Abstract
The Kcnq1ot1 antisense noncoding RNA has been implicated in long-range bidirectional silencing, but the underlying mechanisms remain enigmatic. Here we characterize a domain at the 5′ end of the Kcnq1ot1 RNA that carries out transcriptional silencing of linked genes using an episomal vector system. The bidirectional silencing property of Kcnq1ot1 maps to a highly conserved repeat motif within the silencing domain, which directs transcriptional silencing by interaction with chromatin, resulting in histone H3 lysine 9 trimethylation. Intriguingly, the silencing domain is also required to target the episomal vector to the perinucleolar compartment during mid-S phase. Collectively, our data unfold a novel mechanism by which an antisense RNA mediates transcriptional gene silencing of chromosomal domains by targeting them to distinct nuclear compartments known to be rich in heterochromatic machinery.
Recent in silico-based analyses of the mammalian genome have predicted that approximately 60 to 70% of the genome is transcribed. However, only ∼1.5% of the genome encodes protein; the rest of the mammalian genome appears to be transcribing non-protein-coding RNAs (23). Interestingly, a detailed analysis of the mouse transcriptome by the FANTOM3 consortium indicated that more than 72% of all 43,553 genome-mapped transcription units overlap transcripts encoded from the opposite strand, with the majority of transcripts being noncoding. This suggests that antisense transcription is extremely widespread in mammals (17).
An accumulating weight of new evidence from a variety of model systems indicates that noncoding RNAs play an important role in epigenetically controlled gene expression (2, 29, 32, 40). Noncoding RNAs can be classified into two groups: housekeeping and regulatory noncoding RNAs. Housekeeping noncoding RNAs, which include tRNAs, rRNAs, and snRNAs, have a range of functions that are necessary for cell viability. Regulatory noncoding RNAs have been implicated in phenomena that have an impact on cellular differentiation and development in both plants and animals.
Based on size, regulatory noncoding RNAs can be further grouped into two classes: short and long regulatory noncoding RNAs. Short regulatory noncoding RNAs are specifically 21 to 31 nucleotides in length and include microRNAs, small interfering RNAs, and Piwi-interacting small RNAs, which regulate gene silencing at the transcriptional and/or posttranscriptional level (2). Long regulatory noncoding RNAs, whose lengths range in size from 100 bp to several hundred kilobases, have been shown to participate in several important biological functions. Among the long noncoding RNAs, Xist, an X-inactivation-specific transcript, and Tsix, its antisense counterpart, have been well studied for their functional role in mammalian dosage compensation, an epigenetic process by which levels of X-linked gene products are maintained at an equal ratio between the sexes (13, 19). During early embryonic development, the X inactivation process is initiated with the onset of Xist gene transcription, followed by coating of this RNA all along the future inactive X chromosome. This triggers stepwise recruitment of the heterochromatin machinery, which leads to silencing of the chromosome in cis. However, on the future active X chromosome, Tsix antagonizes Xist-mediated X-chromosome inactivation by epigenetically regulating the Xist locus (25, 31, 34).
One of the salient features of the gene clusters exhibiting parent-of-origin-specific expression patterns is the prevalence of noncoding RNAs at these clusters (26). Of particular interest is that the noncoding antisense RNAs encoded from the differentially methylated imprinting control regions (ICR) are generally long, ranging in size from 50 kb to several hundred kilobases, and are reciprocally imprinted to their sense counterparts. These long antisense transcripts have been functionally implicated in long-range bidirectional control of gene expression at imprinted clusters (22, 30, 33, 37). This unique bidirectional control of the expression of flanking genes by the long antisense RNAs raises the important question of how these RNAs differ in their execution of silencing mechanisms compared to the antisense RNAs, such as Tsix, whose effects are primarily restricted to overlapping genes.
In this investigation to address the functional role of long antisense noncoding RNAs, we have exploited an imprinted cluster located at the distal end of mouse chromosome 7. This cluster is divided into two subexpression domains: the H19/Igf2 domain and the Kcnq1 domain. Imprinting in the H19/Igf2 domain is primarily regulated by a chromatin insulator located at the 5′ end of the H19 gene (1, 11, 14, 35). Imprinted gene regulation in the Kcnq1 imprinted domain seems to require the presence of a long noncoding antisense transcript, Kcnq1ot1, whose expression on the paternal chromosome has been linked to the bidirectional repression in cis of eight maternally expressed genes spread over a megabase region (9, 22, 27, 37). The promoter of the Kcnq1ot1 transcript maps to the Kcnq1 ICR in intron 10 of the Kcnq1 gene. The Kcnq1 ICR is methylated on the maternal chromosome but is unmethylated on the paternal chromosome, which encodes the Kcnq1ot1 antisense transcript. It has been recently documented that the imprinted silencing of the Kcnq1 domain involves histone modifications (16, 20, 38). However, the specific mechanisms underlying the Kcnq1ot1-mediated transcriptional silencing are yet unclear.
Previously we showed that a 3.6-kb Kcnq1 ICR fragment, containing the Kcnq1ot1 antisense promoter and 1.7 kb of downstream sequence, is sufficient to silence flanking genes and, more importantly, that the transcriptional elongation of the 1.7-kb sequence results in the efficient silencing of flanking genes in an episome-based system (16, 36). Based on these observations, we posit that the 1.7-kb Kcnq1ot1 sequence may harbor the crucial information required for bidirectional silencing and that this region silences flanking genes more efficiently when it is part of long transcripts than it does as part of short antisense transcripts. In this investigation, we sought to address the mechanisms by which Kcnq1ot1 executes long-range bidirectional silencing. The data presented here document that Kcnq1ot1 harbors a silencing domain that carries out transcriptional silencing in an orientation-dependent and position-independent manner downstream of the antisense promoter through promoting its interaction with the chromatin. Interestingly, this domain also targets the flanking sequences to the perinucleolar compartment, a distinct nuclear space enriched with factors involved in replication and heterochromatin formation. These results unfold a mechanism by which the silencing domain of Kcnq1ot1 initiates transcriptional silencing by recruiting repressive chromatin machinery and spreading it bidirectionally over flanking chromosomal regions. This is clonally maintained through subsequent cell divisions by targeting the flanking sequences to the perinucleolar compartment.
MATERIALS AND METHODS
Cell culture, transfection, and transcriptional inhibitors.
The human placenta-derived JEG-3 cells were maintained in minimal essential medium (Invitrogen) as previously described (15). Transfection of plasmid DNA into the JEG-3 cells was performed using a standard CaCl2 method.
Plasmid constructs.
PH19 is the parent episomal construct possessing the H19 and hygromycin genes as reporter genes and the simian virus 40 (SV40) enhancer. In the PS4 plasmid, we have inserted the 3.6-kb Kcnq1 ICR at the NotI and XhoI sites in such a way that the Kcnq1ot1 promoter in the 3.6-kb ICR faces the H19 promoter. We constructed PGL3-Basic742, PGL3-Basic1050, and PGL3-Basic1947 by amplifying the 742-, 1,050-, and 1,947-bp Kcnq1 ICR fragments using the Del_519FKpnI, Del_742RBglII, Del_1050RBglII, and Del_1947RBglII primers (Table 1) and cloned them into the KpnI and BglII sites in the PGL-Basic vector. Similarly, we constructed PS742, PS1050, and PS1947 by inserting the PCR-amplified Kcnq1 ICR fragments of 742, 1,050 and 1,947 bp using the Del_519FXhoI, Del_742RNotI, Del_1050RNotI, and Del_1947RNotI primers (Table 1) and cloned them at the XhoI and NotI sites of the PH19 episome. The PS6 episome was made by adding an extra 2.2-kb Kcnq1ot1 sequence, amplified using the 3′FlankP1 primers (Table 1), to the already existing 1.7-kb Kcnq1ot1 sequence in the 3.6-kb Kcnq1 ICR, thus making the total length of the antisense transcription unit in the PS6 plasmid 3.9 kb. The chosen Kcnq1ot1 2.2-kb sequence is contiguous with the 1.7-kb sequence. PS6 was also inserted with the SV40 polyadenylation sequence (amplified as previously described in reference 37) at the unique HpaI site, 11.4 kb downstream from the transcription start site. In the PS6A0 plasmid, the 5.8-kb Kcnq1 ICR was mutated to introduce AgeI restriction sites flanking the 890-bp silencing domain.
TABLE 1.
Primers used for amplifying the Kcnq1ot1 and Xist fragments
| Primer name | Sequence |
|---|---|
| DelF_519XhoI | ATATCTCGAGGGATTCCATGGGACAGCTGA |
| Del_742RNotI | ATATGCGGCCGCAATATGCTGAGGCTGGTGGC |
| Del_1050RNotI | ATATGCGGCCGCGAGAGAACCCAGCAGGCTAA |
| Del_1947RNotI | ATATGCGGCCGCTCCACCCCAAGTTCCCAATC |
| 3′FlankP1F | TTCAACACGCTTCTTTTCCCTCTG |
| 3′FlankP1R | ATATGCGGCCGCCACATACATTTTTGAGGAAGAGATG |
| RepeatSeqPR | TATGGTTGGAGGTCACCACA |
| SalIPF | GTGTTTGGCAGCCACAGA |
| ActinF | CTGTACTGCAGCTTGGGTGA |
| ActinR | AAGGAAGCCCTTCCCAGATA |
| DelF_519KpnI | ATATGGTACCGGATTCCATGGGACAGCTGA |
| Del_742RBglII | ATATAGATCTAATATGCTGAGGCTGGTGGC |
| Del_1050RBglII | ATATAGATCTGAGAGAACCCAGCAGGCTAA |
| Del_1947RBglII | ATATAGATCTTCCACCCCAAGTTCCCAATC |
| Xc_4.4 F | GCCCATCACTCAGCCTATAA |
| Xc_4.4R | GCTACAAAGCACCCTTCACA |
| XistslF | ATATACCGGTATGTTTGCTCGTTTCCCGTG |
| XistslR | ATATACCGGTTGGCGTAACTGGCTCGAGAA |
The PS6A1 to -A4 constructs were PS6 derivatives which have various lengths of deletions in the 3.9-kb Kcnq1ot1 sequence. The A1 to A4 deletions were incorporated into the 3.9-kb Kcnq1ot1 sequence by creating AgeI restriction sites flanking the deletions by site-directed mutagenesis, using the primers PM1 to PM5 (Table 2), followed by AgeI restriction digestion and religation with T4 DNA ligase. The PS6A5 plasmid contains the 890-bp silencing domain in the reverse orientation, made by cutting the 890-bp domain from the PS6A0 plasmid with AgeI and reinserting it in the reverse orientation. The orientation of this domain in PS6A5 was identified by sequencing using the RepeatseqPR primer (Table 1). The construction of PS4polyA4.9, used in this study, was described previously (16). PS4polyA4.9A1, a derivative of PS4polyA4.9, was made by deleting the 890 bp domain from the 4.9-kb Kcnq1ot1 sequence using site-directed mutagenesis and inserting the SV40 poly(A) sequence into the unique NotI site. The PS6A6 to -A9 plasmids were constructed by inserting the 890-bp silencing domain, amplified using SDNotI primers (Table 2), in positive and negative orientations at the unique NotI site, which lies 3.1 kb downstream of the antisense transcription start site of PS6A1 and PS6A5. The PS6A10 plasmid contains a 750-bp neutral sequence, which was PCR amplified using NF750 primers (Table 2), instead of 750 bp of native sequence. It was constructed by inserting the neutral sequence at the unique AgeI site of PS6A1. The PS6A11 and -A12 plasmids were generated by cloning the 890-bp silencing domain, PCR amplified using the SDXhoI primers (Table 2), into the unique XhoI site 2.1 kb upstream of the antisense transcription start site in PS6A1 in both positive and negative orientations. The orientation of the silencing domain at the XhoI site was identified by sequencing using the Sal1PF primer (Table 1). PS6A15 contains a 5.8-kb Kcnq1 ICR that has been mutated at five residues of the A2 conserved motif, using the RP1 primer (Table 2). PS6A16 contains a Kcnq1 ICR that has been mutated at five residues in the A1 conserved motif, using the RP2 primer (Table 2). PS6A13 and PS6A14 were constructed by inserting the PCR-amplified 3.0-kb Kcnq1ot1 fragments with and without the A2 motif, respectively, from the 3′ end of the Kcnq1 ICR into the AgeI-NotI site of PS6A1 (see primers in Table 2).
TABLE 2.
Primers used for site-directed mutagenesis
| Primer name | Sequence |
|---|---|
| PM1F | GTTTTTCTTCAACACCGGTCTTTTCCCTCTGT |
| PM1R | ACAGAGGGAAAAGACCGGTGTTGAAGAAAAAC |
| PM2F | CGT TTT GCA GTT ACC GGT CGG TCC TCG TTT CA |
| PM2R | TGA AAC GAG GAC CGA CCG GTA ACT GCA AAA CG |
| PM3F | CTT TGA GCC CCG AGA CCG GTC GAG TCC CAA |
| PM3R | TTG GGA CTC GAC CGG TCT CGG GGC TCA AAG |
| PM5F | GGGGTGGAGGCCGCACCGGTTGCAGTGGGTTC |
| PM5R | GAACCCACTGCAACCGGTGCGGCCTCCACCCC |
| SDXhoIF | ATATCTCGAGTTAGCCTGCTGGGTTCTCTC |
| SDXhoIR | ATA TCT CGA GAG AAC CCA CTG CAA CCA CTG |
| SDNotIF | ATATGCGGCCGCTTAGCCTGCTGGGTTCTCTC |
| SDNotIR | ATATGCGGCCGCAG AAC CCA CTG CAA CCA CTG |
| NF750R | ATATACCGGTAAAAAGGGAATGAGGAGGAC |
| NF750F | ATATACCGGTGGGGATAGCAAGCAAGAAGA |
| RP1F | CTAGCTCTTGGATTTTCTTTGGGTGTGTGT |
| RP1R | ACACACACCCAAAGAAAATCCAAGAGCTAG |
| RP2F | GTGGGTCTACAGTTTTTTTTAGTCTAGAGTGGAG |
| RP2R | CTCCACTCTAGACTAAAAAAAACTGTAGACCCAC |
| PS6A13F | ATATACCGGTCCGAGTTGGGTGTGTGTATG |
| PS6A13R | ATATGCGGCCGCCTGCTGCTCCCTGGTTTTAG |
| PS6A14F | ATATACCGGTGTTCAAGCCTAAAGGGGTCA |
| PS6A14R | ATATGCGGCCGCTGCTGCTCCCTGGTTTTAGT |
To generate the PS6xc4.4 episome, we PCR amplified a 4.4-kb region from Xist encompassing a region which has previously been shown to have high affinity to chromatin (the using Xc_4.4F and Xc4.4R primers [Table 1]). We inserted the fragment into AgeI (created by site-directed mutagenesis 1.6 kb downstream of the Kcnq1ot1 transcription start site using the PM2F and -R primers) and NotI restriction sites in the PS6 episome. We then added the SV40 poly(A) sequence at the NotI site to truncate the hybrid transcript. Likewise, we generated the PS6A1xc4.4 episome by inserting the PCR-amplified 4.4-kb Xist chromatin localizing region into the AgeI and NotI restriction sites of PS6A1, followed by cloning of the SV40 poly(A) sequence at the NotI site. PS6A1Xsl constructs were generated by inserting the 950-bp Xist silencing domain (amplified using the Xistsl primers [Table 1]) into the AgeI site of PS6A1.
PS6A1Kcnq1ot1Frg1-15 episomal plasmids were constructed by inserting Kcnq1ot1Frg1-15 fragments PCR amplified from mouse genomic DNA using PCR primers listed in Table 3, into the unique AgeI site of the PS6A1 episome. The orientation of these fragments relative to the Kcnq1ot1 antisense promoter was determined by sequencing using the RepeatseqPR primer, listed in Table 1.
TABLE 3.
Primers used for scanning the silencing domain in Kcnq10t1
| Fragment | Primer pair | Sequences | Length of fragment (bp) | Position relative to transcription start site |
|---|---|---|---|---|
| Kcnq1ot1FR1 | Kcnq1ot1F1 | ATATACCGGTGTTCAAGCCTAAAGGGGTCA | 2,688 | 1690-4357 |
| Kcnq1ot1R1 | ATATACCGGTTGCTGCTCCCTGGTTTTAGT | |||
| Kcnq1ot1FR2 | Kcnq1ot1F2 | ATATACCGGTATGGCCAGTGCCACTAGTTC | 2,999 | 4237-7235 |
| Kcnq1ot1R2 | ATATACCGGTAGGAGCTGGGATTCTTCCAT | |||
| Kcnq1ot1FR3 | Kcnq1ot1F3 | ATATACCGGTAACTCTCCTAGCCTCTCCCA | 3,698 | 7121-10818 |
| Kcnq1ot1R3 | ATATACCGGTCCTCCAACTGCTCCAAACCT | |||
| Kcnq1ot1FR4 | Kcnq1ot1F4 | ATATACCGGTGGGCAACAGCCCTGTGTGGA | 3,108 | 9586-12693 |
| Kcnq1ot1R4 | ATATACCGGTTAGGATAGCCTGGGCTGTGCT | |||
| Kcnq1ot1FR5 | Kcnq1ot1F5 | ATATACCGGTCCAGTGTTTGATTTGCAGCC | 3,310 | 12380-15689 |
| Kcnq1ot1R5 | ATATACCGGTCACCCCCATCCCTCATTTTT | |||
| Kcnq1ot1FR6 | Kcnq1ot1F6 | ATATACCGGTAGAACAGCAGGCAGAAAGGG | 3,967 | 15145-19111 |
| Kcnq1ot1R6 | ATATACCGGTAAAACCCAGGTGGCATGAAG | |||
| Kcnq1ot1FR7 | Kcnq1ot1F7 | ATATACCGGTACCTCTAACCCTTTGCCTTG | 3,719 | 18783-22501 |
| Kcnq1ot1R7 | ATATACCGGTTCACAGCTTCTTCTTCCCCA | |||
| Kcnq1ot1FR8 | Kcnq1ot1F8 | ATATACCGGTAAGAGAATGAGTACGGCGTG | 3,946 | 22124-26069 |
| Kcnq1ot1R8 | ATATACCGGTTTATGGTGCCCTACTGGAGA | |||
| Kcnq1ot1FR9 | Kcnq1ot1F9 | ATATACCGGTCCTTCGCCCTACCATCTTCA | 3,654 | 25759-29412 |
| Kcnq1ot1R9 | ATATACCGGTTCACCCCAACTTTTACCCCT | |||
| Kcnq1ot1FR10 | Kcnq1ot1F10 | ATATACCGGTAAAGACACCAAGAGGGGGCA | 3,192 | 29061-32252 |
| Kcnq1ot1R10 | ATATACCGGTTCAACAGGCACGTACCAGCA | |||
| Kcnq1ot1FR11 | Kcnq1ot1F11 | ATATACCGGTCCTTGCCAGTTAATCCCAGT | 3,617 | 31951-35567 |
| Kcnq1ot1R11 | ATATACCGGTACCTGGGGACCAATCAATAA | |||
| Kcnq1ot1FR12 | Kcnq1ot1F12 | ATATACCGGTCTCAAATGTCCACCCCAACC | 4,895 | 35374-40268 |
| Kcnq1ot1R12 | ATATACCGGTCCCAGTTCAGTGTTTTCCCC | |||
| Kcnq1ot1FR13 | Kcnq1ot1F13 | ATATACCGGTTGAATGAGCGTGATGGTAGA | 5,766 | 39556-45321 |
| Kcnq1ot1R13 | ATATACCGGTAGGGAGCCAGTTTGAGTTGA | |||
| Kcnq1ot1FR14 | Kcnq1ot1F14 | ATATACCGGTTGGAGGGTGTGTGCTTGAGAA | 4,996 | 45120-50116 |
| Kcnq1ot1R14 | TATACCGGTGTTCCCAGCCCTTTTTGTGT | |||
| Kcnq1ot1FR15 | Kcnq1ot1F15 | ATATACCGGTGGGAGAGGTGATGGGAGATT | 4,488 | 49284-53772 |
| Kcnq1ot1R15 | ATATACCGGTTTTTCCCCTTGGGTCTTGTG |
Luciferase assays.
JEG-3 cells were transiently transfected with the PGL3-Basic, PGL3-Basic742, PGL3-Basic1050, PGL3-Basic1947, and PGL3-BasicCycB2 vectors. After 36 h of transfection, cells were lysed and luciferase activities in duplicate samples were determined as described by the manufacturer (Promega). The pCH110 vector carrying the β-galactosidase gene under the control of the SV40 promoter was used as an internal control for transfection efficiency. Luciferase values were calculated by dividing the luciferase activity by the β-galactosidase activity.
Episome silencer assay.
The wild-type and mutated versions of the PS6 episomal vectors were transfected into the human placenta-derived JEG-3 cells according to a method previously described (15). In order to assess the effect of various modifications in the 5.8-kb Kcnq1 ICR on the antisense RNA-mediated bidirectional silencing, total RNA was extracted 8 days after transfection and an RNase protection assay (RPA) was performed as previously described (15) using a 365-bp H19 antisense probe, a 320-bp hygromycin antisense probe, a 150-bp GAPDH (glyceraldehyde 3-phosphate dehydrogenase gene) antisense probe as a control, and a 270-bp Kcnq1ot1 probe. Quantification of individual protected fragments was done using a Fuji FLA 3000 phosphorimager. H19 and Kcnq1ot1 expression was corrected with respect to both the internal control (GAPDH) and episome copy numbers as determined by Southern blot analysis of BamHI-restricted DNA, hybridized with the H19 and β-Actin probes (the actin DNA probe was PCR amplified using actin primers, listed in Table 1).
In addition to RPA, hygromycin gene activity was also measured by transfecting equimolar concentrations of episome-based plasmids into the JEG-3 cells. Following transfection, the cells were selected with 150 μg/ml of hygromycin until all the cells died in the control plate, which contained the cells incubated with transfection reagent without any episomal plasmid DNA. Following selection, the drug-resistant colonies were stained with hematoxylin and counted.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed using the protocol described earlier (18) and the following antibodies: H3K9Ac (Upstate), H3K9Me3 (Abcam), RNA polymerase II (Pol II) (Euromedex), and TFIIB (Santa-Cruz).
We performed quantitative real-time PCR measurements for input and immunopurified material using Quantitect Sybr green PCR mix in an iCycler real-time PCR machine (BioRad). Each PCR reaction was run in triplicate to control for PCR variation. We calculated the percentage of immunoprecipitation by dividing the average value of the immunoprecipitation by the average value of the corresponding input. The primers and the PCR conditions used for detecting the Kcnq1ot1, H19, and hygromycin gene promoters have been previously described (16).
For the purification of chromatin-associated RNA, we used histone H3 antibody (Abcam) to affinity purify the chromatin by including appropriate RNase inhibitors (GE Healthcare), followed by DNase I (RQ1; Promega) treatment and reverse transcription (Superscript II; Invitrogen) of the purified chromatin using randomprimer (Invitrogen). Before we proceeded with quantitative and semiquantitative analyses of the purified chromatin, we thoroughly checked for DNA contamination in the extracted RNAs by using PCR. We performed quantitative real-time PCR measurements using the Quantitect Sybr green PCR mix (Invitrogen) in ABI Prisam7900 as described above using primers for Kcnq1ot1 (amplified using primers Kcnq1ot1F [5′ TTGGATTACTTCGGTGGGCT 3′] and Kcnq1ot1R [5′ACACGGATGAAAACCACGCT 3′]) and β-Actin (amplified using primers ActinF [5′TGTTACCAACTGGGACGACA 3′] and ActinR [5′ GGGGTGTTGAGGTCTCAAA 3′).
RNA/DNA FISH.
Human placenta-derived JEG-3 cells, propagated with episomes in both transient and stable conditions, were grown on poly-l-lysine-coated slides overnight. The slides were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) on ice, followed by fixation in 4% formaldehyde-5% acetic acid for 18 min at room temperature and washing with PBS three times at room temperature. The slides were dehydrated in ethanol series and air-dried. We carried out probe hybridization and subsequent procedures as described previously (10). We visualized the episome-encoded Kcnq1ot1 transcripts with digoxigenin-labeled single-stranded DNA probes, which were prepared as described previously (5). The probe used to detect episome-encoded Kcnq1ot1 corresponded to the sequence from 102950 to 109990 (accession no. AP001295). DNA fluorescent in situ hybridization (FISH) was performed on slides containing JEG-3 cells, propagated with episomes either transiently or stably using a protocol described previously (15). The PH19 episome was used as a probe for DNA FISH, labeled with digoxigenin-dUTP or biotinylated dUTP.
Combined RNA/DNA immuno-FISH.
RNA FISH was performed initially as described above, followed by immunodetection with nucleophosmin antibody. After RNA immuno-FISH, the slides were refixed in 4% formaldehyde in PBS for 30 min at room temperature, washed three times with PBS for 5 min each, and treated with 100 μg/ml RNase A in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 h at 37°C. After brief rinse, sections were denatured in 70% formamide-2× SSC for 5 min at 85°C and dehydrated with 70% ethanol for 5 min at −20°C followed by 100% ethanol for 3 min on ice, and cells were air dried prior to hybridization with the DNA FISH probe. Hybridization with the DNA probe and posthybridization and signal detection steps were carried out as described previously (3).
RESULTS
Functional characterization of a silencing domain at the 5′ end of the Kcnq1ot1 antisense RNA.
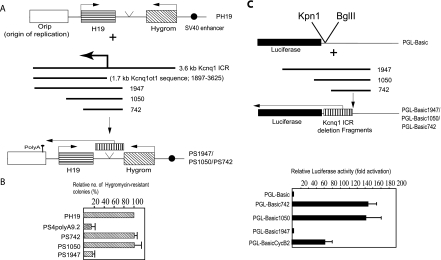
Previous investigations from our lab have implicated a 1.7-kb sequence (1897 to 3625; Kcnq1ot1 transcription start site at 1897), downstream of the Kcnq1ot1 promoter in the 3.6-kb Kcnq1 ICR, in bidirectional silencing of flanking reporter genes. To define the sequences that are critical for bidirectional silencing, we initially generated serial deletions encompassing the region that encodes the 1.7-kb antisense transcription unit in the 3.6-kb Kcnq1 ICR using a PCR-based strategy (Fig. 1A). We inserted all these modified ICRs between the H19 and hygromycin reporter genes in the PH19 episome. In all of these constructs, the ICR is oriented in such a way that the Kcnq1ot1 antisense promoter faces the H19 promoter; thus, the H19 and hygromycin gene promoters become overlapping and nonoverlapping promoters, respectively, to the Kcnq1ot1 promoter (Fig. 1A). We transfected all of the episomal constructs into the human placenta-derived JEG-3 cell line and analyzed the activities of the Kcnq1ot1, H19, and hygromycin genes using an RPA. The hygromycin gene activity was also measured by counting the hygromycin-resistant colonies obtained after selection with hygromycin.
FIG. 1.
Identification of a silencing domain at the 5′ end of Kcnq1ot1 antisense RNA. (A) Schematic diagrams of PH19 and the 3.6-kb Kcnq1 ICR deletion fragments. PH19 is the parent episome construct, which contains H19 (box with long horizontal stripes) and the hygromycin gene (Hygrom; box with diagonal stripes) as reporter genes under the regulation of the SV40 enhancer (filled circle). PS742, -1050, and -1947 are derivatives of the PH19 episome and contain various portions of the 3.6-kb Kcnq1 ICR covering the 1.7-kb Kcnq1ot1 sequence (box with vertical stripes). The arrow represents the transcription start site. The SV40 poly(A) sequence, depicted as a round-headed arrow, is inserted 9.2 kb downstream of the transcription start site. (B) The hygromycin gene activities in the PS742, PS1050, and PS1947 episome constructs was analyzed by counting the hygromycin-resistant cell colonies and are represented as percent expression in relation to that of the parent construct PH19; the data represent means ± standard deviations for a minimum of six independent experiments. (C) Schematic drawings of PGL-Basic and its derivatives, PGL-Basic742, -1050, and -1947. A bar graph showing the relative luciferase activities of the PGL-Basic742, -1050, and -1947 plasmids in JEG-3 cells is shown below. The strengths of the promoter activities of the constructs are presented relative to that for PGL-BasicCycB2. The data represent means ± standard deviations for three independent experiments, each performed in duplicate.
Analysis of the serial deletions indicated that a fragment encompassing the antisense promoter and downstream 600-bp region (see PS1050 in Fig. 1A) could not silence the hygromycin reporter gene. However, hygromycin gene silencing was detected with a fragment encompassing both the antisense promoter and downstream 1.7-kb region (see PS1947 in Fig. 1A), suggesting that a 1.1-kb region (2479 to 3625; Kcnq1ot1 transcription start site at 1897) may play an important role in silencing (Fig. 1B) (see Fig. S1A in the supplemental material).
When we analyzed the fragments that harbored serial deletions for their ability to support promoter activity in a promoter-less luciferase construct, we detected significant luciferase activity from the fragments that did not encompass the 1.1-kb region but not from the fragment that included it (compare PGL_Basic742 and PGL_Basic1050 with PGL_basic1947 in Fig. 1C). Interestingly, however, all of these serial deletions could produce RNA in the episomal context (see Fig. S1B in the supplemental material) and in the luciferase constructs (see Fig. S1C in the supplemental material). The lack of luciferase activity in PGL_Basic1947 could be due to retention of the fused transcript (contains the 1.1-kb portion of the Kcnq1ot1+luciferase gene) in the nuclear compartment and/or interference in the translation of the fused transcript due to occupancy of the bulky heterochromatin complex in the 1.1-kb portion of the fused transcript. We presume that these results collectively indicate that the 1.1-kb region in the 1.7-kb Kcnq1ot1 sequence holds crucial information required for bidirectional silencing.
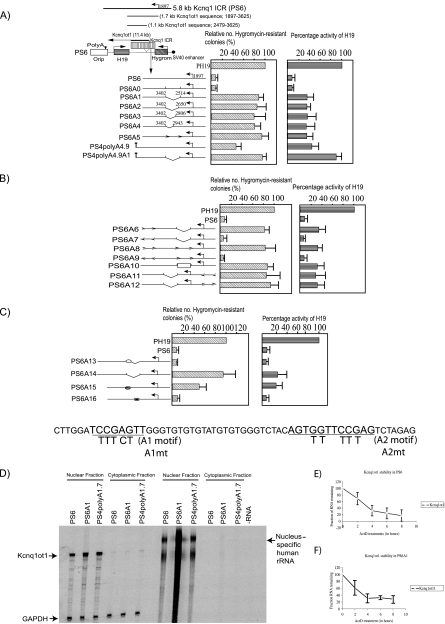
To further define the minimal region required for bidirectional silencing, we created selective internal deletions in the 1.1-kb Kcnq1ot1 sequence as described in Materials and Methods. To incorporate internal deletions into the 1.1-kb sequence, we used a larger version of the Kcnq1 ICR that contained a 3.9-kb Kcnq1ot1 sequence (PS6 in Fig. 2A). In the PS6 episome, the Kcnq1ot1 transcript encoded from the ICR runs on through the H19 coding gene and is truncated at the SV40 poly(A) sequence inserted 11.4 kb downstream of the Kcnq1ot1 start site (Fig. 2A; also data not shown). We inserted each of the modified ICRs, carrying various internal deletions, between the H19 and hygromycin reporter genes in the PH19 episome (Fig. 1A and 2A) and performed a silencing assay as described above.
FIG. 2.
The Kcnq1ot1 SD at the 5′ end of the Kcnq1ot1 antisense RNA silences flanking genes in an orientation- and transcription-dependent manner. (A to C) Bar graphs show the activities of the hygromycin and H19 genes, analyzed for each construct. The maps alongside the bar graphs depict the wild-type and mutant forms of the 5.8-kb Kcnq1 ICR, which were inserted into the PH19 episome between the H19 and hygromycin genes. The activities of reporter genes in each construct are shown as percent expression levels relative to those of control vectors (PH19 for the H19 and hygromycin genes). The expression levels of H19 were quantified by RPA 8 days after transient transfection of wild-type or mutant constructs into the JEG-3 cell line. The levels were normalized against total input RNA (using the GAPDH control) and episome copy numbers (by Southern hybridization of genomic DNA from the transfected cells with an episome-specific probe and an internal β-actin control probe). Hygromycin gene activity was analyzed by counting the hygromycin-resistant cell colonies after selection with hygromycin. RPA results for H19 are expressed as means ± standard deviations for three independent experiments; hygromycin-resistant colony counts are expressed as means ± standard deviations for a minimum of six independent experiments. (D) Episome-encoded Kcnq1ot1 is specifically localized in the nuclear compartment, and selective deletion of the silencing domain from Kcnq1ot1 or truncation of Kcnq1ot1 has no effect on its nuclear localization. (E and F) The half-life of the episome-encoded Kcnq1ot1 transcript is 3.2 h (E), and selective deletion of silencing domain has no significant effect on the stability of Kcnq1ot1 (F).
Selective deletion of an 890-bp sequence (2514 to 3402; PS6A1 in Fig. 2A) from the fine-mapped 1.1-kb region, 617 bp downstream of the antisense transcription start site, resulted in incomplete silencing of the H19 gene, whereas expression of the hygromycin gene was significantly increased. We observed a slight increase in the activity of the Kcnq1ot1 promoter in PS6A1 (see Fig. S2A in the supplemental material). We have further narrowed down the 890-bp silencing domain to 450 bp by creating several deletions of 150 bp starting from the 5′ end of the 890-bp fragment (PS6A2 to PS6A4 in Fig. 2A; also see Fig. S2A in the supplemental material) and found no discernible differences between PS6A1 and PS6A2 to PS6A4, since the hygromycin, H19, and Kcnq1ot1 gene activities more or less remained the same between these constructs (Fig. 2A; also see Fig. S2A in the supplemental material).
If the 890-bp fragment was a silencing domain, one would expect a complete activation of the H19 and hygromycin genes on deletion of this fragment rather than the complete activation only of the hygromycin gene. Previously, by analyzing the kinetics of silencing and heterochromatin formation on the H19 and hygromycin genes in relation to Kcnq1ot1 transcription, we showed that the silencing of the nonoverlapping hygromycin gene occurs primarily because of Kcnq1ot1-mediated heterochromatin formation whereas the overlapping H19 gene silencing occurs by both the Kcnq1ot1 transcription-mediated occlusion of the basal transcription machinery and heterochromatin formation (16). So we presume that specific activation of the hygromycin gene in the 890-bp deletion could be due to the loss of Kcnq1ot1-mediated heterochromatin formation while the incomplete silencing of the H19 gene could result from partial occlusion of the basal transcription machinery due to Kcnq1ot1 transcription.
To further confirm these observations, we deleted the 890-bp Kcnq1ot1 silencing domain (henceforth the 890-bp Kcnq1ot1 silencing domain is known as the Kcnq1ot1 SD) in the PS4polyA4.9 construct [contains a 4.9-kb Kcnq1ot1 sequence followed by an SV40 poly(A) sequence], wherein we have previously shown that the 4.9-kb-long episome-encoded Kcnq1ot1 transcript is capable of inducing moderate silencing of the H19 and hygromycin genes. In this construct, the Kcnq1ot1 transcript does not overlap with the H19 gene (16). Interestingly, we did not see silencing of the H19 and hygromycin genes in PS4polyA4.9A1 compared to results for PS4polyA4.9, suggesting that the incomplete silencing of the H19 gene in PSA1 to PSA4 is mainly due to occlusion of basal transcription machinery (see PS4polyA4.9A1 and PS4polyA4.9 in Fig. 2A; also see Fig. S2A in the supplemental material).
To further reinforce this statement, we used ChIP assays to determine the levels of the major constituents of the preinitiation complex, RNA Pol II and TFIIB, on both the H19 and hygromycin gene promoters. As shown in Fig. S3A in the supplemental material, we found a significant reduction of both RNA Pol II and TFIIB on the H19 promoter but not on the hygromycin promoter in PS6A1. This suggests that moderate silencing of the H19 gene in PS6A1 is primarily due to occlusion of the preinitiation complex from its promoter by the antisense transcription machinery. In addition, we have analyzed two positions in the H19 coding regions (H19c1 and H19c2) to check for the loading efficiency of basal transcription machinery over the nonpromoter regions, which revealed that the basal transcription machinery is specifically loaded over the promoters (see Fig. S3B in the supplemental material).
The Kcnq1ot1 SD induces gene silencing in an orientation- and transcription-dependent but position-independent manner downstream of the Kcnq1ot1 promoter.
We next addressed whether a change in the position and/or orientation of the Kcnq1ot1 SD relative to the Kcnq1ot1 promoter affects its bidirectional silencing property. First, we wanted to investigate whether transcription through the Kcnq1ot1 SD is a prerequisite for the silencing process. To address this issue, we moved the Kcnq1ot1 SD from its natural position to 1.9 kb upstream of the antisense promoter. In this configuration, the Kcnq1ot1 SD is not transcribed by the Kcnq1ot1 promoter (PS6A11 and PS6A12 in Fig. 2B). As can be seen from Fig. 2B (also see Fig. S2B in the supplemental material), the Kcnq1 ICR in this configuration could not silence the hygromycin gene, suggesting that transcription through the Kcnq1ot1 SD is crucial for bidirectional silencing. However, when we moved the Kcnq1ot1 SD from its natural position to 3.3 kb downstream of the antisense promoter in the correct orientation, we observed significant silencing of both the H19 and hygromycin genes, indicating that the Kcnq1ot1 SD has all the information required for bidirectional silencing and that it carries out silencing in a distance-independent manner when present downstream of the antisense promoter (PS6A7 in Fig. 2B; also see Fig. S2B in the supplemental material).
We next addressed the effect of the orientation of the Kcnq1ot1 SD on bidirectional silencing by inverting the Kcnq1ot1 SD at its native position and at other positions. The Kcnq1 ICR with the inverted Kcnq1ot1 SD no longer brought about silencing of the hygromycin gene compared to results with the wild-type Kcnq1 ICR, suggesting that the sequence of the transcribed Kcnq1ot1 SD is crucial for bidirectional silencing (see PS6A5 and PS6A6 in Fig. 2A and B; also see Fig. S2A and B in the supplemental material). Moreover, when we placed the Kcnq1ot1 SD in both orientations, 3.3 kb downstream of the Kcnq1ot1 transcription start site in PS6A5, we observed bidirectional silencing only with its native orientation but not with the opposite orientation (see PS6A5, PS6A8, and PS6A9 in Fig. 2A and B). In addition, when we replaced the Kcnq1ot1 SD with a neutral fragment in the Kcnq1 ICR, we could not detect any silencing by the Kcnq1 ICR (PS6A10 in Fig. 2B; also see Fig. S2B in the supplemental material). Taken together, these experiments provide a strong support for the functional role of the Kcnq1ot1 antisense RNA in bidirectional silencing.
We next wanted to investigate whether the loss of bidirectional silencing upon Kcnq1ot1 SD deletion could be due to a disturbance in its localization or stability of the transcript. To this end, we first characterized the localization of the episome-encoded Kcnq1ot1 transcript with respect to the nuclear and cytoplasmic compartments. We found that the Kcnq1ot1 transcript is exclusively localized in the nuclear compartment. We noted, however, that the deletion of the Kcnq1ot1 SD had no effect on its nuclear localization or on stability of the transcript (Fig. 2D to F), suggesting that the lack of bidirectional silencing in the PS6A1 construct is not due to change in the half-life or nucleus-specific localization of the PS6A1 episome-encoded Kcnq1ot1 transcript.
Functional role of conserved sequence motifs in Kcnq1ot1 antisense RNA.
We next sought to address the functional role of specific sequences within the Kcnq1ot1 SD that are critical for bidirectional silencing. A previous bioinformatic study identified five evolutionarily conserved 30-bp repeat sequences (MD1 repeats) at the 5′ end of the Kcnq1ot1 transcript (21). Interestingly, these repeat sequences map between the Kcnq1ot1 promoter and the Kcnq1ot1 SD identified in this study. Although PS6A1 has all the five 30-bp repeats intact, there was no detectable silencing of the hygromycin gene, suggesting that these repeats do not play any functional role in the silencing process. This observation is consistent with a recently published report that targeted deletion of these conserved repeats from the Kcnq1 ICR in the mouse had no effect on the imprinting of flanking genes (22).
In another recent bioinformatic study, several evolutionarily conserved motifs were identified in the Kcnq1 ICR (28). Interestingly, the A1 (TCCGAGTY) and A2 (YGYGGTTCYGAG) conserved motifs identified in this study map to the 3′ end of the Kcnq1ot1 SD. We decided to explore the functional role of the A1 and A2 motifs in Kcnq1ot1-mediated bidirectional silencing. When we mutated five conserved residues in the A2 motif, we observed a moderate loss of silencing of the hygromycin gene (PS6 and PS6A15 in Fig. 2C; also see Fig. S2C in the supplemental material). However, when we introduced mutations into the conserved residues of the A1 motif (PS6A16 in Fig. 2C), we could not detect any loss of silencing. To gain further insights into the functional role of the A2 motif in bidirectional silencing, we generated episome constructs by inserting Kcnq1ot1 sequences with (PS6A13) and without (PS6A14) the A2 motif into PS6A1. As can be seen from Fig. 2C (also see Fig. S2C in the supplemental material), we noticed significant activation of the hygromycin gene in PS6A14 but not in PS6A13, implicating a critical role for the A2 motif in long-range transcriptional silencing.
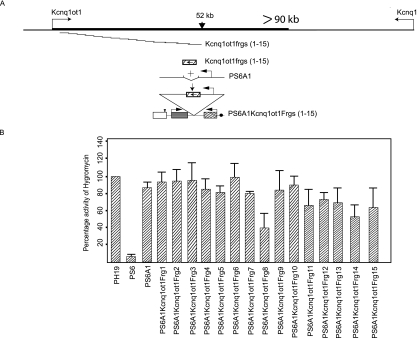
Preliminary results from our lab indicated that Kcnq1ot1 antisense RNA spans more than 90 kb in vivo. A search for the sequence motifs that show homology to the A2 motif in the 90-kb-long Kcnq1ot1 transcription unit revealed none, indicating that A2 could represent the most important functional sequence in the antisense RNA. To address this issue further, we PCR amplified overlapping Kcnq1ot1 sequences spanning about a 52-kb region of Kcnq1ot1 (each fragment ranging in size from 3 to 5 kb in length) and cloned them into PS6A1 (Fig. 3A). We analyzed the effect of these fragments on silencing by measuring hygromycin gene activity. Interestingly, none of the fragments encompassing the 52-kb Kcnq1ot1 transcription unit showed silencing activity comparable to that of the Kcnq1ot1 SD, indicating that the 890-bp sequence could be one of the main functional sequences mediating long-range transcriptional gene silencing (Fig. 3B). We did not see any significant effect on the Kcnq1ot1 promoter activity due to insertion of these fragments in PS6A1 (see Fig. S4 in the supplemental material).
FIG. 3.
The search for a Kcnq1ot1 fragment with bidirectional silencing features over a 52-kb portion of the Kcnq1ot1 transcription unit revealed none of the fragments that have similarities with the Kcnq1ot1 SD. (A) Physical maps depicting the Kcnq1/Kcnq1ot1 locus, overlapping fragments of 3 to 5 kb of the Kcnq1ot1 transcription unit used in episome-silencing assays, and a diagrammatic presentation of the cloning of the Kcnq1ot1 gene fragments into the PS6A1 episome. (B) Bar graph shows the percentage activity of the hygromycin gene in PS6A1Kcnq1ot1Frg1 to -15, analyzed by counting the hygromycin-resistant cell colonies. The data represent means ± standard deviations for a minimum of three independent experiments.
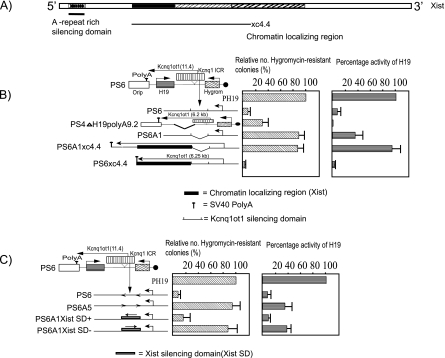
Flanking the Kcnq1ot1 SD with the Xist chromatin attachment region increases efficiency of bidirectional silencing.
Previously, by selectively incorporating deletions into Xist in an embryonic stem cell model system, it was shown that the chromatin localizing property of Xist maps to several regions in a functionally redundant fashion (39) (Fig. 4A). Here we tested whether the efficiency of silencing by the Kcnq1ot1 SD increases if it is flanked with a region of Xist that shows high chromatin localizing activity. We constructed an episomal plasmid wherein a chromatin localizing region of Xist was inserted flanking the Kcnq1ot1 SD and the SV40 poly(A) sequence (PS6xc4.4) was inserted at the end of the Xist chromatin localizing region such that the length of the encoded transcript was around 6.25 kb. Similarly, we also constructed an episomal plasmid containing only the Xist chromatin localizing region and SV40 poly(A) at the end but lacking the Kcnq1ot1 SD (PS6A1xc4.4). In both the PS6xc4.4 and PS6A1xc4.4 episomes, Kcnq1ot1 does not overlap the overlapping H19 reporter gene. We compared the reporter gene activity in PS6xc4.4 with that of a control episomal plasmid, PS4ΔH19polyA9.2, which encodes a length of Kcnq1ot1 similar to that of PS6xc4.4 (see reference 16 for details). As can be seen from Fig. 4A and B (also see Fig. S5A in the supplemental material), the efficiency of bidirectional silencing by the Kcnq1ot1 silencing domain was significantly enhanced when it was flanked with the chromatin localizing region (compare PS6xc4.4 with PS4ΔH19polyA9.2 in Fig. 4B). However, we could not detect any silencing if the chromatin localizing region was transcribed in the absence of the Kcnq1ot1 SD (compare PS6A1xc4.4 with PS6xc4.4 in Fig. 4B) or it was transcribed along with the Kcnq1ot1 SD positioned in an inverted orientation (data not shown), suggesting that the Kcnq1ot1 SD is a critical regulator of bidirectional silencing and its efficiency of silencing significantly increases with the inclusion of the chromatin localizing regions of Xist. This can be compared to the previously demonstrated effect of a 950-bp Xist silencing domain, an A-rich repeat sequence located at the 5′ end of the Xist transcript, which displayed similar functional features in our episome-based system, i.e., it induces bidirectional silencing in an orientation-dependent manner (Fig. 4C; also see Fig. S5B in the supplemental material).
FIG. 4.
High-affinity chromatin attachment regions of the Xist transcript increase the efficiency of bidirectional silencing. (A) A schematic representation of the Xist RNA. The silencing domain containing the A-rich repeats is located at the 5′ end of the transcript. The line labeled xc4.4 indicates the chromatin localizing region that has previously been mapped (39). (B and C) Schematic maps depicted alongside the bar graphs are the wild-type and mutant Kcnq1 ICR fragments (the Kcnq1ot1 SD is either flanked with a high-affinity Xist chromatin localizing region or replaced with the Xist SD) inserted into the PS6 or PS6A1 episome. The schematic map of the PS4ΔH19 polyA9.2 construct alongside the bar graph is shown along with the reporter genes. PS6Xist SD+ and PS6Xist SD- indicate that the Xist SD was inserted in both positive and negative orientations, respectively, in PS6A1. Bar graphs show the percentage activities of the hygromycin and H19 genes in the episomes containing wild-type and mutant Kcnq1 ICR fragments, calculated as described in the legend for Fig. 2. The data represent means ± standard deviations for three independent experiments.
The Kcnq1ot1 SD silences the flanking reporter genes by spreading repressive epigenetic modifications.
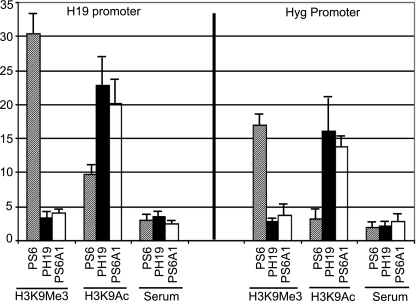
Previously we have documented that Kcnq1ot1 silences the flanking reporter genes through spreading epigenetic modifications in cis (16). In this investigation, we addressed whether the acquisition of epigenetic modifications by the flanking chromatin due to the antisense RNA involves the functional role of the Kcnq1ot1 SD. To this end, we investigated the chromatin structure on both the H19 and hygromycin promoters by analyzing the levels of histone H3 with trimethylated (H3K9me3) and acetylated (H3K9ac) lysine 9, using a ChIP assay with JEG-3 cells transfected with PS6 and PS6A1. Strikingly, both the H19 and hygromycin promoters were enriched in H3K9me3 when fully silenced (Fig. 5, PS6) but not when silencing was lost due to deletion of the Kcnq1ot1 SD (Fig. 5, PS6A1). Interestingly, we observed a marked increase in the levels of H3K9ac on both the H19 and hygromycin promoters in PS6A1 (Fig. 5, PS6A1), suggesting that the Kcnq1ot1 SD silences the flanking genes by spreading epigenetic modifications specific to inactive chromatin.
FIG. 5.
The Kcnq1ot1 SD mediates transcriptional silencing through regulating the chromatin structure. Quantitative analysis of H3K9Me3 and H3K9Ac profiles using ChIP over the H19 and hygromycin gene promoters on cross-linked chromatin obtained from JEG-3 cells transfected with the PS6, PH19, or PS6A1 episomes. Bar graphs shows percent enrichment, and the data were quantified as described in Materials and Methods. The data represent means ± standard deviations for three independent ChIP experiments.
Kcnq1ot1 mediates transcriptional silencing by targeting the episomes to the perinucleolar region.
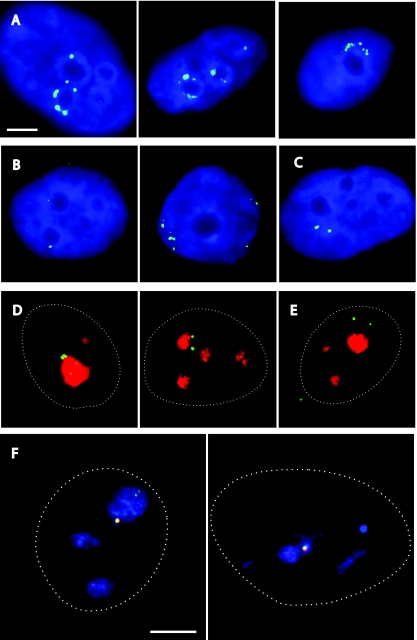
The above observations clearly indicate that the Kcnq1ot1 SD mediates transcriptional silencing of flanking genes by regulating chromatin structure, probably through recruiting heterochromatic machinery. Several studies have found a link between gene silencing and recruitment to nuclear heterochromatin compartments (3, 4, 7, 8, 12). We were therefore keen to examine whether Kcnq1ot1 SD-mediated transcriptional silencing correlates with positioning of the episomal sequences in distinct nuclear compartments. We first wanted to confirm that episome-encoded Kcnq1ot1 RNA can be detected in the vicinity of episomal DNA sequences. To this end, we performed combined RNA/DNA FISH with a Kcnq1ot1 RNA probe and an episomal DNA probe on JEG-3 cells transfected transiently with PS6A13 (with the A2 motif) for 4 days. We found that RNA signals colocalize with DNA signals, indicating that episomal sequences can be visualized by either RNA or DNA FISH (see Fig. S6A and B in the supplemental material). We then evaluated the nuclear localization of episomal sequences in JEG-3 cells propagated with the PS6, PS6A1, PS6A5, PS6A13 and PS6A14 (without A2 motif) episomes by RNA and DNA FISH (Fig. 6). We noted that 4 days after transfection with PS6A13, many transfected cells displayed clusters of episomal signals either surrounding nucleoli or enriched near the nuclear periphery (Fig. 6A). Similarly, after 30 days under hygromycin selection, in many cells carrying the PS6 construct, episomal signals were located in close proximity to nucleoli or the nuclear periphery (Fig. 6B). We scored the number of cells (n = 104) displaying different types of distribution of RNA FISH signals for JEG-3 cells transiently transfected with PS6A13. We found that in 34% of nuclei, the Kcnq1ot1 signals were surrounding nucleoli (Fig. 6A). A further 17% of the nuclei showed RNA signals dispersed at the nuclear periphery. We also assessed the nuclear positioning of the DNA FISH signals in JEG-3 cells stably propagated with PS6 and PS6A1. PS6 episomes were located in the vicinity of nucleoli in 43% of the nuclei in the cellular population (n = 101) (Fig. 6B) or localized at the nuclear periphery in another 29% of nuclei (Fig. 6B). By contrast, in the vast majority of JEG-3 cells stably propagated with PS6A1, the episomal FISH signals did not occupy perinucleolar or perinuclear positions (Fig. 6C). In these cells, association with nucleoli and perinuclear distribution were found only in 11% and 8% of all nuclei, respectively. These results show that silencing of the episomal genes is indeed associated with positioning of the episomes in close proximity to heterochromatin. Importantly, we did not detect specific enrichment of PS6A5 (having the silencing domain in reverse orientation) episomes at the nucleolar periphery (data not shown). This observation indicates that the silencing domain in the Kcnq1ot1 transcript, rather than the DNA sequence, is crucial for nucleolar targeting.
FIG. 6.
The Kcnq1ot1 SD localizes the linked episomal sequences to the perinucleolar and perinuclear regions. (A) RNA FISH with JEG-3 cells transiently transfected with PS6A13 showing perinucleolar localization of episomes. The Kcnq1ot1 probe is detected in green; 4′,6′-diamidino-2-phenylindole (DAPI) staining is in blue. Nucleoli are visible as darker areas in the DAPI staining pattern. Scale bar = 5 μm. (B and C) DNA FISH with JEG-3 cells propagated with PS6 (B) or PS6A1 (C) for 30 days under hygromycin selection. Episomal DNA FISH signals are detected in green; DAPI staining is in blue. The scale is as in panel A. Representative examples of cells with PS6 signals in close proximity to a nucleolus (B) (cell on the left), the nuclear periphery (B) (cell on the right), and PS6A1 signals positioned away from these nuclear compartments (C) are shown. (D and E) RNA immuno-FISH with JEG-3 cells stably propagated with PS6A13 (D) or PS6A14 (E). Green represents episome-encoded RNA; red corresponds to nucleolar marker nucleophosmin. The outlines of the cells are shown as dotted lines. The scale is as in panel A. (F) RNA/DNA immuno-FISH with JEG-3 cells stably propagated with PS6A13. Episome-encoded RNA is in green, DNA is in red, and nucleophosmin staining is in blue. The outlines of the cells are shown as dotted lines. Scale bar = 5 μm.
We wanted to examine the association of Kcnq1ot1 RNA transcripts with the nucleolus in more detail. We analyzed JEG-3 cells stably propagated with the PS6A13 and PS6A14 episomes. We performed RNA FISH in combination with immunostaining for a nucleolar marker, nucleophosmin. Consistent with our earlier findings, we demonstrated that PS6A13-encoded Kcnq1ot1 RNA was in direct contact with nucleophosmin-stained nucleoli (Fig. 6D and E), and the percentage of cells that show nucleolar contact in nucleophosmin-stained cells was more than 40% and was similar to what had been obtained with RNA FISH. We next performed combined RNA/DNA immuno-FISH with PS6A13-stably propagated JEG-3 cells to check whether RNA and DNA signals that are in contact with nucleoli colocalize with each other. In combined RNA/DNA-immuno FISH, following hybridization with a digoxigenin-labeled Kcnq1ot1 probe overnight, RNA FISH signals were visualized by immunodetection of digoxigenin with fluorescent antibodies. After that, the cells are refixed and treated with RNase. The RNA FISH signals survive the RNase treatment, because Kcnq1ot1 RNA was protected by the probe and additional layers of antibodies. Chromatin was then denatured, followed by a second round of hybridization with a biotin-labeled episome probe. This probe hybridizes to episomal DNA sequences and is subsequently visualized by fluorescent detection of biotin. These experiments confirmed that nucleolus-associated DNA and RNA FISH signals colocalize with each other (Fig. 6F). In combination, our results show that the Kcnq1ot1 SD plays a crucial role in targeting the episomes to the vicinity of the nucleolus.
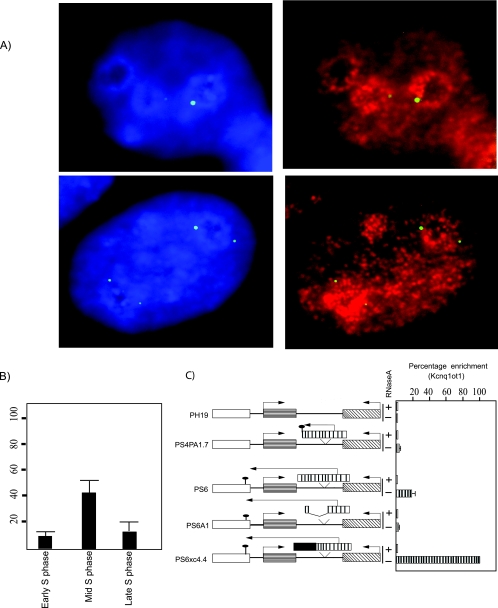
In a recent investigation using BrdU labeling and PCNA immunostaining, it was documented that Xist-mediated inactive X-chromosome contact with the nucleolus occurs predominantly during mid- to late S phase (41). Using the same methodology, we wanted to investigate whether localization of Kcnq1ot1 DNA FISH signals in the nucleolar periphery corresponds to any particular phase of the cell cycle. Interestingly, we found that 41% of Kcnq1ot1 RNA FISH signals were localized in the vicinity of the nucleolus around mid-S phase of the cell cycle, although we do see nucleolar localization during early (8%) and late (14%) S phase but at a lower level than that of mid-S phase (41%), implying that silencing domain-mediated nucleolar contact is dependent on the phase of the cell cycle (Fig. 7A and B).
FIG. 7.
Perinucleolar localization of Kcnq1ot1 occurs during mid-S phase. (A) Simultaneous detection of episomal DNA by DNA FISH (green) and replication foci by immunostaining for PCNA (red). (B) Bar graph showing percent nucleolar localization of PS6A13 episomes in early, mid, and late S phase of the cell cycle. The data represent means ± standard deviations for three independent experiments (n = >100). (C) Kcnq1ot1 interacts with chromatin. Real-time quantification of reverse-transcribed immunopurified chromatin-associated RNA suggests that Kcnq1ot1 interacts with chromatin and this interaction is mediated by the Kcnq1ot1 SD.
Kcnq1ot1 SD mediates chromatin interaction of Kcnq1ot1 antisense transcript.
Our combined RNA/DNA FISH indicated that the RNA and DNA signals were colocalized with each other, giving an indication that the transcribed Kcnq1ot1 RNA might be in direct contact with chromatin. To address whether Kcnq1ot1 RNA is in direct contact with chromatin, we have used chromatin-associated RNA immunoprecipitation. In this technique, we immunopurified the chromatin using a histone H3 antibody and extracted RNA from the immunopurified chromatin, followed by DNase I treatment and reverse transcription of the purified RNA. We found that chromatin purified from cells transfected with PS6xc4.4 showed a five- to sixfold enrichment of Kcnq1ot1 in comparison to chromatin purified from cells transfected with PS6. This suggests that Kcnq1ot1 encoded by PS6 associates with chromatin, albeit with a lower affinity than Kcnq1ot1 containing chromatin-localizing signals from Xist (compare PS6 with PS6xc4.4 in Fig. 7C). Surprisingly, we found that the chromatin association of Kcnq1ot1 encoded by PS6 was dependent on the Kcnq1ot1 SD, since the loss of this region from the transcript resulted in a significant decrease in the level of its association with chromatin (compare PS6A1 with PS6 in Fig. 7C). Moreover, when we truncated Kcnq1ot1 to a 1.7-kb length, we also found a significant decrease in Kcnq1ot1 RNA levels in the purified chromatin, suggesting that the length of Kcnq1ot1 determines its association with the chromatin (compare PS4polyA1.7 with PS6 in Fig. 7C).
DISCUSSION
Several models have been proposed to explain the mode of action of long noncoding RNAs. For example, for Drosophila melanogaster, it has been documented that expression of noncoding transcripts from trithorax response elements has been linked to the recruitment of the trithorax protein Ash1 to the downstream cis regulatory elements (32). Similarly, onset of Xist expression, followed by the coating of one of the two X chromosomes with it during early female embryonic development, coincides with the recruitment of transcriptional repressors and inactivation of the X chromosome. It has been shown that an A-repeat-rich silencing domain at the 5′ end of Xist and chromatin localizing domains, spread in a redundant fashion all over the gene body of Xist, play a crucial role in Xist-mediated X-chromosome inactivation (39). On the other hand, mechanisms underlying the antisense noncoding RNA functions have not been investigated in greater detail. It is as yet unclear whether antisense RNA itself or its transcriptional process as such plays an important role in transcriptional silencing. However, recent studies of Tsix have provided some important insights into antisense RNA-mediated transcriptional silencing. It has been shown that the production of Tsix antisense RNA has been linked to Xist promoter regulation of the future active X chromosome. Using RNA immunoprecipitation and chromatin immunoprecipitation experiments, it has been demonstrated that Tsix associates with DNMT3B and that the promoter region of Xist is also enriched with DNMT3B, indicating that Tsix may be involved in Xist promoter regulation through DNMT3B deposition (34). Interestingly, the effects of Tsix antisense RNA are restricted to its overlapping sense counterpart, Xist. Unlike Tsix, the other long functional antisense RNAs, such as Kcnq1ot1 and Air, are implicated in the transcriptional silencing of not only overlapping genes but also nonoverlapping genes. However, the mechanisms underlying the mode of action of these RNAs have so far not been investigated in greater detail. We discuss the possible implications of our findings in understanding the mechanisms underlying Kcnq1ot1-mediated long-range transcriptional silencing.
One of the most important observations of the present investigation is the identification of a Kcnq1ot1 silencing domain (Kcnq1ot1 SD) at the 5′ end of the Kcnq1ot1 antisense transcript. This domain silences genes in an orientation-dependent and position-independent manner only when it is located downstream of the antisense promoter but not when it is placed upstream of the antisense promoter. A lack of silencing by the Kcnq1ot1 SD upstream of the antisense promoter rules out the possibility that the cis-acting DNA sequences of the Kcnq1ot1 SD, independently and/or in coordination with the antisense promoter, may take part in silencing and confirms that antisense transcription through the Kcnq1ot1 SD is a prerequisite to inducing long-range bidirectional silencing. The orientation-dependent and position-independent silencing by the Kcnq1ot1 SD downstream of the antisense promoter suggests that the primary sequence of the transcribed RNA is crucial for the silencing process and that it has all the information required for inducing the silencing process. Interestingly, a scan for sequences over 52 kb of the more than 90-kb-long Kcnq1ot1 revealed no sequences that show silencing activity in a manner similar to that of the Kcnq1ot1 SD. Based on our observations, we rule out the possible role of transcriptional interference in the Kcnq1ot1-mediated silencing pathway in the episomal context, since we found significant activation of the hygromycin gene despite increased expression of mutant Kcnq1ot1 in PS6A1 compared to that in PS6. In addition, the latter data also rule out the functional role of double-stranded-RNA-mediated RNA interference in the bidirectional silencing pathway. To our knowledge, the uncovering of functional sequences in Kcnq1ot1 represents the first characterization of functional sequences in an antisense RNA.
Another important observation that we provide in these investigations is the Kcnq1ot1 SD-mediated localization of the episomal DNA sequences in the vicinity of the nucleolar compartment. Perinuclear and perinucleolar compartments have long been suggested to harbor protein factors involved in repressive chromatin formation. Interestingly, the perinucleolar localization of the episomes by the silencing domain occurred as early as 4 to 5 days after transient transfection, and their colocalization with the nucleolus increased in the cell lines containing stably propagated episomes, suggesting that the perinucleolar localization of the episomes may be crucial for both establishment and maintenance of Kcnq1ot1 silencing domain-mediated transcriptional silencing. The observations that (i) RNA and DNA signals colocalize in the perinucleolar region, (ii) the silencing domain mediates transcriptional silencing through spreading repressive chromatin structures by promoting chromatin interaction, and (iii) the lack of perinucleolar contacts of the episomes and the loss of repressive chromatin structures over neighboring chromatin regions of the episomes when Kcnq1ot1 was transcribed in the absence of the Kcnq1ot1 SD suggest that the Kcnq1ot1 SD establishes transcriptional silencing through recruiting ribonucleoprotein complexes and promoting chromatin interaction, and it maintains the repressive chromatin further through successive cell divisions by translocating the linked episomal sequences to the nucleolar periphery. This specific localization of the episomes by the Kcnq1ot1 SD occurs during mid S phase, indicating that contact of the Kcnq1ot1 SD with the nucleolus is regulated in a cell cycle-dependent manner.
Interestingly, however, the functional features of the Kcnq1ot1 SD resemble closely those of the Xist silencing domain (Xist SD), which was shown to mediate transcriptional silencing on the inactive X chromosome in an orientation- and position-independent manner with an ex vivo embryonic stem cell model system (39). Moreover, the Xist SD also displays similar silencing features in the episome-based system, documenting the specificity and reliability of using an episome-based system in addressing the intricate molecular pathways underlying the function of long antisense noncoding RNAs. Most importantly, perinucleolar targeting of episomal sequences by the Kcnq1ot1 SD is reminiscent of Xist-mediated perinucleolar localization of the inactive X chromosome. It has been shown that the Xic transgene maintains the transcriptional silencing of flanking autosomal genes by targeting them to the nucleolar periphery (41). It is not yet clear whether the Xist SD has any functional role in perinucleolar targeting of the inactive X chromosome. However, it has been shown that the Xist SD is required for relocating the linked genes into the Xist RNA silent domain (6), indicating that the Xist SD may also have a functional role in perinucleolar targeting of X-linked genes. Both the Xist and Kcnq1ot1 silencing domains hardly show any homology at the primary sequence level, indicating similarity at the functional level and/or target-specific interactions.
Yet another interesting observation of the current investigation is the characterization of the functional role of conserved motifs in the Kcnq1ot1-mediated silencing process. Mutation or selective exclusion of the previously characterized highly conserved motif A2 results in a relaxation of the silencing of flanking genes, indicating that the A2 motif plays a critical role in Kcnq1ot1-mediated silencing. More importantly, sequences encompassing the 12-bp A2 motif also play a crucial role in targeting the linked episomes to the nucleolar periphery. m-fold predictions on a sequence containing the wild-type A2 motif revealed a putative stem-loop structure, with the stem mainly formed by the A2 motif. However, we did not observe a proper stem-loop structure with the sequence containing the mutant A2 motif, indicating that the secondary structure attained by the A2 motif may play a crucial role in the silencing process (data not shown). Although in silico-based m-fold predictions emphasize the functional role of RNA secondary structures in the silencing process, we cannot exclude the possibility that the crucial cis-acting elements within the A2 motif may be involved in the formation of repressive chromatin through recruiting transcription factors. We could not, however, detect any sequence motif that shows homology with the A2 motif in the 90-kb-long Kcnq1ot1 transcript, suggesting that the A2 motif itself may be playing a crucial role in Kcnq1ot1-mediated transcriptional silencing.
It is interesting to note that when we flanked the Kcnq1ot1 SD with a chromatin-localizing region of Xist, we observed not only a significant increase in the silencing activity but also increased chromatin interaction of Kcnq1ot1. These observations will have implications in understanding why Xist transcript-mediated transcriptional silencing on the inactive X chromosome is spread over most of the chromosome, in comparison to Kcnq1ot1-mediated silencing, whose effects are restricted only to a 900-kb region. We presume that the limited silencing by Kcnq1ot1 could be due to the lack of high-affinity chromatin localizing regions, such as those seen in Xist. Although the Kcnq1ot1 SD mediates chromatin localization, it seems that the affinity is insufficient for chromosome-wide silencing. The Xist SD, on the other hand, mediates chromosome-wide transcriptional silencing with the help of chromatin localizing regions which are distributed throughout the transcript in a functionally redundant fashion (39).
From the above observations, it appears that the transcriptional silencing that we see in the PS6 episome is mediated by the antisense RNA per se and that in part it mimics the mode of action of Xist. In this model, the Kcnq1ot1 antisense RNA is expressed and coats the chromatin of flanking sequences in cis, followed by the recruitment of heterochromatinization machinery, which in turn targets the linked chromosomal regions to the perinucleolar vicinity in order to maintain the repressive chromatin through successive cell divisions (Fig. 8). Although we have shown that repressive chromatin marks over the flanking sequences correlate with its chromatin localizing activity, the chromatin-associated RNA immunoprecipitation methodology cannot resolve whether the RNA localizes the entire episome in cis or it remains attached at the site of antisense transcription through the Kcnq1ot1 SD. Interestingly, a recent report using high-resolution fiber RNA FISH and RNA tagging and recovery of associated proteins demonstrated that the human KCNQ1OT1/LIT1 transcript coats the neighboring regions of chromatin containing the SLC22A18/IMPT1 and CDKN1C/p57KIP2 genes (24). Taken together, these results show that Kcnq1ot1-mediated transcriptional silencing mimics in part the Xist-mediated X-chromosome inactivation.
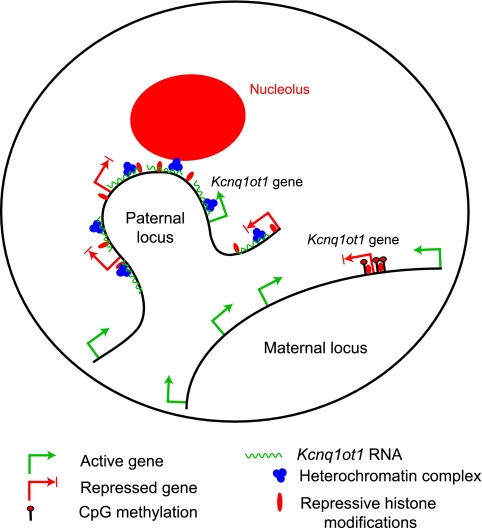
FIG. 8.
The transcriptional silencing by Kcnq1ot1 requires the linked sequences at the perinucleolar region. Production of Kcnq1ot1 on the paternal chromosome results in the recruitment of heterochromatic machinery by the Kcnq1ot1 SD. Subsequently, the heterochromatic machinery associated with the antisense RNA (shown in green squiggles) mediates the chromatin interaction in a manner analogous to that of the Xist transcript, resulting in the spreading of heterochromatic histone modifications (red blobs) in a Kcnq1ot1-dependent manner in cis. Then, the Kcnq1ot1 transcript and associated heterochromatic machinery guides the linked chromosomal domain to the perinucleolar region during mid-S phase in order to maintain bidirectional transcriptional silencing of flanking genes (red and green arrows indicate silenced and active status of a gene, respectively) through successive cell divisions.
Taken together, here we provide evidence that demonstrates a critical role for the antisense RNA itself. In sum, mechanistic insights provided in this study represent a first step in understanding the multilayered silencing pathway mediated by the long noncoding antisense RNAs.
Supplementary Material
Acknowledgments
We gratefully acknowledge Wolf Reik for providing the Kcnq1ot1 RNA probe for our studies. We greatly appreciate Joanne Whitehead for critical review of the manuscript. We thank Rolf Ohlsson, Dept. of Development and Genetics, Uppsala University, Sweden; Johan Ericsson, Ludwig Institute for Cancer Research, Uppsala, Sweden; and Thierry Grange, Institut Jacques Monod, Paris, France, for help and suggestions.
This work was supported by grants from the Swedish Research Council (VR-NT) and the Swedish Cancer Research foundation (Cancerfonden) to C.K. C.K. is a Senior Research Fellow supported by the Swedish Medical Research Council (VR-M).
Footnotes
Published ahead of print on 25 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, E., and C. D. Allis. 2005. RNA meets chromatin. Genes Dev. 191635-1655. [DOI] [PubMed] [Google Scholar]
- 3.Brown, K. E., S. Amoils, J. M. Horn, V. J. Buckle, D. R. Higgs, M. Merkenschlager, and A. G. Fisher. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3602-606. [DOI] [PubMed] [Google Scholar]
- 4.Brown, K. E., J. Baxter, D. Graf, M. Merkenschlager, and A. G. Fisher. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3207-217. [DOI] [PubMed] [Google Scholar]
- 5.Chakalova, L., D. Carter, and P. Fraser. 2004. RNA fluorescence in situ hybridization tagging and recovery of associated proteins to analyze in vivo chromatin interactions. Methods Enzymol. 375479-493. [DOI] [PubMed] [Google Scholar]
- 6.Chaumeil, J., P. Le Baccon, A. Wutz, and E. Heard. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 202223-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csink, A. K., and S. Henikoff. 1996. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381529-531. [DOI] [PubMed] [Google Scholar]
- 8.Dernburg, A. F., J. W. Sedat, and R. S. Hawley. 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86135-146. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with targeted deletion of KvDMR1. Nat. Genet. 32426-431. [DOI] [PubMed] [Google Scholar]
- 10.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5377-386. [DOI] [PubMed] [Google Scholar]
- 11.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 12.Harmon, B., and J. Sedat. 2005. Cell-by-cell dissection of gene expression and chromosomal interactions reveals consequences of nuclear reorganization. PLoS Biol. 3e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heard, E., P. Clerc, and P. Avner. 1997. X-chromosome inactivation in mammals. Annu. Rev. Genet. 31571-610. [DOI] [PubMed] [Google Scholar]
- 14.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C.-F. Qi, A. Wolffe, R. Ohlsson, and A. Lobanenkov. 2000. Functional interaction of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10853-856. [DOI] [PubMed] [Google Scholar]
- 15.Kanduri, C., C. Holmgren, G. Franklin, M. Pilartz, E. Ullerås, M. Kanduri, L. Liu, V. Ginjala, E. Ulleras, R. Mattsson, and R. Ohlsson. 2000. The 5′-flank of the murine H19 gene in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 10449-457. [DOI] [PubMed] [Google Scholar]
- 16.Kanduri, C., N. Thakur, and R. R. Pandey. 2006. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J. 252096-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 3091564-1566. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, M., and C. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19425-433. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. 2003. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol. 13R242-R254. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 361291-1295. [DOI] [PubMed] [Google Scholar]
- 21.Mancini-DiNardo, D., S. J. Steele, R. S. Ingram, and S. M. Tilghman. 2003. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum. Mol. Genet. 12283-294. [DOI] [PubMed] [Google Scholar]
- 22.Mancini-DiNardo, D., S. J. Steele, J. M. Levorse, R. S. Ingram, and S. M. Tilghman. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 201268-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattick, J. S., and I. V. Makunin. 2006. Non-coding RNA. Hum. Mol. Genet. 15R17-R29. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, K., M. Oshimura, and H. Kugoh. 2007. Suggestive evidence for chromosomal localization of non-coding RNA from imprinted LIT1. J. Hum. Genet. 52926-933. [DOI] [PubMed] [Google Scholar]
- 25.Navarro, P., S. Pichard, C. Ciaudo, P. Avner, and C. Rougeulle. 2005. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 191474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill, M. J. 2005. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum. Mol. Genet. 14R113-R120. [DOI] [PubMed] [Google Scholar]
- 27.Pandey, R. R., M. Ceribelli, P. B. Singh, J. Ericsson, R. Mantovani, and C. Kanduri. 2004. NF-Y regulates the antisense promoter, bidirectional silencing, and differential epigenetic marks of the Kcnq1 imprinting control region. J. Biol. Chem. 27952685-52693. [DOI] [PubMed] [Google Scholar]
- 28.Paulsen, M., T. Khare, C. Burgard, S. Tierling, and J. Walter. 2005. Evolution of the Beckwith-Wiedemann syndrome region in vertebrates. Genome Res. 15146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasanth, K. V., and D. L. Spector. 2007. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2111-42. [DOI] [PubMed] [Google Scholar]
- 30.Rougeulle, C., and E. Heard. 2002. Antisense RNA in imprinting: spreading silence through Air. Trends Genet. 18434-437. [DOI] [PubMed] [Google Scholar]
- 31.Sado, T., Y. Hoki, and H. Sasaki. 2005. Tsix silences Xist through modification of chromatin structure. Dev. Cell 9159-165. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Elsner, T., D. Gou, E. Kremmer, and F. Sauer. 2006. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science 3111118-1123. [DOI] [PubMed] [Google Scholar]
- 33.Sleutels, F., R. Zwart, and D. P. Barlow. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415810-813. [DOI] [PubMed] [Google Scholar]
- 34.Sun, B. K., A. M. Deaton, and J. T. Lee. 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21617-628. [DOI] [PubMed] [Google Scholar]
- 35.Szabó, P., S.-H. Tang, A. Rentsendorj, G. Pfeifer, and J. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10607-610. [DOI] [PubMed] [Google Scholar]
- 36.Thakur, N., M. Kanduri, C. Holmgren, R. Mukhopadhyay, and C. Kanduri. 2003. Bidirectional silencing and DNA methylation sensitive methylation spreading properties of the KcnqI ICR map to the same regions within the KcnqI imprinting control region. J. Biol. Chem. 2739514-9519. [DOI] [PubMed] [Google Scholar]
- 37.Thakur, N., V. K. Tiwari, H. Thomassin, R. R. Pandey, M. Kanduri, A. Gondor, T. Grange, R. Ohlsson, and C. Kanduri. 2004. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 247855-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umlauf, D., Y. Goto, R. Cao, F. Cerqueira, A. Wagschal, Y. Zhang, and R. Feil. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 361296-1300. [DOI] [PubMed] [Google Scholar]
- 39.Wutz, A., T. P. Rasmussen, and R. Jaenisch. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30167-174. [DOI] [PubMed] [Google Scholar]
- 40.Yang, P. K., and M. I. Kuroda. 2007. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell 128777-786. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, L. F., K. D. Huynh, and J. T. Lee. 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129693-706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.