Abstract
A disease-causing G-to-T transversion at position +6 of BRCA1 exon 18 induces exclusion of the exon from the mRNA and, as has been suggested by in silico analysis, disrupts an ASF/SF2-dependent splicing enhancer. We show here using a pulldown assay with an internal standard that wild-type (WT) and mutant T6 sequences displayed similar ASF/SF2 binding efficiencies, which were significantly lower than that of a typical exonic splicing enhancer derived from the extra domain A exon of fibronectin. Overexpression or small interfering RNA (siRNA)-mediated depletion of ASF/SF2 did not affect the splicing of a WT BRCA1 minigene but resulted in an increase and decrease of T6 exon 18 inclusion, respectively. Furthermore, extensive mutation analysis using hybrid minigenes indicated that the T6 mutant creates a sequence with a prevalently inhibitory function. Indeed, RNA-protein interaction and siRNA experiments showed that the skipping of T6 BRCA1 exon 18 is due to the creation of a splicing factor-dependent silencer. This sequence specifically binds to the known repressor protein hnRNPA1/A2 and to DAZAP1, the involvement of which in splicing inhibition we have demonstrated. Our results indicate that the binding of the splicing factors hnRNPA1/A2 and DAZAP1 is the primary determinant of T6 BRCA1 exon 18 exclusion.
Pre-mRNA splicing is an essential step in the gene expression process, connecting DNA transcription to protein translation. The splicing process is catalyzed by the spliceosome, a dynamic complex of five small nuclear ribonucleoproteins (snRNPs) and a large number of additional proteins (19, 30). During pre-mRNA splicing, exon coding sequences must be precisely distinguished from noncoding intronic sequences and joined together to form mature mRNA (2). To achieve this, the splicing machinery requires essential signals located around the splice site junctions: 5′ and 3′ splice sites, polypyrimidine tracts, and branch site sequences (2). However, these splice signals alone provide only a part of the information required by the spliceosome machinery for an efficient splicing process (23, 39). In fact, correct exon definition requires positive and negative accessory elements broadly referred to as exonic splicing enhancers (ESEs) and silencers (ESSs). Most ESEs are known to interact with members of the serine/arginine-rich family, which are splicing factors essential for promoting spliceosome assembly at the correct splice sites (3, 17, 36). In contrast, ESS sequences have often been found to bind specific trans-acting factors belonging to the heterogeneous nuclear ribonucleoprotein (hnRNP) family, which plays a role in splicing repression (13, 22, 35). In addition, enhancers or silencers may in some cases coexist in composite exonic splicing regulatory elements, giving rise to complex effects of natural and site-directed mutants on splicing (32, 34). Mutations in exonic splicing regulatory elements that result in splicing alterations are a common event in human pathology (6, 7, 16, 31). The analysis of several gene systems, such as the ATM (40), NF1 (1), CFTR (34), SMN (26), and BRCA1 (27) systems, has shown that exonic mutations can affect, independently from their effect on the amino acid sequence, splicing regulatory sequences and induce different splicing defects, including exon skipping.
A clear identification of the nature and location of exonic splicing regulatory elements is fundamental to understand the effect of genomic variants on splicing and consequently develop appropriate therapeutic strategies. To identify the composition of exonic splicing regulatory sequences and thus predict the splicing phenotype of exonic mutations in human genes, several in silico programs have been developed (10, 14, 15, 41, 42). However, this approach does not always correlate with the splicing phenotype observed, presenting severe limitations concerning its use in clinical genetics (12, 32, 34).
Notwithstanding the relatively high occurrence of defective splicing due to exonic mutations in several disease-related genes, the underlying mechanism is still controversial. Two different models have been proposed to explain exon skipping due to exonic mutations. The enhancer loss model suggests the disruption of sequences with enhancer properties, with ASF/SF2 being the main binding factor involved. On the contrary, the silencer gain-of-function model implies the creation of a silencer sequence by the mutation, which is mostly recognized by members of the hnRNP family. These two models have been extensively explored in the SMN1/2 systems, in which a critical C-to-T synonymous substitution in position 6 induces exon 7 skipping (8, 9, 20, 21, 37). The natural G-to-T mutation at position +6 of BRCA1 exon 18 (E1694X) presents common features with the SMN system. This mutation occurs at the same exonic position of SMN2; it has been previously reported to be associated with exon skipping (27), and both computer-assisted analysis and in vitro splicing analysis of a limited number of mutants have suggested that it disrupts a putative binding site for ASF/SF2 (25). However, this binding has never been experimentally proven, although the BRCA1 exon 18 system has been used for the in vitro characterization of new ASF/SF2-specific exonic splicing enhancers by systematic evolution of ligands by exponential enrichment (SELEX) (38). Small interfering RNA (siRNA) of ASF/SF2 has been recently shown not to have any functional effect on wild-type (WT) BRCA1 exon 18 splicing, whereas the BRCA1 E1694X mutant was found to bind hnRNPA1/A2 (21). However, as depletion of both hnRNPA1 and hnRNPA2 had no effect on mutant BRCA1 splicing, it was suggested that the disruption of enhancer binding to an unknown splicing factor contributes to mutant exon skipping (21).
Our work evaluates extensively the composition of the splicing-regulatory element affected by the natural G-to-T transversion at position +6 in BRCA1 exon 18. The analysis we performed includes systematic site-directed mutagenesis in hybrid minigenes, in vitro binding experiments, and siRNA treatment of the splicing factors involved. We find no preferential binding of ASF/SF2 to the WT sequence compared with the T6 mutant. On the contrary, the BRCA1 exon 18 T6 substitution creates a sequence with silencer properties that binds to hnRNPA1/A2, a known repressor protein, and deleted in azoospermia-associated protein 1 (DAZAP1). Altogether, our data support the gain-of-function model for defective splicing due to the +6 G-to-T exonic mutation in BRCA1 exon 18.
MATERIALS AND METHODS
Hybrid minigene constructs.
The BRCA1 genomic region containing the last 85 bp of exon 17 and first 1,408 bp of intron 17 was amplified from normal DNA using BRC90BstEII Dir (5′-CTGGTACCAAGTTTGCCAGAAAACACCACATCACTTTAACTAATC-3′) and BRC1566 Rev (5′-AACACCCAGAGGTCTCCTGTATTACACAAG-3′) primers and blunt ligated in the SmaI site of pBluescript KS to generate pBS BRA17. Similarly, the last 334 bp of intron 18 along with 58 bp at the beginning of exon 19 was amplified with BRC735 Dir (5′-TAGCAATGTAGCATATGAGCTAGGGATTTA-3′) and BRC4451 Rev (5′-AACATCAAGTACTTACCTCATTCAGC-3′) primers and blunt ligated in the SmaI site of pBluscript KS to generate pBS BRA19. Subsequently, pBS BRA19 was digested with NdeI and KpnI and cloned in the corresponding sites of pBS BRA17 to generate pBS BRA17-19, with NdeI as a unique site. The BstEII-BstEII fragment contained in pBS BRA17-19 was then inserted in the unique BstEII site of a modified pTB α-globin minigene to obtain pBRAint. The BRCA1 genomic region encompassing exon 18 and adjacent intronic sequences (289 bp of intron 17, full-length exon 18, and 255 bp of intron 18) was amplified with BRC138 Dir (5′-GGCATATGGAGATCTATAGCTAGCCTTGGCGTCTAGAAGATGG-3′) and BRC760 Rev (5′-AATCCCTAGCTCATATGCTAACATTGCTAGG-3′) primers and cloned in the unique NdeI site of pBRAint. On this resulting construct, an EcoRI site was inserted in intron 18 (49 bases downstream of the 5′ splice site of exon 18) by PCR-mediated site-directed mutagenesis, creating the final hybrid minigene, named pBRAwt. The natural PstI and the artificial EcoRI unique sites create a cassette system, which facilitates subsequent cloning procedures. The final minigene lacks the central part of intron 17 (1,960 bp), and the α-globin and BRCA1 17, 18, and 19 exons are in frame. pBRA minigene variants were generated by PCR site-directed mutagenesis replacing the pBRAwt PstI-EcoRI cassette with the mutated sequence. The constructs containing the serial deletions (∼10 bp) within exon 18 were created by overlapping PCR mutagenesis using suitable primers on pBRA WT or T6 mutant minigenes as templates. The identity of each minigene was verified by sequence analysis. The pSMN2 construct has been previously reported (8).
Cell culture, transfections, and reverse transcription-PCR (RT-PCR) analysis.
Human cell lines (Hep3B and HeLa) were cultured in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen), in standard conditions. The DNA used for transfection was purified using JetStar columns (Genomed). Hep3B cells (3 × 105) were transfected with 1.5 μg of each reported plasmid construct, employing the DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) liposomal transfection reagent (Alexis Biochemicals) according to the manufacturer's instructions. After 12 h, the transfection medium was replaced with fresh medium, and 24 h later the total RNA was extracted using TRIreagent solution (Ambion). Retrotranscription was performed with random primers and Moloney murine leukemia virus enzyme (Invitrogen). To amplify only the messengers derived from the transfected plasmid, PCR was performed with BRC90BsteII Dir and Glo800Rev (5′-GCTCACAGAAGCCAGGAACTTGTCCAGG-3′), which hybridize to BRCA1 exon 17 and α-globin exon 3 sequences, respectively. The conditions used for the PCRs were the following: 94°C for 3 min for the initial denaturation; 94°C for 45 s, 54°C for 45 s, and 72°C for 45 s for 30 cycles; and 72°C for 10 min for the final extension. PCRs were optimized to be in the exponential phase of amplification, and products were routinely fractionated in 2% (wt/vol) agarose gels. For protein overexpression experiments, 1.5 μg of the minigene was cotransfected with 1 μg of the ASF/SF2 coding sequence cloned into a pCG vector (a kind gift from J. Caceres). Amplification of the pSMN2 minigene was performed with the previously reported oligonucleotides (8). The results of all the transfections are representative of at least three independent experiments. ImageJ 1.38 software (http://rsb.info.nih.gov/ij/) was used in order to quantify the proportion of exon 18 skipping.
Affinity purification of RNA binding proteins.
Two synthetic RNA oligonucleotides, exon 18 WT (UGCAGAUGCUGAGUUUGUGU) and U6 (UGCAGAUGCUUAGUUUGUGU), were generated by Integrated DNA Technologies and used as targets for pulldown assays. Twelve micrograms of target BRCA1 RNA oligonucleotides was placed in 400 μl of reaction mixture (100 mM sodium acetate [NaOAC], pH 5.0, and 5 mM sodium m-periodate Σ), incubated for 1 h in the dark at room temperature, ethanol precipitated, and finally resuspended in 100 μl of 100 mM NaOAc (pH 5.0). Approximately 400 μl of adipic acid dehydrazide-agarose beads (50% slurry; Sigma) previously equilibrated with 100 mM NaOAC (pH 5.2) was added to each periodate-treated RNA, and the mixture was incubated for 12 h at 4°C on a rotator.
The beads with the bound RNA were then washed two times with 1 ml of 2 M NaCl and equilibrated in 1× washing buffer (5.2 mM HEPES, pH 7.5, 1 mM MgCl2, 0.8 mM Mg acetate). Then the beads were incubated, in a final volume of 500 μl, with 0.5 mg of HeLa cell nuclear extract (C4; Biotech), 1× binding buffer (5.2 mM HEPES, pH 7.9, 1 mM MgCl2, 0.8 mM Mg acetate, 0.52 mM dithiothreitol, 3.8% glycerol, 0.75 mM ATP, 1 mM GTP), and heparin (final concentration, 1 μg/μl) for 30 min on a rotator at room temperature. The beads were then washed four times with 1.5 ml of washing buffer before addition of sodium dodecyl sulfate (SDS) sample buffer and loading on SDS-10% polyacrylamide gels.
Proteins were visualized by Coomassie brilliant blue staining. Protein sequence analysis of the bands excised from the gel was performed using an electrospray ionization mass spectrometer (LCQ DECA XP; ThermoFinnigan). Protein bands were digested with trypsin, and the resulting peptides were extracted with water and 60% acetonitrile-1% trifluoroacetic acid. Fragments were then analyzed by mass spectrometry, and proteins were identified by analysis of the peptide tandem mass spectrometry data with Turbo SEQUEST (ThermoFinnigan) and MASCOT (Matrix Science).
In vitro transcription and Western blot analysis.
Plasmids for in vitro transcription were generated by annealing the sense (5′-CTGTGTGTGTGTGTTTTTTGCAGATGCTKAGTTTGTGTG-3′) and the antisense (5′-GATCCACACAAACTMAGCATCTGCAAAAAACACACACACACAGAGCT-3′) oligonucleotides, purchased from Sigma, for both WT and T6 mutant exon 18 sequences. This was followed by direct cloning into SacI-BamHI-digested pBluescript vector, under the control of the T7 RNA promoter.
Part of the fibronectin extra domain A (EDA) sequence (5′-CTGTGTGTGTGTGTTTTTTGCACCTGATGGTGAAGAAGACACTGCAGAGC-3′) was also cloned by following the same procedure and used for pulldown analyses, acting as internal control for ASF/SF2 binding.
Plasmids were first linearized by HindIII digestion and then in vitro transcribed with T7 RNA polymerase (Promega) according to standard procedures. Approximately 12 to 15 μg of transcribed and purified RNA was placed in a 400-μl reaction mixture of 100 mM NaOAc, pH 5.0, and 5 mM sodium m-periodate (Sigma) and processed as described above for pulldown assays. After the final centrifugation, 60 μl of SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer was added to the samples, followed by heating for 5 min at 90°C before loading onto a 12% SDS-PAGE gel. The gel was electroblotted on a polyvinylidene difluoride membrane according to standard protocols (Amersham Biosciences), and the membrane was blocked with phosphate-buffered saline-5% skimmed milk. Proteins were identified using different antibodies, and Western blot signals were detected with a chemiluminescence kit (ECL; Pierce Biotechnology).
Rabbit anti-hnRNPA1 serum was generously provided by R. Klima, and polyclonal antibodies against hnRNPA2 and TDP43 were provided by E. Buratti (ICGEB, Trieste, Italy). Purified glutathione S-transferase (GST)-DAZAP1 protein was used to immunize a rabbit (New Zealand strain) according to standard protocols to obtain polyclonal anti-DAZAP1 antibodies. Anti-ASF/SF2 monoclonal antibody 96 was purchased from Zymed Laboratories Inc., and antitubulin monoclonal antibody was kindly provided by F. Porro (ICGEB, Trieste, Italy).
Electromobility shift assay (EMSA).
RNA synthetic oligonucleotides (200 ng) were labeled by phosphorylation with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C, precipitated, and resuspended in 200 μl of water. Each binding reaction was made by mixing the purified protein with the labeled RNA oligonucleotide in 1× binding buffer (5.2 mM HEPES, pH 7.9, 1 mM MgCl2, 0.8 mM Mg acetate, 0.52 mM dithiothreitol, 3.8% glycerol, 0.75 mM ATP, 1 mM GTP) to a 20-μl final volume. The reaction mixture was left at room temperature for 15 to 20 min before loading the sample on a native polyacrylamide gel (5%), which was run at 100 to 120 V at 4°C. The protein-nucleic acid complexes were visualized using Biomax MS films (Kodak). The cDNA of each target protein was amplified and cloned in pGEX-3X plasmid (Pharmacia) and then expressed in BL21(DE3) bacteria (Novagen) under the induction of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The resulting recombinant proteins were purified with glutathione S-Sepharose 4B beads (Pharmacia) according to the manufacturer's instructions using imidazole buffers for protein elution.
siRNA transfections.
siRNA transfections were performed in HeLa cells using Oligofectamine reagent (Invitrogen). The sense strands of RNA interference oligonucleotides (Dharmacon) used for silencing the different target proteins were the following: human hnRNPA1, CAGCUGAGGAAGCUCUUCA, and human hnRNPA2, GGAACAGUUCCGUAAGCUC (20); human DAZAP1, GAGACUCUGCGCAGCUACU; human ASF/SF2, ACGAUUGCCGCAUCUACGU (8); and luciferase no. 2 gene control, GCCAUUCUAUCCUCUAGAGGAUG.
HeLa cells were plated at 2.5 × 105 cells per well in 60-mm plates to achieve 40 to 50% confluence. The next day, 6 μl Oligofectamine was combined with 24 μl of Opti-MEM medium (Invitrogen) and 5 to 10 μl of 40 μM siRNA duplex oligonucleotides was diluted in a final volume of 400 μl of Opti-MEM medium. The two mixtures were combined and left for 20 min at room temperature. Finally, this mixture was added to the cells, which were maintained in 1.6 ml of Opti-MEM. After 24 h a second round of siRNA transfection was performed as described above. Six to 8 h later Opti-MEM was exchanged with Dulbecco's modified Eagle medium and the cells were transfected with the minigene of interest (1 μg) using Qiagen Effectene transfection reagents. On the third day, HeLa cells were harvested and divided in two parts for protein and RNA extractions. RT-PCR from total RNA was performed as for the transfection protocol described above. Whole-protein extracts were obtained by cell sonication in lysis buffer (15 mM HEPES, pH 7.5, 250 mM NaCl, 0.5% NP-40, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride) and analyzed for hnRNPA1 and -A2, DAZAP1, and ASF/SF2 endogenous protein expression by immunoblotting using the antibodies described above. Tubulin was used as protein loading control. Each siRNA treatment experiment was repeated at least three times.
RESULTS
ASF/SF2 affects exon 18 splicing independently from the T6 mutation.
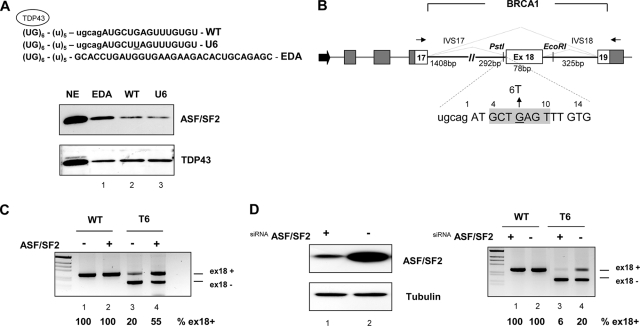
To evaluate the role of ASF/SF2 in the regulation of BRCA1 splicing, we set up a modified pulldown assay where exon 18 WT or U6 RNAs transcribed in vitro were linked through a (U)5 spacer to a sequence composed by six UG dinucleotide repeats (Fig. 1A, top). The UG repeats, which specifically bind TDP43 (4), served as an internal standard to normalize the amount of RNAs pulled down. An ASF/SF2 purine-rich enhancer, already described regarding the alternative splicing of the fibronectin EDA exon (29), was used as a positive control to estimate the binding efficiency of the BRCA1 sequences. The pulldown assay was followed by Western blotting and probing with ASF/SF2 and TDP34 antibodies. Relative to the binding of TDP43, used as a normalization reference, the WT and U6 mutant RNAs bind to ASF/SF2 with a similar, low efficiencies in comparison with the binding of ASF/SF2 to the strong EDA enhancer (Fig. 1A, bottom, lanes 2, 3, and 1, respectively). The low binding efficiencies of both WT and mutant sequences suggest a common, minor splicing enhancing effect of ASF/SF2. These results were confirmed in classical pulldown experiments using RNA oligonucleotides without the UG tail (data not shown).
FIG. 1.
ASF/SF2 affects exon 18 splicing independently from the T6 mutation. (A) Western blot after pulldown analysis of WT and U6 sequences. Shown is a schematic representation of the RNA sequence transcribed in vitro (top) that contains (UG)6 repeats, a (U)5 spacer, part of intron 17 (lowercase), and the first part of WT and T6 exon 18 (uppercase). The mutant nucleotide U6 is underlined. The bottom lane shows the EDA sequence used as a control for ASF/SF2 binding. TDP43 was used to normalize the assay for the amount of RNA. The nuclear extract sample (NE) corresponds to 1/20 of the amount used for the pulldown assay. (B) Schematic representation of the pBRA hybrid minigene system. Dark gray and white boxes represent α-globin and BRCA1 exons, respectively. Thick lines are introns (IVS), and the large black arrow at the 5′ end indicates the simian virus 40 promoter. The lengths of BRCA1 exon 18 and flanking introns are shown. The primers used for RT-PCR are indicated by thin arrows, and the two alternative splicing possibilities, exon 18 inclusion and exclusion, are indicated as dotted lines. The positions of the unique PstI and EcoRI restriction sites used for subsequent cloning are shown. At the bottom, a few bases from intron 17 (lowercase) and exon 18 (from position +1 to +14) are shown, divided in codon triplets (uppercase). The putative ASF/SF2 binding site predicted by the ESE finder program (25) is boxed in light gray. The G-to-T natural mutation (E1694X) is indicated (27). (C) In vivo overexpression of ASF/SF2 promotes exon 18 T6 inclusions. HeLa cells were transfected with the WT or T6 pBRA minigene as indicated, along with the ASF/SF2-coding plasmid (+) or with the empty vector (−). Splicing patterns were analyzed by separating the RT-PCR products on 2% agarose gel and staining with ethidium bromide. The identity of the transcripts, i.e., including exon 18 (ex18+) and lacking exon 18 (ex18−), is indicated at the right side of the gel. The percentages of exon 18 inclusion, obtained from three independent transfections, are indicated below the lane numbers. (D) In vivo depletion of ASF/SF2 induces complete T6 exon 18 skipping. The left panel shows the Western blot analysis of extracts from HeLa cells treated with either ASF/SF2 siRNA (+) or a control luciferase siRNA (−). The amount of the protein extract was normalized using an antibody against tubulin. The right panel shows the RT-PCR results after transfection of the WT or T6 pBRA plasmid minigenes into ASF/SF2 siRNA-treated (lanes 1 and 3) and luciferase control-treated (lanes 2 and 4) HeLa cells, and the percentages of BRCA1 exon 18 inclusion are indicated.
To better evaluate the effect of ASF/SF2 on human BRCA1 exon 18 splicing, we prepared a pBRA hybrid minigene. The pBRA minigene system contains most of the genomic sequences of the BRCA1 gene from exon 17 to 19, cloned in frame within the α-globin pTB minigene (Fig. 1B) (33). We tested the in vivo splicing pattern of the pBRA WT and T6 constructs by transient transfection of HeLa cells followed by RT-PCR analysis of the total RNA. The two minigenes spliced in different ways: while WT exon 18 was efficiently included (Fig. 1C, lane 1), the T6 mutated exon was predominantly skipped, with only 20% of it being included (Fig. 1C, lane 3). Cotransfection experiments using a plasmid coding for ASF/SF2 showed an increase in the percentage of exon 18 inclusion for the T6 mutant minigene (Fig. 1C, lane 4), whereas the enhancing effect was not detectable for the WT minigene since exon 18 is already fully included in the absence of ASF/SF2 overexpression (Fig. 1C, lane 2).
We further investigated the in vivo role of ASF/SF2 by siRNA treatment of HeLa cells. In this case the WT and T6 pBRA constructs were transfected in siRNA-treated cells. After 24 h cells were collected and analyzed for the level of endogenous ASF/SF2 by Western blotting. We observed a strong reduction in ASF/SF2 expression in cells transfected with siRNA but not in luciferase siRNA-treated cells, used as control (Fig. 1D, left). RT-PCR analysis of the total RNA showed that ASF/SF2 depletion induced a nearly complete exclusion of T6 exon 18 from the mature mRNA (Fig. 1D, right, lane 3). In contrast, WT BRCA1 exon 18 did not respond to siRNA treatment (Fig. 1D, right, lane 1). Overall, these results do not support a direct role for ASF/SF2 in the aberrant splicing of T6 mutant exon 18.
The natural G-to-T transition in BRCA1 exon 18 creates a sequence with silencer properties.
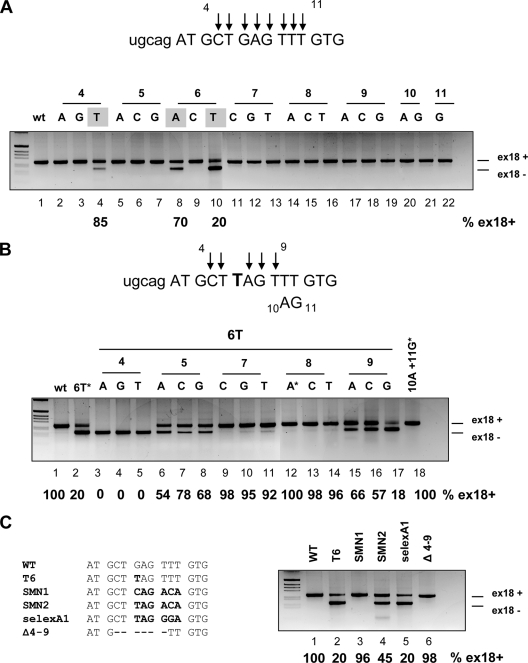
To understand the composition of the splicing-regulatory element and the effect of the G6T natural mutation, we performed systematic site-directed mutagenesis. A series of single point mutations from position +4 to position +11 were introduced in the pBRA hybrid minigene and evaluated by means of the functional-splicing assay. Interestingly, out of 21 substitutions only the natural T6 mutation resulted in a very low level of exon inclusion (20%) (Fig. 2A, lane 10). Two additional mutations, in positions 4 (T4) and 6 (A6), partially affected the splicing pattern, causing 85% and 70% exon inclusion, respectively (Fig. 2A, lanes 4 and 8). As the widely spanning mutagenesis analysis seemed more compatible with the creation of a negative element by the T6 variant, we decided to test this putative silencer by introducing in the context of the T6 substitution consecutive point mutations from position +4 to +9. As shown in Fig. 2B, the majority of the double site-directed mutations flanking position 6 increased the splicing efficiency compared with that for the single T6 variant. In particular, all of the substitutions in positions 7 and 8 restored almost completely exon inclusion (Fig. 2B, lanes 9 to 14), and three mutations in position 5 and two in position 9 (T6A9 and T6C9) increased significantly the splicing efficiency (Fig. 2B, lanes 6 to 8, 15, and 16), whereas T6G9 had no effect (Fig. 2B, lane 17). In contrast, mutations at position +4 negatively affected splicing, resulting in complete exon 18 skipping (Fig. 2B, lanes 3 to 5). Thus, double substitutions that restore normal exon 18 inclusion disrupt the mutant “TAG” sequence at the 5′ end of exon 18. The “TAG” sequence therefore appears to be the core silencer element involved.
FIG. 2.
The G-to-T transition in BRCA1 exon 18 creates a sequence with silencer properties. (A) Single-point-mutation analysis of the exon 18 putative regulatory region. Parts of the intron 17 and BRCA1 exon 18 sequences are shown in lowercase and uppercase, respectively. The arrows indicate the nucleotide positions where all three possible single point mutations were substituted, from nucleotide 4 to 11. The constructs were transfected into Hep3B cells, and the splicing pattern was analyzed by RT-PCR. The splicing products corresponding to exon 18 inclusion (ex18+) and exclusion (ex18−) are indicated. The gray boxes indicate the mutants that showed exon skipping (lanes 4, 8, and 10). Their percentages of exon 18 inclusion are indicated below the lanes. (B) Double-point-mutation analysis. The upper panel shows part of the sequence of intron 17 and exon 18. The positions of single point mutations, introduced in the context of the 6T variant, are indicated by arrows. The position of the 10AG11 double mutant is indicated. The substitutions that create a stop codon are highlighted by an asterisk (lanes 2, 12, and 18). The percentages of BRCA1 exon 18 inclusion, calculated as for panel A, are indicated below the lanes. (C) Analysis of silencer sequences in the BRCA1 exon 18 context. The left panel shows WT and mutant sequences of BRCA1 exon 18 from position +1 to +14. The mutant elements are in boldface, and the deletion is represented as a dashed line (Δ4-9). The splicing profile was analyzed as described for panel A. The percentages of BRCA1 exon 18 inclusion are indicated.
On the basis of this assumption, we evaluated splicing-regulatory sequences derived from the SMN1 and SMN2 genes (20) and a SELEX A1 silencer sequence (5) in the pBRA minigene context. These sequences include or do not include the “TAG” core element (Fig. 2C, left). Exon 18 splicing was affected by the insertion of the SELEX A1 sequence at the same level as with the T6 mutant (Fig. 2C, lane 5). The SMN2 sequence in the pBRA minigene showed a lower but still negative effect, with 50% exon 18 inclusion, whereas SMN1, which does not contain TAG, did not affect mRNA processing (Fig. 2C, lanes 4 and 3, respectively). Finally, a deletion from position +4 to +9 in the pBRA Δ4-9 construct did not change the splicing pattern in comparison with WT pBRA, further ruling out the presence of a strong enhancer at the 5′ end of exon 18 (Fig. 2C, lane 6).
An indirect effect of nonsense-mediated decay on splicing in this system was excluded, as two nonsense-coding minigenes, T6A8 and 10A11G, and the out-of-frame Δ4-9 mutant showed, unlike the T6 variant (E1694X), complete exon inclusion (Fig. 2B, lanes 12 and 18, and C, lane 6).
Identification of nuclear proteins binding to the splicing-regulatory element of the BRCA1 exon 18 T6 mutant.
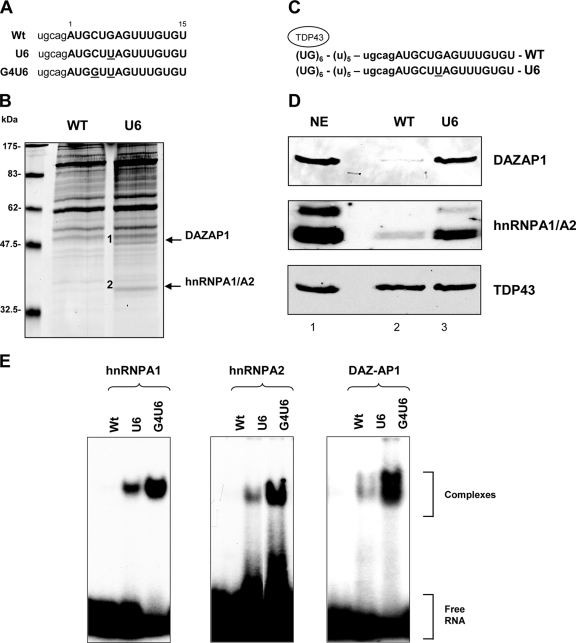
To identify the trans-acting factor(s) differentially binding to BRCA1 exon 18 WT and mutant sequences, we performed a pulldown analysis using two synthetic RNA oligonucleotides containing either the WT or the natural U6 mutated sequence (Fig. 3A). Agarose bead-linked RNAs were incubated with HeLa nuclear extracts, and the associated proteins were analyzed by SDS-PAGE followed by Coomassie blue staining. Comparison of the patterns of binding proteins showed two specific bands of about 50 and 38 kDa that were specifically pulled down by the U6 but not by the WT RNA (Fig. 3B). Sequence analysis of the excised bands and a database search revealed that the band of 50 kDa is DAZAP1, also named proline-rich RNA-binding protein, and that the band of 35 kDa corresponds to both hnRNPA1 and its close homologue hnRNPA2 factor.
FIG. 3.
Identification of nuclear proteins binding to the splicing-regulatory element of the BRCA1 exon 18 T6 mutant. (A) Sequences of the BRCA1 RNA oligonucleotides. Intronic and exonic sequences are shown in lowercase and uppercase, respectively, and the mutated nucleotides are underlined. (B) Coomassie-stained gel of a pulldown analysis where HeLa nuclear extracts were incubated with adipic dehydrazide beads derivatized with the target RNAs. The arrows indicate two bands of about 50 and 38 kDa, present only in the U6 lane. The proteins in these bands were sequenced and identified as DAZAP1 (1) and hnRNPA1/A2 (2), respectively. (C) Schematic representation of the RNA sequence transcribed in vitro that contains (UG)6 repeats, a (U)5 spacer, and the target BRCA1 sequences. The mutagenized nucleotide is underlined. (D) Binding analysis of hnRNPA1 and -A2 and DAZAP1 performed by Western blotting after pulldown assays. After in vitro transcription, RNA was pulled down and the bound proteins were analyzed by Western blotting with anti-hnRNPA1, -hnRNPA2, -DAZAP1, and -TDP43 polyclonal antibodies. The nuclear extract sample corresponds to 1/20 of the amount used for the pulldown assay. (E) Interaction of purified human GST-hnRNPA1 and -A2 and DAZAP1 proteins with WT, U6, and G4U6 RNA oligonucleotides monitored by EMSA. Labeled RNAs were incubated with different GST fusion proteins as indicated, and the complexes were resolved on a 5% polyacrylamide gel. The free RNA and the formed complex are indicated on the right.
To investigate in more detail the differential binding efficiency of the splicing-regulatory sequences of BRCA1 exon 18, we prepared in vitro T7-transcribed RNAs that contained a fixed (UG)6 repeat sequence linked through a poly(U) linker to WT and U6 exon 18 (Fig. 3C). After pulldown and SDS-PAGE, the proteins were blotted onto polyvinylidene difluoride filters and evaluated using specific antibodies. In the presence of comparable amounts of pulled down TDP43, the U6 mutant but not the WT bound very efficiently to DAZAP1 and hnRNPA1/A2 (Fig. 3D). These results were also confirmed by classical pulldown experiments using synthetic RNA oligonucleotides (data not shown).
In order to test the specificity of the three different splicing factors for binding to BRCA1, we performed EMSA with purified hnRNPA1, hnRNPA2, and DAZAP1 GST-tagged proteins. For the EMSA experiments we selected the WT and the U6 RNAs and an additional double mutant, G4U6, which induced complete exon skipping in a splicing assay (Fig. 2B, lane 4). Labeled WT and mutant RNAs were incubated with recombinant proteins, and the RNA-protein complexes were resolved on a native acrylamide gel. A band of shifted material with the RNAs carrying the T6 and G4T6 mutations is present, whereas no complexes are formed by the WT oligonucleotide (Fig. 3E). This difference in binding efficiency is visible for all the three proteins analyzed, and it is in agreement with the percentage of exon skipping detected in the minigene splicing assay.
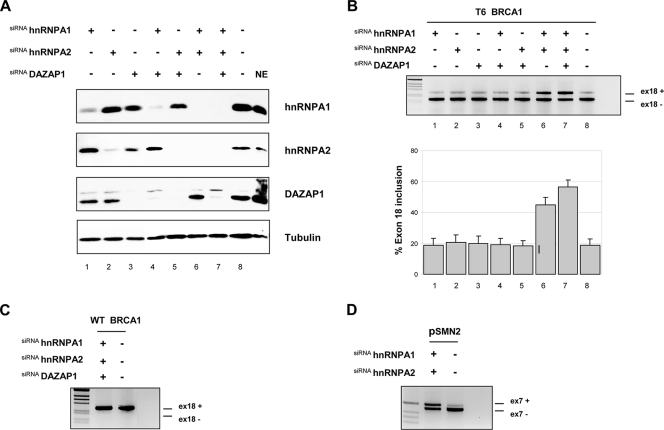
siRNA against hnRNPA1 and -A2 and DAZAP1 affects BRCA1 T6 exon 18 splicing.
hnRNPA1 and the related hnRNPA2 protein are well-known splicing-inhibitory factors, whereas no functional role for DAZAP1 in splicing regulation has been reported. In order to evaluate the inhibitory role of hnRNPA1 and -A2 and DAZAP1 in the in vivo splicing of BRCA1 exon 18, we used single or combined siRNA against these splicing factors. We transiently transfected HeLa cells, and the silencing effect was analyzed by Western blotting using antibodies against each target protein. At the same time, the splicing pattern was evaluated by RT-PCR using total RNA. HeLa cells transfected with the corresponding siRNAs showed a strong reduction in hnRNPA1 and -A2 and DAZAP1 levels (Fig. 4A), whereas no effect was detected in the luciferase control protein extract (Fig. 4A, lane 8). Unexpectedly, DAZAP1 showed a small reduction in the amount of hnRNPA1 and hnRNPA2 (Fig. 4A, lane 3), possibly related to an in vivo physical interaction (24) or to a regulatory effect of DAZAP1 on hnRNPA1/A2 splicing and/or stability. In the T6 mutant, the combined siRNA-mediated knockdown of the three splicing-inhibitory factors increased the percentage of BRCA1 exon 18 inclusion from 20 to about 55% (Fig. 4B, lane 7). A slightly smaller effect was observed for the siRNA-mediated knockdown of hnRNPA1/A2 (Fig. 4B lane 6), with about 45% exon inclusion, whereas the individual and double (hnRNPA1/DAZAP1 and hnRNPA2/DAZAP1) siRNAs did not change the pattern of splicing (Fig. 4B, lanes 1 to 5). As a control for hnRNPA1/A2 silencing efficiency we cotransfected the pBRCA1T6 mutant with the pSMN2 minigene in the double-siRNA-treated cells. siRNA-mediated silencing of hnRNPA1 and hnRNPA2 showed, as previously reported (8, 21), significant rescue of the SMN2 splicing defect (Fig. 4D). Exon 18 inclusion in the WT was not affected by siRNA-mediated depletion of any of the three splicing-inhibitory factors (Fig. 4C).
FIG. 4.
In vivo depletion of hnRNPA1 and -A2 and DAZAP1 by siRNA treatment partially rescues BRCA1 exon 18 inclusion. (A) Western blot analysis of HeLa cells treated (+) with the indicated different siRNAs. NE, nuclear extract. Lane 8 corresponds to the control luciferase siRNA. (B) siRNA-treated (+) and untreated (−) cells were transfected with the pBRA T6 minigene plasmid. Total RNA was prepared and analyzed by RT-PCR. The splicing products obtained were separated on 2% agarose gels and stained with ethidium bromide. The identities of the transcripts including exon 18 (ex18+) and lacking exon 18 (ex18−) are indicated. The graph is the quantification of three independent experiments expressed as means ± standard deviations. (C) Effect of siRNA against hnRNPA1, hnRNPA2, and DAZAP1 on the WT pBRCA1 minigene. (D) Effect of double siRNA against hnRNPA1 and hnRNPA2 on the splicing pattern of the control pSMN2 minigene. pSMN2 and pBRCA1 T6 minigenes were cotransfected in the siRNA hnRNPA1- and hnRNPA2-treated cells and analyzed with specific primers for pSMN2. The identity of the transcript with inclusion or exclusion of the SMN2 exon 7 is indicated.
BRCA1 exon 18 contains additional splicing-regulatory sequences that are functional only in the context of the T6 mutant.
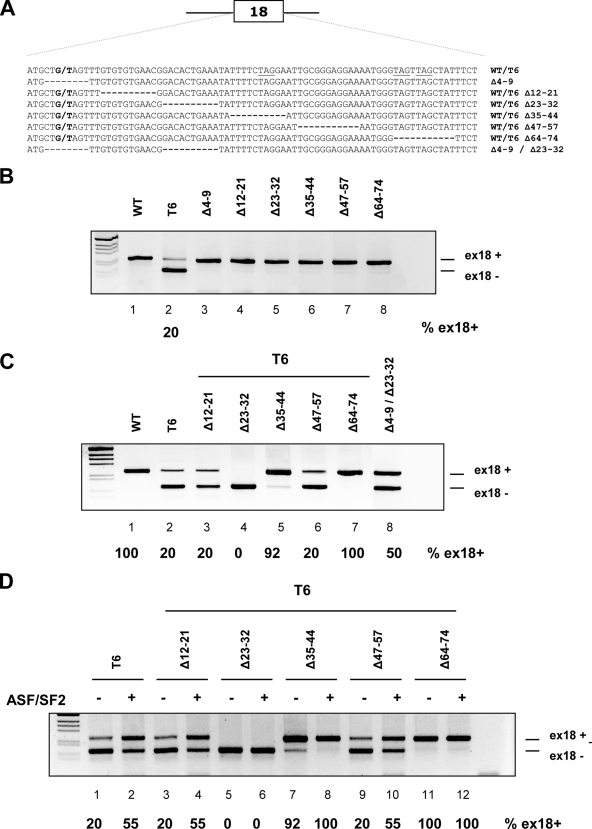
To identify additional exonic splicing-regulatory elements that modulate T6 aberrant skipping, we performed several consecutive deletions within BRCA1 exon 18. Those deletions analyzed in the context of the WT pBRA had no effect on normal exon 18 inclusion, indicating the absence of strong enhancer sequences in the natural exon (Fig. 5B, lanes 3 to 8). On the other hand, some deletions analyzed in the context of the T6 mutant resulted in changes in the pattern on splicing. In comparison to the T6 mutant, T6Δ35-44 and T6Δ64-74, which disrupt “TAG” elements, increased the percentage of exon inclusion up to 90% and 100%, respectively (Fig. 5C, lanes 5 and 7), indicating the presence of two downstream silencers at positions 35 to 44 and 64 to 74. On the other hand, deletion of the sequences between positions 23 and 32 in the T6Δ23-32 minigene induced complete exon skipping (Fig. 5C, lane 4), indicating the presence of a weak enhancer, which is functionally relevant only in the context of the T6 mutant. In a similar manner we evaluated also the combined effect of the two deletions in positions 4 to 9 and 23 to 32. These deletions alone had no effect on splicing but, when combined (Δ4-9 plus Δ23-32), are able to induce partial exon skipping (Fig. 5C, lane 8). These data suggest the presence of two nearby weak enhancer sequences, which may contribute in a synergic manner to the definition of the WT exon. However, in the context of the Δ23-32 deletion the extent of splicing inhibition mediated by the single T6 substitution is more severe than the splicing inhibition mediated by the Δ4-9 deletion. In fact, the T6Δ23-32 minigene induced complete exon exclusion, whereas Δ4-9-Δ23-32 showed only partial exon skipping, with 50% exon inclusion. This result is consistent with the strong inhibitory effect mediated by the binding of hnRNPA1/A2 and DAZAP1 on the T6 mutant.
FIG. 5.
Mapping the exonic regulatory elements within the BRCA1 exon 18 sequence. (A) Schematic representation of the BRCA1 exon 18 deletion mutants. The G or T substitution at position +6 and the deleted nucleotides are indicated, while the TAG motives are underlined. (B and C) Exon 18 WT (B) and T6 (C) serial deletion analysis. The indicated minigenes were transfected into Hep3B cells, and the splicing pattern was analyzed by RT-PCR. The two resulting splicing products corresponding to exon 18 inclusion (ex18+) and exclusion (ex18−) are indicated, and the percentage of exon inclusion is reported. (D) Overexpression of ASF/SF2 in T6 deletion mutant minigenes. The indicated constructs were cotransfected with the ASF/SF2 coding plasmid (+) or with the empty vector (−).
To identify potential splicing-regulatory sequences that could mediate the ASF/SF2 observed enhancing effect on the T6 mutant (Fig. 1C), we also evaluated in cotransfection experiments the effect of this splicing factor on the different minigenes with single deletions. Overexpression of ASF/SF2 increased the percentage of exon inclusion in the single T6 mutant and in three deletion mutants, T6Δ12-21, T6Δ35-44, and T6Δ47-54 (Fig. 5D, lanes 4, 8, and 10), and had no effect on the already fully included T6Δ64-74 minigene (Fig. 5D, lane 12). On the other hand, overexpression of this splicing factor did not induce exon inclusion in the T6Δ23-32 minigene (Fig. 5D, lane 6). Thus, the ESE located between nucleotides 23 and 32 in exon 18 stimulates BRCA1 exon 18 splicing through ASF/SF2 only in the context of the T6 mutant.
DISCUSSION
In this paper we provide biochemical and functional evidence that the natural G-to-T transversion at position +6 of BRCA1 exon 18, previously suggested to disrupt an ASF/SF2-dependent splicing enhancer, induces defective splicing through the creation of a sequence with splicing-inhibitory function. We show, in a series of systematic site-directed mutagenesis and pulldown RNA and EMSA experiments, that the mutation creates a splicing silencer element that interacts specifically with three splicing-inhibitory factors: hnRNPA1, hnRNPA2, and DAZAP1. By means of siRNA experiments, we found that hnRNPA1/A2 and DAZAP1 are involved in the generation of a defective splicing of T6 BRCA1 exon 18. In addition, we report a novel role for DAZAP1 in splicing regulation.
Computer-assisted analysis originally suggested that the natural G6T transversion in BRCA1 exon 18 disrupts a putative binding site for ASF/SF2 and accordingly affects an exonic splicing enhancer. Based on this prediction and the functional evaluation of a limited number of site-directed mutants in in vitro splicing assays, a general model of defective splicing due to exonic mutation was proposed consistent with the disruption of an ESE-dependent ASF/SF2 binding site (25). Using a modified pulldown technique followed by Western blot analysis, we unexpectedly found that both WT and T6 mutants bind equally to ASF/SF2 (Fig. 1A, lanes 2 and 3). Most importantly, compared with the classical GAA-rich fibronectin EDA enhancer (Fig. 1A, lane 1), the WT and mutant BRCA1 sequences show a diminished interaction with ASF/SF2, which is unlikely to be functional. The efficiency of this binding was carefully evaluated by linking BRCA1 RNA targets to a (UG) tail which specifically binds TDP43 and allows an accurate normalization of the amount of proteins pulled down. Consistent with the absence of a strong ASF/SF2-dependent enhancer at the 5′ end of the exon disrupted by the mutation, and in accordance with recently published data (21), siRNA-mediated depletion of ASF/SF2 did not induce exon exclusion in the WT minigene (Fig. 1D, lane 1), indicating that ASF/SF2 is not essential for BRCA1 exon 18 splicing. Similarly, the lack of a strong enhancer at the 5′ end of BRCA1 is also indicated by site-directed mutagenesis analysis. In fact, most point substitutions along the 5′ end of BRCA1 exon 18 (Fig. 2A) and the complete deletion of the putative enhancer sequence (Δ4-9 pBRA construct) have no effect on the splicing pattern (Fig. 2C, lane 6).
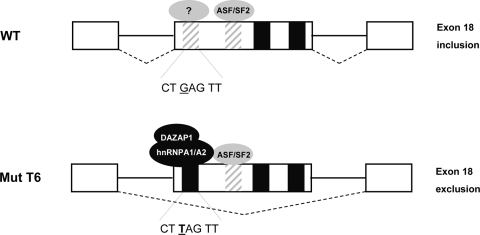
On the other hand, we found that the T6 mutant responds to siRNA-mediated depletion (Fig. 1D, right, lane 3) or overexpression of ASF/SF2 (Fig. 1C, lane 4) by reducing or increasing, respectively, the percentage of exon inclusion. A small negative effect by depletion of ASF/SF2 has been also recently observed (21). In contrast to what was found for the BRCA1 exon T6 mutant, the defective splicing of the related SMN2 exon 7 gene was not affected by either overexpression or depletion of ASF/SF2 (20, 21), suggesting different roles for this splicing factor in the two systems. Indeed, in a series of short exonic deletions in the WT and T6 BRCA1 minigene contexts we identify a series of splicing-regulatory sequences, which are functionally relevant only in the presence of the T6 mutation (Fig. 5). In particular, the enhancer sequence in position 23 to 32 mediated ASF/SF2-dependent splicing activation (Fig. 5D). This evidence suggests that the T6 BRCA1-specific response to ASF/SF2 in overexpression and siRNA experiments may be due to the presence of a binding site for this splicing factor at position 23 to 32. This ASF/SF2 ESE stimulates BRCA1 exon 18 splicing, inducing exon inclusion only in the context of the T6 mutant (Fig. 6).
FIG. 6.
Splicing-regulatory elements in BRCA1 exon 18 and creation of an ESS by the T6 mutant. WT exon 18 contains two weak ESEs (striped box) and two ESSs (black boxes), and its inclusion is largely dependent on 3′ and 5′ splice site definition. The G-to-T mutation at position +6 creates a strong silencer element at the 5′ end of the exon binding to hnRNPA1/A2 and DAZAP1 and may also disrupt a weak splicing enhancer. The ASF/SF2 ESE located in the middle of exon 18 stimulates BRCA1 exon 18 splicing only in the context of the T6 mutant.
Several lines of evidence indicate that the natural mutation studied here creates an exonic splicing silencer. The double-site-directed mutation experiments indicate that the majority of point mutations affecting the “TAG” core sequence between positions +6 and +8 completely restore normal splicing (Fig. 2B). The mutant sequence, different from the WT, binds to three different proteins in pulldown assays and EMSA: hnRNPA1 and the related hnRNPA2 factor and DAZAP1. hnRNPA1 and hnRNPA2 are well-characterized splicing-inhibitory factors involved in a variety of cellular and viral systems (11) and have been identified as splicing repressors in the regulation of SMN2 exon 7 (20, 21). In contrast to hnRNPA1/A2, few data are available regarding the functional properties of DAZAP1, which has been mainly implicated in mRNA transport and stability (24, 28). Interestingly, SELEX experiments have shown two conserved sequences for the mouse homologue, AAUAG and GU1-3AG (18), which contain the same UAG splicing sequence as the BRCA1 T6 mutant. By means of siRNA experiments, we provide functional evidence that DAZAP1 and hnRNPA1/A2 are involved and probably have a redundant function in the splicing regulation of the defective BRCA1 exon 18. Treatment with hnRNPA1/A2 siRNA resulted in a significant recovery of the defective splicing, which was further enhanced by DAZAP1 depletion (Fig. 4B). The requirement of multiple siRNAs for an efficient rescue of defective T6 splicing and the EMSA experiments with purified proteins suggest that hnRNPA1/A2 and DAZAP1 specifically recognize the inhibitory sequence. This is further supported by the fact that DAZAP1 colocalizes and physically interacts with the splicing repressor proteins hnRNPA1 and hnRNPC1 (24). However, triple treatment with siRNA for hnRNPA1/A2 and DAZAP1 did not completely restore BRCA1 exon inclusion. This can be explained by the presence of residual amounts of the splicing factors, not completely depleted by the siRNAs, or to the binding of an additional splicing-inhibitory factor(s) at the silencer element not revealed by the pulldown analysis. Even if our results clearly indicate that the T6 mutant creates a strong silencer binding to hnRNA1/A2 and DAZAP1, we cannot exclude the simultaneous disruption of a weak enhancer, as recently suggested (21). In fact, we observed that the combined deletion of the 5′ end of BRCA1 exon 18 along with the downstream enhancer (Δ4-9-Δ23-32) induced partial exon skipping. Thus, with the limitation provided by the gross deletion analysis, which can interfere with RNA secondary structure and acceptor site accessibility or reduce the exon definition below a critical threshold, the 5′ end on BRCA1 exon 18 might contain a weak enhancer, which becomes evident only when the downstream enhancer is deleted. This weak enhancer in the WT sequence might bind to an unidentified splicing factor(s) or regulate RNA secondary structure constraints.
In agreement with our findings, the BRCA1 T6 mutant was recently found to bind to hnRNPA1, but its aberrant splicing was not corrected by hnRNPA1 and hnRNPA2 silencing (21). We do not have a clear explanation of these different findings, but culture cell lines may show variable concentrations of splicing-inhibitory factors like DAZAP1. These factors can efficiently replace hnRNAPA1/A2 at the T6 mutant silencer or can act on other splicing-regulatory elements modulating siRNA rescue splicing efficiency. Consistent with this hypothesis, we observed a different cell-type-dependent rescue efficiency of the splicing pattern in siRNA experiments (data not shown).
In summary, our results indicate that the primary determinant of the T6 BRCA1 exon 18 exclusion is not the disruption of an ASF/SF2-dependent enhancer but the creation of a new silencer element recognized specifically by the splicing-inhibitory factors hnRNPA1/A2 and DAZAP1 (Fig. 6). These act as repressors that bind to the silencer and inhibit exon recognition during the splicing process. We have clearly established that the basic mechanism affecting splicing due to mutations is not always obvious and cannot be simply derived from in silico predictions. The creation of exonic splicing silencers may be a common event in human pathology and has to be taken into account in order to develop therapeutic strategies aimed at correcting splicing defects.
Acknowledgments
This work was supported by the Associazione Italiana Ricerca sul Cancro and Italian Cystic Fibrosis Foundation to F.P. and E.G.
We thank Cristiana Stuani and Erica Bussani for technical assistance, Adrian Krainer for the pSMN2 minigene, Rodolfo Garcia for helpful discussion, and F. E. Baralle for continuous support and suggestions.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Ars, E., E. Serra, J. Garcia, H. Kruyer, A. Gaona, C. Lazaro, and X. Estivill. 2000. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 9237-247. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeois, C. F., F. Lejeune, and J. Stevenin. 2004. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 7837-88. [DOI] [PubMed] [Google Scholar]
- 4.Buratti, E., T. Dork, E. Zuccato, F. Pagani, M. Romano, and F. E. Baralle. 2001. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 201774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 131197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18186-193. [DOI] [PubMed] [Google Scholar]
- 7.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3285-298. [DOI] [PubMed] [Google Scholar]
- 8.Cartegni, L., M. L. Hastings, J. A. Calarco, E. de Stanchina, and A. R. Krainer. 2006. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 7863-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartegni, L., and A. R. Krainer. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30377-384. [DOI] [PubMed] [Google Scholar]
- 10.Cartegni, L., J. Wang, Z. Zhu, M. Q. Zhang, and A. R. Krainer. 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 313568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabot, B., C. LeBel, S. Hutchison, F. H. Nasim, and M. J. Simard. 2003. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog. Mol. Subcell. Biol. 3159-88. [DOI] [PubMed] [Google Scholar]
- 12.Disset, A., C. F. Bourgeois, N. Benmalek, M. Claustres, J. Stevenin, and S. Tuffery-Giraud. 2006. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum. Mol. Genet. 15999-1013. [DOI] [PubMed] [Google Scholar]
- 13.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3195-205. [DOI] [PubMed] [Google Scholar]
- 14.Fairbrother, W. G., R. F. Yeh, P. A. Sharp, and C. B. Burge. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 2971007-1013. [DOI] [PubMed] [Google Scholar]
- 15.Fairbrother, W. G., G. W. Yeo, R. Yeh, P. Goldstein, M. Mawson, P. A. Sharp, and C. B. Burge. 2004. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 32W187-W190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17419-437. [DOI] [PubMed] [Google Scholar]
- 17.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 61197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hori, T., Y. Taguchi, S. Uesugi, and Y. Kurihara. 2005. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 33190-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 20.Kashima, T., and J. L. Manley. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34460-463. [DOI] [PubMed] [Google Scholar]
- 21.Kashima, T., N. Rao, C. J. David, and J. L. Manley. 2007. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 163149-3159. [DOI] [PubMed] [Google Scholar]
- 22.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11363-371. [DOI] [PubMed] [Google Scholar]
- 23.Lim, L. P., and C. B. Burge. 2001. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. USA 9811193-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Y. T., and P. H. Yen. 2006. A novel nucleocytoplasmic shuttling sequence of DAZAP1, a testis-abundant RNA-binding protein. RNA 121486-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, H. X., L. Cartegni, M. Q. Zhang, and A. R. Krainer. 2001. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 2755-58. [DOI] [PubMed] [Google Scholar]
- 26.Lorson, C. L., and E. J. Androphy. 2000. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 9259-265. [DOI] [PubMed] [Google Scholar]
- 27.Mazoyer, S., N. Puget, L. Perrin-Vidoz, H. T. Lynch, O. M. Serova-Sinilnikova, and G. M. Lenoir. 1998. A BRCA1 nonsense mutation causes exon skipping. Am. J. Hum. Genet. 62713-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton, S., H. T. Yang, N. Moleleki, D. G. Campbell, P. Cohen, and S. Rousseau. 2006. Phosphorylation of the ARE-binding protein DAZAP1 by ERK2 induces its dissociation from DAZ. Biochem. J. 399265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muro, A. F., M. Caputi, R. Pariyarath, F. Pagani, E. Buratti, and F. E. Baralle. 1999. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 192657-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 251147-1149. [DOI] [PubMed] [Google Scholar]
- 31.Pagani, F., and F. E. Baralle. 2004. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 5389-396. [DOI] [PubMed] [Google Scholar]
- 32.Pagani, F., E. Buratti, C. Stuani, and F. E. Baralle. 2003. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 27826580-26588. [DOI] [PubMed] [Google Scholar]
- 33.Pagani, F., E. Buratti, C. Stuani, M. Romano, E. Zuccato, M. Niksic, L. Giglio, D. Faraguna, and F. E. Baralle. 2000. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 27521041-21047. [DOI] [PubMed] [Google Scholar]
- 34.Pagani, F., C. Stuani, M. Tzetis, E. Kanavakis, A. Efthymiadou, S. Doudounakis, T. Casals, and F. E. Baralle. 2003. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 121111-1120. [DOI] [PubMed] [Google Scholar]
- 35.Pozzoli, U., and M. Sironi. 2005. Silencers regulate both constitutive and alternative splicing events in mammals. Cell. Mol. Life Sci. 621579-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen, H., and M. R. Green. 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16363-373. [DOI] [PubMed] [Google Scholar]
- 37.Singh, N. N., E. J. Androphy, and R. N. Singh. 2004. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 315381-388. [DOI] [PubMed] [Google Scholar]
- 38.Smith, P. J., C. Zhang, J. Wang, S. L. Chew, M. Q. Zhang, and A. R. Krainer. 2006. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 152490-2508. [DOI] [PubMed] [Google Scholar]
- 39.Sun, H., and L. A. Chasin. 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 206414-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teraoka, S. N., M. Telatar, S. Becker-Catania, T. Liang, S. Onengut, A. Tolun, L. Chessa, O. Sanal, E. Bernatowska, R. A. Gatti, and P. Concannon. 1999. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 641617-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, X. H., and L. A. Chasin. 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 181241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, X. H., T. Kangsamaksin, M. S. Chao, J. K. Banerjee, and L. A. Chasin. 2005. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 257323-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]