S. H., Goto, H., Inagaki, M., and Dong, D. (2003) Oncogene 22 5291-5297 [DOI] [PubMed] [Google Scholar]
- 44.Ashburner, B. P., Westerheide, S. D., and Baldwin, A. S. (2001) Mol. Cell. Biol. 21 7065-7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, L., Scolnick, D. M., Trievel, R. C., Zhang, H. B., Marmorstein, R., Halazonetis, T. D., and Berger, S. L. (1999) Mol. Cell. Biol. 19 1202-1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, X., Yue, P., Khuri, F. R., and Sun, S. Y. (2004) Cancer Res. 64 5078-5083 [DOI] [PubMed] [Google Scholar]
- 47.Chen, L. F., and Greene, W. C. (2003) J. Mol. Med. 81 549-557 [DOI] [PubMed] [Google Scholar]
- 48.Kang, Y. C., Kim, K. M., Lee, K. S., Namkoong, S., Lee, S. J., Han, J. A., Jeoung, D., Ha, K. S., Kwon, Y. G., and Kim, Y. M. (2004) Cell Death Differ. 11 1287-1298 [DOI] [PubMed] [Google Scholar]
- 49.Sautin, Y. Y., Crawford, J. M., and Svetlov, S. I. (2001) Am. J. Physiol. 281 C2010-C2019 [DOI] [PubMed] [Google Scholar]
- 50.Yang, S. H., Vickers, E., Brehm, A., Kouzarides, T., and Sharrocks, A. D. (2001) Mol. Cell. Biol. 21 2802-2814 [DOI] [PMC free article] [PubMed] [Google Scholar]