Abstract
The telomeres and mating-type loci of budding yeast adopt a condensed, heterochromatin-like state through recruitment of the silent information regulator (SIR) proteins SIR2p, SIR3p, and SIR4p. In this study we characterize the chromatin binding determinants of recombinant SIR3p and identify how SIR3p mediates chromatin fiber condensation in vitro. Purified full-length SIR3p was incubated with naked DNA, nucleosome core particles, or defined nucleosomal arrays, and the resulting complexes were analyzed by electrophoretic shift assays, sedimentation velocity, and electron microscopy. SIR3p bound avidly to all three types of templates. SIR3p loading onto its nucleosomal sites in chromatin produced thickened condensed fibers that retained a beaded morphology. At higher SIR3p concentrations, individual nucleosomal arrays formed oligomeric suprastructures bridged by SIR3p oligomers. When condensed SIR3p-bound chromatin fibers were incubated in Mg2+, they folded and oligomerized even further to produce hypercondensed higher-order chromatin structures. Collectively, these results define how SIR3p may function as a chromatin architectural protein and provide new insight into the interplay between endogenous and protein-mediated chromatin fiber condensation pathways.
The genome of the budding yeast Saccharomyces cerevisiae contains transcriptionally silent, heterochromatin-like regions that are both stable throughout the cell cycle and inherited epigenetically. A specific complex of proteins, the silent information regulators (SIRs), is thought to mediate the assembly and maintenance of this silenced chromatin. Genetic and biochemical studies have identified the constituents of the SIR complex (SIR1p, -2p, -3p, and -4p) and have described the molecular relationships within the SIR complex and between the SIRs and the histone components of chromatin (6, 36; for reviews, see references 9, 50, and 51). The SIR protein that binds to chromatin fibers and affects higher-order chromatin structure is SIR3p. Overexpression of SIR3p in vivo leads to “spreading” of silenced chromatin outside the natural boundaries, and this “extended” silent chromatin is depleted of the other SIR complex proteins (27, 49, 59). When studied in vitro, purified, recombinant SIR3p efficiently binds to DNA, mononucleosomes, and nucleosomal arrays and forms complexes with highly reduced electrophoretic mobility on agarose gels (20). On the basis of these properties, SIR3p has been placed in a growing group of proteins that can directly condense chromatin fibers into specific higher-order secondary and tertiary chromatin structures. Such proteins have been termed chromatin architectural proteins (CAPs) (38).
CAPs that have been characterized biochemically to date include human MeCP2 (1, 21, 44), avian MENT (23, 40), and Drosophila PCC complex (18). CAPs have multiple DNA- and/or histone-binding sites that participate in local fiber folding in cis and fiber-fiber “bridging” in trans. Some CAPs, such as MeCP2, act as monomers (1, 44), while others, such as MENT, are dimers (57), and the PCC complex is a hetero-oligomer (18). The biochemical mechanisms for achieving protein-mediated chromatin fiber condensation clearly are diverse. In the present study we focused on the possible relationships between SIR3p self-association and its function as a CAP. Self-association of SIR3p in solution has been widely reported (16, 36, 42). The self-association behavior of the full-length protein has recently been characterized in detail and shown to be quite complicated (39). The most stable form of SIR3p is an 8S dimer. However, in low-micromolar-concentration solutions and under physiological salt conditions, the 8S SIR3p dimers are able to self-associate indefinitely in a reversible, concentration-dependent manner to produce unusually large oligomers that sediment in excess of 70 to 80S (39). While such oligomerization properties could help explain SIR3p-dependent assembly and the spreading of silent chromatin architecture, the biochemical properties and morphology of SIR3p-bound chromatin fibers have yet to be investigated in detail.
Nucleosomal arrays in the absence of other bound proteins are intrinsically dynamic; under physiological salt conditions, nucleosomal arrays are in equilibrium between unfolded monomers (i.e., beads-on-a-string structures), folded monomers (i.e., “30-nm” fibers), and oligomeric suprastructures (2, 5, 14, 17, 26, 48, 52, 53, 56, 61, 66). All four of the unmodified core histone N-terminal “tail” domains (NTD) contribute to intrinsic fiber condensation (22), and the H3 (31, 68) and H4 tails (13, 55) appear to play particularly important roles. In this regard, it is well established that there are both genetic and physical interactions between SIR3p and the H3 and H4 tails (7, 20, 27, 29, 36). This raises a key question. When SIR3p binds to the H3 and H4 tails and condenses chromatin fibers, does this process inhibit the intrinsic condensation pathway dependent on the same tails? To investigate this question and determine the structure and physicochemical properties of SIR3p-chromatin fiber complexes, we incubated purified recombinant SIR3p with long DNA molecules, nucleosome core particles (NCPs), and nucleosomal arrays (NA) and characterized the resulting nucleoprotein complexes by electrophoretic mobility shift assay (EMSA), analytical ultracentrifugation, and electron microscopy (EM). The results demonstrate that SIR3p binds to both naked DNA and the nucleosomal components of chromatin fibers and, acting through reversible concentration-dependent SIR3p-SIR3p self-association, assembles condensed structures with novel morphologies. Strikingly, exposure of the condensed SIR3p-bound chromatin fibers to Mg2+ led to further array folding into hypercondensed chromatin structures. The ramifications of these results for SIR-dependent silencing and chromatin fiber structural dynamics are discussed.
MATERIALS AND METHODS
SIR3p purification.
Sf9 insect cells were infected with a recombinant baculovirus expressing full-length rSIR3p fused to six tandem C-terminal histidines. The protein was purified by conventional ion-exchange chromatography as previously described (39). The resulting protein was >95% pure, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie staining. The extinction coefficient used for protein concentration determination was 83,835 M−1 cm−1 at 276 nm (ProtParam).
DNA, NCP, and nucleosomal array preparation.
208-12 DNA composed of 12 208-bp ribosomal DNA repeats was digested and purified as previously described (8). Purified pUC19 plasmid DNA was purified and linearized by digestion with EcoRI. DNA was quantified with a Beckman DU 600 spectrophotometer using the absorbance at 260 nm. NCPs were prepared as previously described (15) using the sea urchin 5S ribosomal DNA 146-bp fragment and hypoacetylated recombinant Xenopus laevis core histone octamers. Chromatin fibers were prepared using the 5S 208-12, 5S 172-12, and linearized pUC-19 DNA templates and purified hypoacetylated chicken erythrocyte core histone octamers (19, 24) or unacetylated recombinant Xenopus laevis core histone octamers (22). The level of saturation of the DNA templates by nucleosomes was determined by using sedimentation velocity ultracentrifugation (25).
EMSA.
Purified proteins, DNA, mononucleosomes, and chromatin fibers were mixed in TEN (10 mM Tris [pH 7.5; 22°C], 0.25 mM EDTA, 2 mM NaCl) at the specified molar ratios. The reactions were incubated for 30 min at room temperature, and glycerol was added from a 50% (vol/vol) solution to a final concentration of 5% (vol/vol). The samples were loaded onto a 1% agarose gel prepared and run in 1× TAE buffer at room temperature (21 to 23°C) at 5 V/cm for 2 to 2.5 h. The gels were stained with ethidium bromide and visualized by using a UV transilluminator. Images were recorded on a Gel Logic 200 (Kodak) and are inverted for clarity. The molecular weight marker is a BstEII digest of λ DNA (New England Biolabs).
EM.
DNA and NA samples in 5 mM HEPES (pH 8.0)-0.5 mM EDTA and the desired amounts of NaCl were placed in minidialyzer units (Pierce) and dialyzed for 4 h at 4C against buffer containing fresh 0.1% EM-grade glutaraldehyde, followed by overnight dialysis against buffer alone. Samples were prepared for EM essentially as described previously (44, 67). We used 2% uranyl acetate as a negative stain, or, after the grids were rinsed with water, as a positive stain. Some samples were shadowed with platinum at an angle of 10° after drying from glycerol (21). Grids were examined in a Tecnai 12 electron microscope (FEI Corp.) at 100 kV using a LaB6 filament, and images were recorded on a charge-coupled device camera (2,048 × 2,048; Tietz Gmbh, Gauting, Germany).
Analytical ultracentrifugation.
Sedimentation velocity measurements were performed using either a Beckman XL-I or Beckman Proteome XL-A analytical ultracentrifuge, using the absorbance optical system. Two-sector, charcoal-filled Epon centerpieces were filled with 400 μl of sample and 420 μl of reference (buffer) solutions. Samples were centrifuged in either a Beckman An60Ti 4-hole rotor or An50Ti 8-hole rotor at 21°C. Velocity data were edited and analyzed by using the boundary analysis method of Demeler and van Holde (11) as implemented in UltraScan (version 7.3 for Windows). All sedimentation coefficients are reported in Svedberg units (S), where 1 S = 10−13 s, and corrected to that of water at 20°C (s20,w). The partial specific volume of SIR3p (V-bar, 0.7472 cm3/g at 20°C) and solvent densities (ρ) were calculated within UltraScan. The predicted sedimentation coefficient of an extended 12-mer nucleosome array with one bound SIR3p dimer per nucleosome was modeled by using the Kirkwood Theory (34) as implemented in UltraScan version 2.88. Values of 65 ± 5 nm for the Stokes radius and 14S for the sedimentation coefficient of a Sir3 dimer-NCP complex (see Fig. 2B) were used and returned a predicted value of 42S ± 2S for the extended SIR3p-bound array.
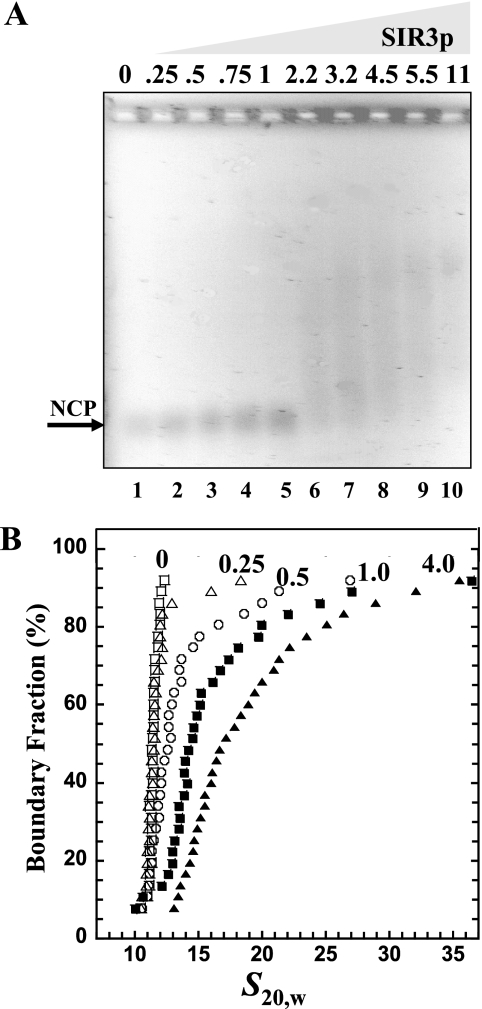
FIG. 2.
Binding of SIR3p to NCPs. (A) EMSA. NCP 200 ng (1 pmol) of 146-bp NCPs was incubated with r[SIR3p]2 = 0, 0.25, 0.5, 0.75, 1, 2.25, 3.25, 4.5, 5.5, and 11 (12.5 to 550 nM SIR3p dimer, respectively) prior to electrophoresis on a 1% agarose gel. The gel was stained with ethidium bromide and visualized by UV transillumination. The arrow indicates the position of unbound nucleosomes. (B) Sedimentation velocity. A total of 10 μg (50 pmol) of NCP was incubated alone (□) or with r[SIR3p]2 of 0.25 (▵), 0.5(○), 1 (▪), or 4 (▴) (31, 62.5, 125, and 500 nM SIR3p dimer, respectively) prior to sedimentation velocity. The resulting integral distribution of S [corrected for water at 20°C (s20,w)] is shown.
Differential centrifugation.
The differential centrifugation assay for self-association was performed as described previously (53, 54, 64). Briefly, nucleosomal arrays (A260, ∼1.4) were mixed with SIR3p and incubated for 15 min. An equal volume of a 2× MgCl2 solution as added to achieve the desired final salt concentration. After a 5-min incubation at 20°C, the samples were centrifuged (13,000 × g) for 10 min at 20°C in a microcentrifuge. The absorbance at 260 nm of the soluble fraction was measured, and the percentage of the nucleosomal array sample remaining in the supernatant was calculated as described previously (53, 54, 64).
RESULTS
SIR3p-DNA interactions.
Our previous work demonstrated that SIR3p is able to bind nonspecifically to DNA, and the results from EMSA suggested that DNA binding may be cooperative (20). However, neither the mechanism of SIR3p-DNA interactions nor the structure of SIR3p-DNA complexes was examined in that study. Consequently, in the present study our primary goal was to characterize SIR3p-DNA interactions by EM. EMSA experiments were first performed as controls. Purified SIR3p was incubated with 208-12 DNA (∼2.5 kb) in increasing molar equivalents of protein to 208-bp DNA repeat, and the mixtures were analyzed by EMSA using 1% agarose gels (Fig. 1A, lanes 1 to 5). Because SIR3p minimally is a dimer under the conditions of our assays (39; see also the Discussion), the molar ratio of protein to DNA is expressed throughout this study as moles of SIR3p dimer to moles of 208-bp DNA (r[SIR3p]2). The results indicate that increased SIR3p binding progressively retards the mobility of the DNA until complexes are formed that fail to migrate into the gel. At r[SIR3p]2 of 0.5 (i.e., 6 SIR3p dimers added per 208-12 DNA template), the mobility of SIR3p-bound 5S DNA was only slightly retarded. Between r[SIR3p]2 of 1 and 2 the bands underwent a dramatic decrease in mobility, and by r[SIR3p]2 of 2, all of the DNA molecules were in some form of shifted nucleoprotein complex. The diffuse nature of the bands is consistent with previous results (20) and suggests that SIR3p formed multiple complexes with different sizes, shapes, and/or charges under these conditions. At r[SIR3p]2 of 4, we observed both a defined, slowly migrating band and a significant fraction of complexes that were unable to migrate through the gel. When this experiment was repeated using linearized pUC19 DNA the same general gel shifts trends were observed although, compared to 5S DNA, the pUC19 DNA was shifted to equivalent levels at lower r values (Fig. 1A, lanes 6 to 10). Thus, the SIR3p-DNA interactions seen in Fig. 1 may exhibit sequence dependence.
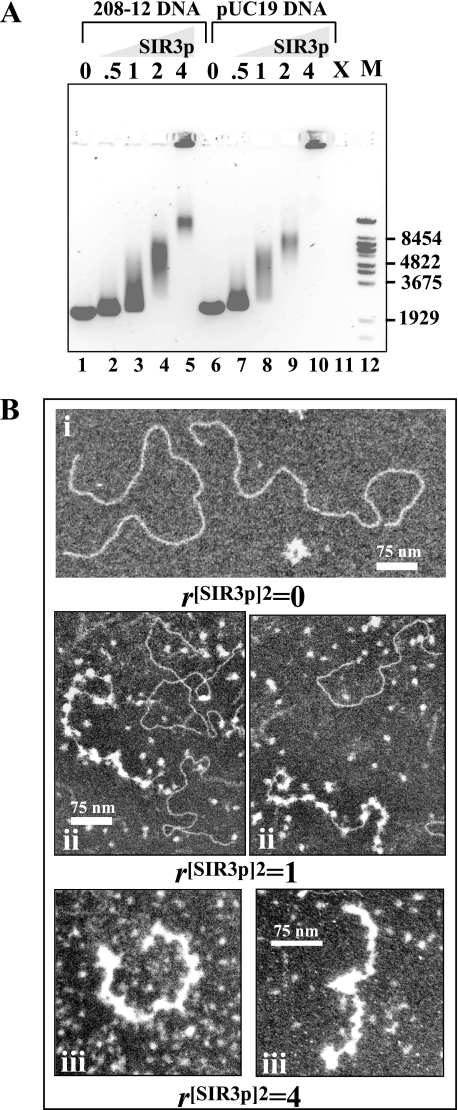
FIG. 1.
Interaction of SIR3p with DNA. (A) EMSA of SIR3p and DNA. Linear 208-12 DNA (lanes 1 to 5) or linear pUC19 DNA (lanes 6 to 10) (0.5 μg; 3.6 pmol) was incubated at r[SIR3p]2 of 0.5, 1, 2, and 4 (90, 180, 360, and 720 nM SIR3p dimer, respectively) prior to electrophoresis. The positions of the markers (M, λ BstEII digest fragments) are shown at right. Lane 11 was not loaded (X). (B) EM of the interaction of SIR3p with 208-12 DNA. (i) Appearance of DNA alone after positive staining. (ii) 208-12 DNA with r[SIR3p]2 of 1 dimer per 208 unit. Some DNA molecules are studded with bound SIR3p dimers, while others have little or none. Unbound dimers are seen in the background. (iii) At r[SIR3p]2 of 4, most DNA molecules are coated with protein, and the complexes have a significantly shorter contour length. Some (apparently) free DNA is also seen. Scale bars represent 75 nm: the higher-magnification images show greater detail within the complexes.
The SIR3p-DNA complexes from Fig. 1A next were visualized by EM to determine their morphologies. Representative images obtained at r[SIR3p]2 of 0, 1, and 4 are shown in Fig. 1B. At r[SIR3p]2 of 0, the naked linear 208-12 DNA was in an extended form on the grid (Fig. 1Bi). At r[SIR3p]2 of 1, we observed either naked DNA molecules or DNA that was nearly saturated with SIR3p (Fig. 1Bii). In the latter case, quantitation of these results indicated an average of 23 bound SIR3p particles per 208-12 molecule, which equates to one SIR3p dimer bound per ∼110 bp of DNA. The SIR3p molecules appeared as bright, nearly uniform ellipsoids, both in the background and when bound to DNA, and were roughly the same size as nucleosomes. These data demonstrate that SIR3p binding to long DNA at r[SIR3p]2 of 1 is a cooperative process that produces relatively regular arrays of SIR3p-DNA complexes, independent of DNA sequence. The length and sinuous morphology of the SIR3p-bound DNA at r[SIR3p]2 of 1 was comparable to that of the naked DNA, indicating that SIR3p binding did not condense the DNA under these conditions. At r[SIR3p]2 of 4 most DNA molecules were essentially completely coated with SIR3p, and it was not possible to determine stoichiometries by counting particles. When the contour lengths were quantitated, protein-saturated complexes had a mean length of 264 nm (standard error [SE] = 17 nm), ca. 40% shorter than DNA alone (413 nm, SE = 6.5 nm), presumably due to SIR3p-SIR3p interactions. The SIR3p-coated molecules are assumed to correspond to the relatively narrow band present in lane 5 of Fig. 1A. We did not observe large SIR3p-DNA complexes corresponding to the material in the wells in lane 5 of Fig. 1A. Such large aggregates tend to precipitate and are not readily imaged. As will be discussed below, the data in Fig. 1 suggest a role for double-stranded DNA interactions in chromatin fiber condensation.
SIR3p-NCP interactions.
Because the biological function of the SIR proteins is to silence chromatin, the ultimate goal of the present study was to investigate the structure of SIR3p-bound chromatin fibers. The next level in the hierarchical organization of chromatin is the NCP, which consists of 147 bp of DNA wrapped around a histone octamer. NCPs with more than 147 bp of DNA are called mononucleosomes, and the presence of the extranucleosomal DNA is thought to mimic the linker DNA between nucleosomes in a chromatin fiber. We have previously shown that SIR3p binds to mononucleosomes containing ∼60 bp of extranucleosomal DNA (20). Given the robust binding of SIR3p to DNA documented in Fig. 1, we sought to determine how well SIR3p bound to NCPs lacking this extranucleosomal linker DNA. In anticipation of obtaining heterogeneous samples (20), EMSA experiments were first performed as controls and subsequently correlated with definitive sedimentation velocity analyses. As expected, the EMSA data were complex (Fig. 2A). Titration of SIR3p at first led to little apparent binding as judged by the lack of shift of the NCP band (lanes 2 to 5). Higher SIR3p concentrations produced extensive smearing and no well-defined, slower-migrating bands, although all of the NCP band had been shifted (lanes 6 to 10). This result suggests that SIR3p binding to NCPs produced small amounts of many different nucleoprotein species, a finding consistent with the extensive smearing seen in our earlier EMSA experiments (20). To test this interpretation using a definitive solution-state technique, we analyzed the SIR3p-NCP complexes by sedimentation velocity in the analytical ultracentrifuge. NCPs and SIR3p were mixed at r[SIR3p]2 of 0.25 − 4, and the resulting boundaries were analyzed by using the Demeler-van Holde method (11), which produces a plot of the integral distribution of sedimentation coefficients in the sample, G(s) (Fig. 2B). A vertical plot is indicative of the presence of a single homogeneous species, while a sloping plot results from a heterogeneous sample (10). If the sample is heterogeneous, the relative amounts of each species can be determined from the G(s) plot (10). Free NCPs sedimented as a homogenous 11S species, as expected from many previous studies (3, 12, 58). At r[SIR3p]2 of 0.25, ∼90% of the NCPs sedimented at 11S, and the remaining ∼10% sedimented at between 14S and 18S. At [SIR3p]2 of 0.5, ∼65% of the NCPs sedimented at 11S, and the remaining 35% sedimented at between ∼14 and 25S. At r[SIR3p]2 of 1, ∼60% of the NCPs sedimented at 14S, while the remainder of the sample sedimented at between 14 and 37S. The homogenous 14S component of the sample most likely corresponds to the NCP-SIR3p dimer complex, while the faster-sedimenting species must be some type of higher-order complexes. This same SIR3p/NCP ratio when analyzed by EMSA showed only smearing above the NCP band (Fig. 2A, lane 5). By r[SIR3p]2 of 4, ∼30% of the sample sedimented at 14S and the remaining 70% sedimented from 14 to 37S. The observed 14 to 37S range of complexes is indicative of the presence of a range of nucleoprotein complexes with multiple SIR3p dimers and/or multiple nucleosomes. Consistent with this conclusion, the sample, when analyzed by EMSA (Fig. 2A, lane 8), exhibited a broad smear with no distinct banding. Together, the sedimentation velocity and EMSA data are internally consistent and show that SIR3p avidly binds to NCPs and assembles them into a population of higher-order nucleoprotein complexes. These results also indicate that the ability to bind to nucleosomes is not compromised by the lack of extranucleosomal DNA.
SIR3p-dependent spreading of condensed chromatin fiber structures in vitro.
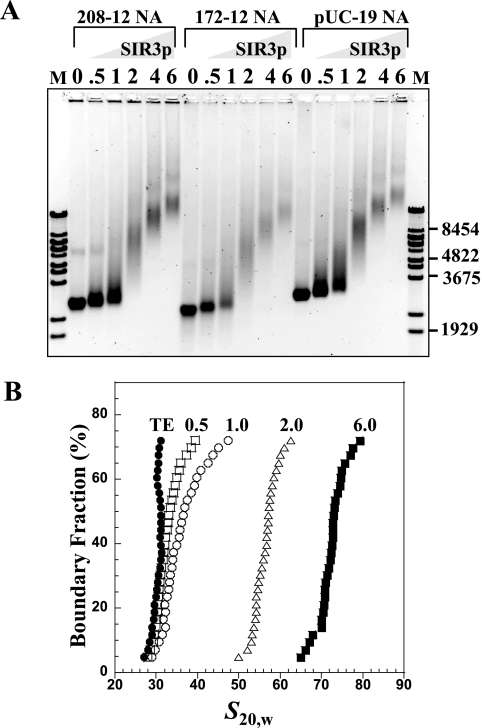
We next examined SIR3p binding to nucleosomal arrays and SIR3p-dependent changes in chromatin fiber morphology. Low salt conditions were used in the binding reactions so that the nucleosomal arrays were in the extended beads-on-a-string conformation, i.e., binding was not influenced by the endogenous folding pathway. Three types of nucleosomal arrays were used for these experiments: 208-12 nucleosomal arrays with positioned nucleosomes and long nucleosome repeats, 172-12 arrays with positioned nucleosomes and much shorter nucleosome repeat lengths closer to those of yeast chromatin in vivo (∼165 bp) (35, 43, 62), and pUC19 arrays that have randomly positioned nucleosomes and widely variable linker-DNA lengths. These three templates were reconstituted using native histone octamers to nearly identical saturation, as determined by sedimentation velocity (data not shown). Increasing molar ratios of SIR3p were mixed with the nucleosomal arrays and analyzed by EMSA using 1% agarose gels (Fig. 3). The banding patterns were quite complex. In the case of all three chromatin templates, increasing the r[SIR3p]2 from 0.5 to 6 led to a continuous retardation and smearing of the chromatin band and produced a broad continuum of slower-migrating species. Importantly, the banding patterns of all three chromatin samples were essentially identical, indicating that, as judged by EMSA, DNA sequence, linker DNA length and nucleosome spacing have little influence on SIR3p binding to nucleosomal arrays.
FIG. 3.
Binding of SIR3p to nucleosomal arrays. (A) EMSA. A chromatin array with 0.5 μg (3.6 pmol) of 208-12 (lanes 2 to 7), a 172-12 chromatin array (lanes 8 to 13), and linear, assembled pUC19 DNA chromatin arrays (lanes 14 to 19) were incubated at r[SIR3p]2 of 0.5, 1, 2, 4, and 6 (per DNA repeat or per 208 bp for pUC19, net SIR3p dimer concentrations of 90, 180, 360, 720, and 1,080 nM, respectively) prior to electrophoresis on a 1% agarose gel. The gel was stained with ethidium bromide and visualized by UV transillumination. The positions of the markers (M, λ Bst-E II digest fragments) are shown at the right. (B) Sedimentation velocity. A total of 10 μg (73 pmol) of chromatin array was incubated with r[SIR3p]2 of 0 (•), 0.5 (□), 1 (○), 2 (▵), and 6 (▪) of SIR3p dimers (net SIR3p dimer concentrations of 91, 182, 365, 720, and 1,095 nM, respectively) prior to sedimentation velocity. The resulting integral distribution of S [corrected for water at 20°C (s20,w)] over the bottom 75% of the boundary is shown (the top 25% of the boundary consisted of unproductive aggregates).
The complexes formed between SIR3p and 208-12 nucleosomal arrays also were examined by sedimentation velocity. The 208-12 arrays were used because their intrinsic physicochemical properties are well established (17, 26). The nucleosomal arrays in low salt sedimented between 28 and 32S (Fig. 3B), indicating that they were saturated with octamers (i.e., an average of 12 octamers per template). At r[SIR3p]2 of 0.5, roughly half of the arrays had bound SIR3p, as indicated by the increase in sedimentation coefficient above ∼30S. As the input was increased to r[SIR3p]2 of 1, the major effect was further shifting of the sedimentation coefficients of the SIR3p-bound fraction without markedly influencing the fraction of free arrays. This result was reminiscent of binding of SIR3p to NCPs (Fig. 2B). By r[SIR3p]2 of 2, all of the chromatin fibers were bound by SIR3p and sedimented from 50 to 60S. Importantly, at r[SIR3p]2 of 6, the complexes sedimented at 70 to 80S, indicating that SIR3p loading onto the fibers did not reach a plateau under the conditions studied.
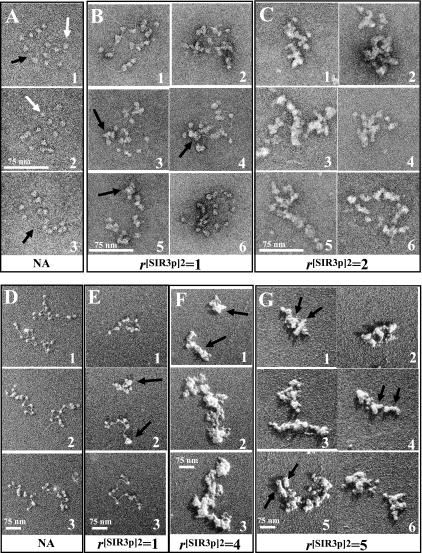
The increase in sedimentation coefficient from ∼30S to 50 to 80 S that occurs due to SIR3p binding in low salt likely reflects increases in mass and, probably, changes in fiber structure, given that an extended 12-mer array of SIR3p dimer-nucleosome complexes is predicted to sediment at only ∼42S (see Materials and Methods) To test this interpretation and visualize the morphology of SIR3p-bound chromatin fibers, samples assembled at r[SIR3p]2 of 0, 1, and 2 were analyzed by EM. Negatively stained images are shown in Fig. 4A to C. As expected, the parent 208-12 chromatin fibers in low salt appeared as extended “beads on a string” structures with 11 to 12 nucleosomes. Individual nucleosomes (∼10 nm in diameter) were well defined (white arrows), and the linker DNA segments between many of the nucleosomes were visible (black arrows). At r[SIR3p]2 of 1, some of the fibers had the same morphology as the parent nucleosomal arrays (Fig. 4B1 and 6), while others showed significant SIR3p binding, as indicated by the increased electron density that appeared to be coincident with the nucleosomes on the fiber (Fig. 4B, 3 to 5, black arrows). SIR3p-bound chromatin fibers assembled at r[SIR3p]2 = 2 were consistently thicker than the other samples (Fig. 4C). Intrafiber nucleosome-nucleosome contacts leading to fiber folding were obvious in all samples at r[SIR3p]2 of 2, a finding consistent with the 50 to 60S sedimentation coefficients of the chromatin fibers in solution. These contacts presumably were mediated by interactions between SIR3p dimers and either the nucleosomal and/or linker DNA components of the same chromatin fiber. Interestingly, the thickened fibers seen at r[SIR3p]2 of 2 maintained a beaded morphology, again indicating that much of the SIR3p binding was to the nucleosomes of the chromatin fiber. However, quantitation revealed that the fibers with r[SIR3p]2 of 1 had an average of 17 particles per array, suggesting some linker DNA binding also was occurring under these conditions.
FIG. 4.
EM of SIR3p-chromatin fiber complexes. (A) Negatively stained 208-12 nucleosomal arrays show 11 to 12 separated nucleosomes with linker DNA occasionally visible (arrows). (B) At r[SIR3p]2 of 1, most nucleosomes appear larger, and the total number of particles averages 17 per array, suggesting that the complexes consist of nucleosomes bound with SIR3p and SIR3p bound to linker DNA. (C) At r[SIR3p]2 of 2, arrays are coated with protein, and individual nucleosomes no longer resolved. (D to F) Glycerol dried, shadow-contrasted NAs. (D) Nucleosomes and linker DNA are clearly seen in untreated NAs. (E) At r[SIR3p]2 of 1, the population includes some that appear unchanged (E3), some that are partially compacted with fewer than 12 separate particles resolved (D1 and 2), and some that are completely compacted (D2). (F) At r[SIR3p]2 of 4, NAs appear as thickened, compact complexes (arrows) which tend to self-associate. (G) At r[SIR3p]2 of 5, thickening and self-association of arrays continues, and here arrows denote putative domains representing individual NAs. Note that with the glycerol drying technique, unbound SIR3p is separated from NAs. Scale bars represent 75 nm.
Images of preparations dried from glycerol and platinum shadowed were also collected to better understand the topography of the complexes (Fig. 4D to G). The free nucleosomal arrays in low salt (Fig. 4D) appeared similar to the structures in the negative stained images from Fig. 4A, except that the shadowing technique more clearly revealed the nucleosomes and linker DNA segments. The samples with r[SIR3p]2 of 1 showed a mixture of unbound arrays (Fig. 4E3) and SIR3p-bound fibers that were somewhat thickened and compacted, a finding consistent with both the negative staining (Fig. 4B) and the sedimentation velocity results (Fig. 3B). The shadowed images also suggested that SIR3p causes clustering of nucleosomes (panel 2, black arrows). Diverse and complex nucleoprotein structures were assembled by SIR3p at higher protein concentrations. Figure 4F1 (r[SIR3p]2 = 4) shows two single chromatin fibers that were condensed and appeared to be coated with SIR3p, while Fig. 4F2 and 3 show oligomeric suprastructures formed by fiber-fiber bridging. The structures in Fig. 4G1 to 6 revealed domains that may represent individual condensed 12-nucleosome arrays (black arrows) in end-to-end or side-to-side contact. Importantly, the EM data, like the sedimentation velocity and EMSA results, show that multiple chromatin fiber species are assembled with differently condensed structures as the SIR3p concentration is increased. As will be discussed below, these properties are likely related to the mechanism of SIR3p spreading when overexpressed in vivo.
SIR3p-bound chromatin fibers undergo salt-dependent folding to form hypercondensed secondary and tertiary chromatin structures.
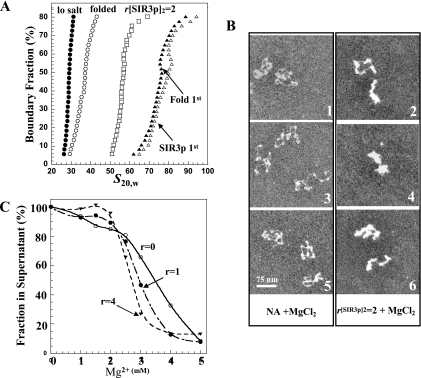
All of the experiments examining chromatin fiber structures thus far (Fig. 3 and 4) were performed in low-salt buffers to prevent induction of the endogenous folding and oligomerization pathways governed by salt concentration (26). Given that both the SIR3p-driven and endogenous condensation pathways appear to share some of the same determinants (e.g., the H3 and H4 tails) under physiological salt conditions, we wanted to test whether the two pathways are in competition. To examine this question, we performed order-of-addition experiments in which chromatin fibers were incubated in 2 mM MgCl2 to induce folding prior to SIR3p binding or in which SIR3p was prebound to fibers that were subsequently incubated with 2 mM MgCl2. These samples were then subjected to sedimentation velocity analysis to determine the sedimentation coefficient distributions of the resulting SIR3p-chromatin fiber complexes. The data obtained for chromatin fibers with r[SIR3p]2 of 2 are shown in Fig. 5A. As seen previously (Fig. 3B), incubation of the parent nucleosomal arrays with SIR3p in low salt increased the G(s) distribution from ∼30S to 50 to 60S. Surprisingly, incubation of these arrays in 2 mM MgCl2 led to further increases in the G(s) distribution to 60 to 80S. In the reverse experiment, incubation of the parent nucleosomal arrays in 2 mM MgCl2 resulted in the 30 to 55S G(s) distribution characteristic of endogenous folding into 30-nm fibers (17, 26, 53). When these folded chromatin fibers were incubated with SIR3p at r[SIR3p]2 of 2, the G(s) distribution increased to 60 to 80S and was essentially the same as the distribution of SIR3p-bound fibers that were subsequently exposed to salt. Identical results were obtained with r[SIR3p]2 of 5 chromatin, except that they formed faster-sedimenting complexes (data not shown). We next used EM to visualize the influence of salt on the structure of chromatin fibers with r[SIR3p]2 of 2 (Fig. 5B). Chromatin fibers in 2 mM MgCl2 in the absence of SIR3p (Fig. 5B1 to 3) exhibit the partially condensed morphology expected (5). The addition of SIR3p led to both a thickening of the fiber and hypercondensation, i.e., fibers that were significantly more condensed than those obtained with SIR3p or salt alone (Fig. 5B4 to 6), further illustrating that the endogenous and SIR3p-dependent condensation pathways function independently and additively. We also examined MgCl2-dependent chromatin fiber oligomerization (55) at r[SIR3p]2 of 1 and 4 (Fig. 5C). The results indicated that the SIR3p bound chromatin fibers oligomerized at slightly lower MgCl2 concentrations than the parent nucleosomal arrays. However, as with folding, the most important aspect of this result is that extensive binding of SIR3p to the chromatin fibers did not prevent salt-dependent self-association.
FIG. 5.
Further condensation of SIR3p-containing chromatin upon addition of MgCl2. (A) Sedimentation velocity. A total of 10 μg of nucleosomal arrays (•) was incubated with 2 mM MgCl2 for 30 min (“folded” [○]) or with SIR3p at r[SIR3p]2 of 2 for 30 min (□). The nucleosomal arrays in salt were then incubated at r[SIR3p]2 of 2 for 30 min (▵), while the chromatin fibers with r[SIR3p]2 of 2 were incubated in 2 mM MgCl2 for 30 min (▴). The resulting integral distribution of S (corrected for water at 20°C) over the bottom 80% of the boundary is shown (the top 20% of the boundary consisted of unproductive aggregates). (B) EM. Negatively stained preparations (1, 3, and 5) of nucleosomal arrays fixed in 2 mM MgCl2 are partially condensed (compare to Fig. 4D4 to 6). In the presence of r[SIR3p]2 of 2, additional condensation occurs (2, 4, and 6). Scale bars represent 100 nm. (C) Self-association of SIR3p-bound chromatin arrays. Nucleosomal arrays were incubated with r[SIR3p]2 of 0 (○), 1 (•), and 4 (▾) prior to the addition of MgCl2. The differential sedimentation assay was performed as described in Materials and Methods.
DISCUSSION
Our previous work suggested that SIR3p may have the ability to assemble chromatin fibers into uniquely condensed secondary and tertiary chromatin structures (20). Here, we have characterized the molecular determinants of SIR3p-chromatin fibers interactions and defined the sequence of biochemical events involved in SIR3p-dependent changes in chromatin fiber morphology. In our experimental design, purified recombinant SIR3p at ∼0.02 to 1 μM was added to naked DNA, NCPs, and nucleosomal arrays under various solution conditions, and the resulting SIR3p-bound complexes were examined by the complementary methods of EMSA, sedimentation velocity, and EM. To properly interpret the resulting biochemical and EM data, one must take into account the complex quaternary structure of SIR3p when free in solution. The 113-kDa full-length SIR3p monomer sediments at 6S (39). Under physiological salt conditions and at submicromolar protein concentrations, the 6S monomer forms a stable 8S dimer (39), most likely arranged in an antiparallel orientation (33). When the SIR3p concentration is raised into the micromolar range, the dimers reversibly and specifically self-associate into 13S tetramers and a series of higher-order oligomers that sediment from 16 to >80S. Extrapolation of these results to the present experimental conditions suggests that 8S (∼226 kDa) dimers were the predominant species in solution in the EM experiments and that mixtures of dimers and higher-order oligomers were present at the higher Sir3p input ratios used in some of the EMSA and sedimentation velocity experiments. In support of this conclusion, free SIR3p in EM images (Fig. 1) had a spherical shape that was roughly the same size as a ∼205-kDa NCP. Another important ramification is that within any given experiment, increasing the SIR3p concentration will shift the equilibrium in favor of more SIR3p-SIR3p interactions.
In view of the oligomerization properties of the free protein, we wondered how the propensity of SIR3p to self-associate is related to the way it interacts with and condenses chromatin fibers. Several lines of evidence indicate that both nucleosomal subunits and linker DNA are targets for SIR3p binding to chromatin fibers. In the former case, SIR3p binds readily to NCPs lacking free DNA (Fig. 2A). Further, SIR3p and nucleosomes mostly colocalize in EM images, producing structures that are thickened but retain the beaded morphology of the underlying chromatin fibers (Fig. 4). In the case of linker DNA, Fig. 1 demonstrates cooperative binding of SIR3p to naked DNA, and the stepwise loading of SIR3p observed in the EM experiments shows a gradual “filling in” of the internucleosomal linker DNA as the amount of SIR3p is increased (Fig. 4D to G). Also note that the EM results detected more than 12 SIR3p molecules bound to each 12-mer nucleosomal array at higher SIR3p concentrations. Taken together, our results indicate that interaction of SIR3p with chromatin fibers is not simple and that each SIR3p dimer is likely to be engaged in many different protein-protein and protein-DNA interactions, including interactions with itself.
Another goal of the present study was to examine the effects of SIR3p binding on chromatin fiber morphology using biochemical (Fig. 3) and EM approaches (Fig. 4). Both types of data are in excellent agreement, and together they suggest a multistep pathway leading to chromatin fiber condensation by SIR3p. The initial step involves binding of SIR3p dimers to the nucleosomal and linker DNA sites on single fibers, as discussed above. As more SIR3p dimers are added they saturate the remainder of the primary nucleosomal sites, followed by additional binding to the SIR3p-nucleosome complexes to yield the thickened beaded 50 to 60S structures seen at r[SIR3p]2 = 2 (Fig. 4C). At r[SIR3p]2 of both 1 and 2, SIR3p-nucleosome interactions lead to local nucleosome clustering and fiber compaction. We believe this is most likely reflective of bridging interactions between SIR3p dimers and its nucleosomal and linker DNA binding sites on the same fiber. Although we obtained no evidence for intermolecular fiber-fiber interactions at r[SIR3p]2 of ≤2, as r[SIR3p]2 was raised to 4 to 5 the shadowed EM images showed oligomeric suprastructures whose morphology appeared to display interfiber interactions. Assembly of these oligomers represents the final step in the condensation pathway in vitro, although in our studies the size of the oligomers continued to increase with increasing SIR3p. Ultimately, our studies show that SIR3p oligomerization remains concentration dependent and indefinite, even when SIR3p is bound to chromatin fibers (Fig. 3 and 4) or DNA (Fig. 1). Moreover, these results directly document concentration-dependent “spreading” of SIR3p oligomers on chromatin fibers in vitro under conditions that mimic in vivo overexpression experiments where SIR3p spreading is known to occur (47, 49, 59; see also below).
In addition to revealing how SIR3p functions as a CAP, these studies have also yielded new information about the interplay between endogenous and protein-mediated chromatin condensation pathways. Endogenous, salt-induced chromatin fiber condensation has been widely studied for over 30 years. With increasing salt, chromatin fibers first fold into a fiber ∼30 nm in diameter based on a zigzag two start geometry (52). Endogenous folding is heavily dependent on the H4 NTD and the contacts it makes with the surface of adjacent nucleosomes (13, 35, 64). As the salt is increased, chromatin fibers cooperatively self-associate through a process that involves all four core histone NTDs (22). Recent cross-linking studies show that the H3 NTDs make intrafiber and interfiber contacts with DNA during self-association (32, 68). Much attention has been paid to the role of the H3 and H4 NTDs in SIR-dependent silencing (7, 27-30, 32, 36, 45, 63). Thus, the endogenous condensation pathways clearly share many of the same macromolecular determinants as the SIR3p-mediated pathway. Hence, we wondered whether the two pathways are mutually exclusive and were surprised to find that they were instead additive and independent (Fig. 5). One possible explanation for this result is that one of the common binding determinants (e.g., H4 NTD) is available to both pathways, and only partial engagement of the binding sites is needed to drive condensation. In other words, in salt solutions, endogenous folding occurs before all of the H4 NTDs are engaged with neighboring nucleosomal surfaces, leaving some H4 NTDs available for binding to SIR3p. In this regard, the maximum extent of H3 NTD cross-linking to chromatin during salt-dependent oligomerization is only ∼30% (31, 68). Alternatively, SIR3p may mediate chromatin condensation exclusively through non-tail (e.g., linker DNA and nucleosome surface [4, 46, 60, 65]) interactions, leaving the tails available for the endogenous pathway. While the endogenous and SIR3p-dependent pathways together produced hypercondensed chromatin fibers, i.e., fibers that were significantly more condensed than obtained with SIR3p or salt alone, the linker DNA was freely accessible to digestion by EcoRI (20; data not shown). Thus, another important ramification of the present study is that extensive chromatin fiber condensation does not automatically correlate with binding site inaccessibility and thus repressed function. Although more work is needed to understand how multiple condensation pathways can coexist, these results suggest that the specific nucleoprotein structure of any given region of the genome is dictated by both endogenous nucleosome-nucleosome interactions and effects contributed by the specific architectural protein(s) in that region.
How do these results relate to SIR-dependent silencing in yeast in vivo? SIR-dependent silencing at telomeres relies on at least three SIR proteins, SIR2p, SIR3p, and SIR4p, which are believed to act in a complex (19, 41, 50). However, the existence of a stable SIR2p-SIR3p-SIR4p complex has yet to be documented biochemically in vitro. Thus, SIR-dependent silencing may involve regulated interaction of SIR3p with a SIR2p/SIR4p complex, leaving some SIR3p bound within the silent chromatin. When SIR3p is overexpressed in vivo, transcriptionally silent regions near the telomeres are extended beyond their normal boundaries, and these regions are depleted of SIR4p and SIR2p (28, 47, 59). This phenomenon has been referred to as SIR3p-dependent spreading of silent chromatin. The absence of SIR2p and SIR4p, together with the increasing molar ratios of SIR3p to nucleosome used in our experiments, likely reproduces many elements of the extended silent architecture in vivo. Unless interactions with SIR2p or SIR4p abolish the extensive SIR3p self-association that occurs when free in solution and when bound to DNA or nucleosomal templates, our observations are likely to be relevant to the role of SIR3p in establishment of the chromatin architecture at native telomeres as well.
Acknowledgments
This study was supported by NIH grants GM66834 to J.C.H. and GM43786 to C.L.W.
Footnotes
Published ahead of print on 24 March 2008.
REFERENCES
- 1.Adams, V. H., S. J. McBryant, P. H. Wade, C. Woodcock, and J. C. Hansen. 2007. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J. Biol. Chem. 28215057-15064. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, N. L., M. Watts, and P. T. Georgel. 2004. To the 30-nm chromatin fiber and beyond. Biochim. Biophys. Acta 167712-23. [DOI] [PubMed] [Google Scholar]
- 3.Ausio, J., D. Seger, and H. Eisenberg. 1984. Nucleosome core particle stability and conformational change: effect of temperature, particle and NaCl concentrations, and cross-linking of histone H3 sulfhydryl groups. J. Mol. Biol. 17677-104. [DOI] [PubMed] [Google Scholar]
- 4.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311856-861. [DOI] [PubMed] [Google Scholar]
- 5.Bednar, J., R. A. Horowitz, S. A. Grigoryev, L. M. Carruthers, J. C. Hansen, A. J. Koster, and C. L. Woodcock. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA 9514173-14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bose, M. E., K. H. McConnell, K. A. Gardner-Aukema, U. Müller, M. Weinreich, J. L. Keck, and C. A. Fox. 2004. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 24774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 2774778-4781. [DOI] [PubMed] [Google Scholar]
- 8.Carruthers, L. M., C. Tse, K. P. Walker, and J. C. Hansen. 1999. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 30419-35. [DOI] [PubMed] [Google Scholar]
- 9.Connelly, J. J., P. Yuan, H. C. Hsu, Z. Li, R. M. Xu, and R. Sternglanz. 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 263256-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demeler, B., H. Saber, and J. C. Hansen. 1997. Identification and interpretation of complexity in sedimentation velocity boundaries. Biophys. J. 72397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demeler, B., and K. E. van Holde. 2004. Sedimentation velocity analysis of highly heterogeneous systems. Anal. Biochem. 335279-288. [DOI] [PubMed] [Google Scholar]
- 12.Dieterich, A. E., R. Axel, and C. R. Cantor. 1979. Salt-induced structural changes of nucleosome core particles. J. Mol. Biol. 129587-602. [DOI] [PubMed] [Google Scholar]
- 13.Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 32785-96. [DOI] [PubMed] [Google Scholar]
- 14.Dorigo, B., T. Schalch, A. Kulangara, S. Duda, R. R. Schroeder, and T. J. Richmond. 2004. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 3061571-1573. [DOI] [PubMed] [Google Scholar]
- 15.Dyer, P. N., R. S. Edayathumangalam, C. L. White, Y. Bao, S. Chakravarthy, U. M. Muthurajan, and K. Luger. 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 37523-44. [DOI] [PubMed] [Google Scholar]
- 16.Enomoto, S., S. D. Johnston, and J. Berman. 2000. Identification of a novel allele of SIR3 defective in the maintenance, but not the establishment, of silencing in Saccharomyces cerevisiae. Genetics 155523-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher, T. M., and J. C. Hansen. 1996. The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr. 6149-188. [DOI] [PubMed] [Google Scholar]
- 18.Francis, N. J., R. E. Kingston, and C. L. Woodcock. 2004. Chromatin compaction by a polycomb group protein complex. Science 3061574-1577. [DOI] [PubMed] [Google Scholar]
- 19.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 2791-16. [DOI] [PubMed] [Google Scholar]
- 20.Georgel, P. T., M. A. Palacios DeBeer, G. Pietz, C. A. Fox, and J. C. Hansen. 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA 988584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgel, P. T., R. A. Horowitz-Scherer, N. Adkins, C. L. Woodcock, P. A. Wade, and J. C. Hansen. 2003. Chromatin compaction by human MeCP2: assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 27832181-32188. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, F., K. Luger, and J. C. Hansen. 2005. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 28033701-33706. [DOI] [PubMed] [Google Scholar]
- 23.Grigoryev, S. A., J. Bednar, and C. L. Woodcock. 1999. MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J. Biol. Chem. 2745626-5636. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, J. C., J. Ausio, V. H. Stanik, and K. E. van Holde. 1989. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry 289129-9136. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, J. C., K. E. van Holde, and D. Lohr. 1991. The mechanism of nucleosome assembly onto oligomers of the sea urchin 5 S DNA positioning sequence. J. Biol. Chem. 2664276-4282. [PubMed] [Google Scholar]
- 26.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31361-392. [DOI] [PubMed] [Google Scholar]
- 27.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80583-592. [DOI] [PubMed] [Google Scholar]
- 28.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 38392-96. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, L. M., P. S. Kayne, E. S. Kahn, and M. Grunstein. 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 876286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, L. M., G. Fisher-Adams, and M. Grunstein. 1992. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 112201-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kan, P. Y., X. Lu, J. C. Hansen, and J. J. Hayes. 2007. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 272084-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 5527-39. [DOI] [PubMed] [Google Scholar]
- 33.King, D. A., B. E. Hall, M. A. Iwamoto, K. Z. Win, J. F. Chang, and T. Ellenberger. 2006. Domain structure and protein interactions of the silent information regulator sir3 revealed by screening a nested deletion library of protein fragments. J. Biol. Chem. 28120107-20119. [DOI] [PubMed] [Google Scholar]
- 34.Kirkwood, J. G. 1957. The forces between protein molecules in solution: a summary. J. Cell Physiol. 49(Suppl. 1)59-62. [DOI] [PubMed] [Google Scholar]
- 35.Krajewski, W. A., and J. Ausió. 1996. Modulation of the higher-order folding of chromatin by deletion of histone H3 and H4 terminal domains. Biochem. J. 316(Pt. 2)395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz, and D. Moazed. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121515-527. [DOI] [PubMed] [Google Scholar]
- 37.Lohr, D., and G. Ide. 1979. Comparison of the structure and transcriptional capability of growing phase and stationary yeast chromatin: a model for reversible gene activation. Nucleic Acids Res. 61909-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBryant, S. J., V. H. Adams, and J. C. Hansen. 2006. Chromatin architectural proteins. Chromosome Res. 1439-51. [DOI] [PubMed] [Google Scholar]
- 39.McBryant, S. J., C. Krause, and J. C. Hansen. 2006. Domain organization and quaternary structure of the Saccharomyces cerevisiae silent information regulator 3 protein, Sir3p. Biochemistry 4515941-15948. [DOI] [PubMed] [Google Scholar]
- 40.McGowan, S., A. M. Buckle, J. A. Irving, P. C. Ong, T. A. Bashtannyk-Puhalovich, W. T. Kan, K. N. Henderson, Y. A. Bulynko, E. Y. Popova, A. I. Smith, S. P. Bottomley, J. Rossjohn, S. A. Grigoryev, R. N. Pike, and J. C. Whisstock. 2006. X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation. EMBO J. 253144-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moazed, D., A. D. Rudner, J. Huang, G. J. Hoppe, and J. C. Tanny. 2004. A model for step-wise assembly of heterochromatin in yeast. Novartis Found. Symp. 25948-56. [PubMed] [Google Scholar]
- 42.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 82257-2269. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, R. G., and W. L. Fangman. 1979. Nucleosome organization of the yeast 2-micrometer DNA plasmid: a eukaryotic minichromosome. Proc. Natl. Acad. Sci. USA 766515-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikitina, T., X. Shi, R. P. Ghosh, R. A. Horowitz-Scherer, J. C. Hansen, and C. L. Woodcock. 2007. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 27864-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, E. C., and J. W. Szostak. 1990. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol. 104932-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, J. H., M. S. Cosgrove, E. Youngman, C. Wolberger, and J. D. Boeke. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32273-279. [DOI] [PubMed] [Google Scholar]
- 47.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 71133-1145. [DOI] [PubMed] [Google Scholar]
- 48.Robinson, P. J., and D. Rhodes. 2006. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 16336-343. [DOI] [PubMed] [Google Scholar]
- 49.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 132207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 51.Santos-Rosa, H., A. J. Bannister, P. M. Dehe, V. Géli, and T. Kouzarides. 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 27947506-47512. [DOI] [PubMed] [Google Scholar]
- 52.Schalch, T., S. Duda, D. F. Sargent, and T. J. Richmond. 2005. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436138-141. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz, P. M., and J. C. Hansen. 1994. Formation and stability of higher order chromatin structures: contributions of the histone octamer. J. Biol. Chem. 26916284-16289. [PubMed] [Google Scholar]
- 54.Schwarz, P. M., A. Felthauser, T. M. Fletcher, and J. C. Hansen. 1996. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry 354009-4015. [DOI] [PubMed] [Google Scholar]
- 55.Shogren-Knaak, M., and C. L. Peterson. 2006. Switching on chromatin: mechanistic role of histone H4-K16 acetylation. Cell Cycle 51361-1365. [DOI] [PubMed] [Google Scholar]
- 56.Simpson, R. T., F. Thoma, and J. M. Brubaker. 1985. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42799-808. [DOI] [PubMed] [Google Scholar]
- 57.Springhetti, E. M., N. E. Istomina, J. C. Whisstock, T. Nikitina, C. L. Woodcock, and S. A. Grigoryev. 2003. Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J. Biol. Chem. 27843384-43393. [DOI] [PubMed] [Google Scholar]
- 58.Stein, A. 1979. DNA folding by histones: the kinetics of chromatin core particle reassembly and the interaction of nucleosomes with histones. J. Mol. Biol. 130103-134. [DOI] [PubMed] [Google Scholar]
- 59.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1183-93. [DOI] [PubMed] [Google Scholar]
- 60.Suto, R. K., M. J. Clarkson, D. J. Tremethick, and K. Luger. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 71121-1124. [DOI] [PubMed] [Google Scholar]
- 61.Thoma, F., T. Koller, and A. Klug. 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83403-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas, J. O., and V. Furber. 1976. Yeast chromatin structure. FEBS Lett. 66274-280. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369245-247. [DOI] [PubMed] [Google Scholar]
- 64.Tse, C., and J. C. Hansen. 1997. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry 3611381-11388. [DOI] [PubMed] [Google Scholar]
- 65.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. 109745-756. [DOI] [PubMed] [Google Scholar]
- 66.Woodcock, C. L., L. L. Frado, and J. B. Rattner. 1984. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J. Cell Biol. 99(Pt. 1)42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woodcock, C. L., and R. A. Horowitz. 1998. Electron microscopic imaging of chromatin with nucleosome resolution. Methods Cell Biol. 53167-186. [DOI] [PubMed] [Google Scholar]
- 68.Zheng, C., X. Lu, J. C. Hansen, and J. J. Hayes. 2005. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J. Biol. Chem. 28033552-33557. [DOI] [PubMed] [Google Scholar]