Abstract
Steroid hormone receptors regulate gene expression, interacting with target DNA sequences but also activating cytoplasmic signaling pathways. Using the human 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2) gene as a model, we have investigated the contributions of both effects on a human progesterone-responsive promoter in breast cancer cells. Chromatin immunoprecipitation has identified two different mechanisms of hormone-induced progesterone receptor (PR) recruitment to the 11β-HSD2 promoter: (i) direct PR binding to DNA at the proximal promoter, abrogated when PR contains a mutated DNA binding domain (DBD), and (ii) STAT5A (signal transducer and activator of transcription 5A)-mediated recruitment of PR to an upstream distal region, impaired by AG490, a JAK/STAT pathway inhibitor. The JAK/STAT inhibitor, as well as expression of dominant-negative STAT5A, impairs hormone induction of 11β-HSD2. On the other hand, the DBD-mutated PR fully supports 11β-HSD2 expression. These results, along with data from a deletion analysis, indicate that the distal region is crucial for hormone regulation of 11β-HSD2. We show active RNA polymerase II tracking from the distal region upon PR and STAT5A binding, concomitant with synthesis of noncoding, hormone-dependent RNAs, suggesting that this region works as a hormone-dependent transcriptional enhancer. In conclusion, coordination of PR transcriptional effects and cytoplasmic signaling activation, in particular the JAK/STAT pathway, are critical in regulating progestin-induced endogenous 11β-HSD2 gene expression in breast cancer cells. This is not unique to this promoter, as AG490 also alters the expression of other progesterone-regulated genes.
Steroid hormone receptors (SHRs) are considered nuclear transcription factors that, upon activation by binding with their corresponding ligands, regulate the expression of different target genes. Ligand-activated SHRs act either by binding as dimers to their hormone-responsive elements (HREs) at promoters or by interaction with other DNA-bound factors. In both cases, the process results in the recruitment of coregulators, chromatin remodeling complexes, and the general transcriptional machinery (7). However, SHRs also modulate gene expression by activation of cytoplasmic signaling pathways (nongenomic actions) (22).
The estrogen receptor (ER) binds to c-Src and to the phosphoinositol 3-kinase (PI3K) regulatory subunit, activating the Src/Ras/Erk and PI3K/Akt pathways, respectively (15, 41). These rapid effects triggered by hormones have been associated with their proliferative role. Ligand-activated progesterone receptor (PR) activates the Src/Ras/Erk pathway indirectly via an interaction with ER in the absence of estrogens (5), although direct interaction and activation of c-Src by PR has also been reported (11).
The relationship between SHRs' direct transcriptional effects and those mediated by activation of cytoplasmic kinase cascades in the hormone-inducible mouse mammary tumor virus (MMTV) promoter was recently investigated (62). After progesterone treatment, Erk and Msk1 are activated and recruited with phosphorylated PR to the promoter, where histone H3 is phosphorylated and acetylated locally. These H3 modifications seem to be a key switch for the exchange of a repressive complex containing HP1γ by coactivators, chromatin remodeling complexes, and RNA polymerase II (RNAP II). Thus, rapid kinase activation by progestin may participate in induction of PR direct target genes by preparing the chromatin for transcription, indicating that both PR actions cross talk to each other.
In breast cancer cells, progestin also induces activation of the JAK/STAT (signal transducer and activator of transcription) pathway and the subsequent phosphorylation of STAT proteins (47). The activation of the JAK/STAT pathway is initiated by cytokines or growth factors binding to their specific membrane-associated receptor. Receptor dimerization leads to JAK activation, which sequentially autophosphorylates and phosphorylates the receptor and STAT proteins. STAT proteins dimerize, translocate to the nucleus, and bind to DNA sequences at target promoters (18, 20, 28). The receptors involved in JAK activation may be those with intrinsic Tyr kinase activity (e.g., epidermal growth factor receptor) as well as those receptors lacking intrinsic kinase activity but to which JAKs are noncovalently associated (e.g., prolactin receptor). Also, it has been reported that, in breast cancer cells, progestin activates the JAK/STAT pathway by means of ligand-bound PR activation of the Tyr kinase c-Src (47). Additionally, it has been reported that progestin stimulates the association between PR and STAT proteins and their translocation to the nucleus (47, 49), as happens with other SHRs (14, 54, 55, 67), but the significance of this interaction in gene expression has not been addressed in detail. Moreover, STAT5A and -B have been implicated in the regulation of the Bcl-X gene by the glucocorticoid and PRs (50, 64).
To further explore the effects of SHRs on endogenous genes, we chose the human 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2) gene as a model system, since it has been identified in previous microarray studies as being among the strongest progestin- and glucocorticoid-induced genes in breast cancer cells (65). The 11β-HSD2 gene codifies an enzyme involved in the metabolism of glucocorticoids, particularly in the conversion of the active ligand cortisol to the inactive agonist cortisone (48). Due to the inability of cortisone to induce the antiproliferative effect of glucocorticoid receptor activation, 11β-HSD2 expression has been associated with cell proliferation. In accordance with this, 11β-HSD2 is overexpressed in many neoplasic tissues and cell lines compared to that in their healthy, differentiated tissues (19, 27, 48).
The 11β-HSD2 promoter spans ∼1.8 kb upstream of the transcription start site (TSS) (1). However, transcription factor binding sites have been mostly characterized in the ∼400 bp of the proximal promoter and part of the first exon (44). The cis-acting elements characterized include Sp1, IkB, and NF-1 binding sites (3, 37, 44). The mechanism by which steroid hormones control 11β-HSD2 promoter activity has not been explored in detail. Therefore, our aims were to characterize the promoter regions responsible for hormone responsiveness and to analyze the signaling pathways involved in hormone induction.
Here, we report that progestin regulates 11β-HSD2 gene expression in breast cancer cells by hormone-dependent PR binding to proximal and distal regions of its promoter. PR binding to the distal promoter region depends on the JAK/STAT pathway activation by progestin and on the recruitment of STAT5A to the same region, while PR recruitment to the proximal promoter region involves DNA direct receptor binding. Interfering with JAK/STAT activation completely abrogates 11β-HSD2 response to progestin. The distal region of the 11β-HSD2 promoter is the main region responsible for the hormone responsiveness of this promoter, acting as an entry site for RNAP II, which then tracks to the main TSS in the proximal promoter. These results provide new insights into the role of rapid nongenomic signaling of steroid receptors in mediating gene expression through activated signaling pathways coupling with transcription factors.
MATERIALS AND METHODS
Reagents.
R5020 was purchased from PerkinElmer Life Sciences; RU486 (RU) and ovine prolactin were from Sigma. ICI182780 (ICI) was from Tocris, and PD98059 (PD), Wortmannin (WM), and AG490 (AG) inhibitors were from Calbiochem. The anti-11β-HSD2 protein antibody was purchased from Binding Site; anti-FLAG-tag antibody was from Sigma; anti-STAT5A was from Santa Cruz; anti-phospho-STAT5A/B (Tyr694/6999), clone 8-5-2, was from Upstate; anti-phospho-JAK2 (Tyr1007/1008) was from Cell Signaling; anti-polymerase II (RNAP II) and phospho-polymerase II (RNAP II S5p) antibodies were from Covance; anti-AcH4 and pS10-H3 antibodies were from Upstate; anti-3MeK4-H3 and SRC-1 were from Abcam; anti-tubulin was from Sigma.
Cell lines, culturing conditions, and hormone treatment.
T47D breast cancer cells and T47D-MTVL cells (carrying one stably integrated copy of the luciferase [Luc] reporter gene driven by the MMTV promoter [59]) were routinely grown in Dulbecco's modified Eagle's medium and RPMI 1640 medium, respectively, supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin.
T47D-YV cells (PR-negative clonal derivative cell line of T47D [30, 53]) were used to generate TYML cells (T47D-YV-derived cell lines with one integrated copy of MMTV-Luc) expressing either the wild-type (WT) PR isoform B (PRB) or PRB mutants (DNA binding domain [DBD] or AF2) from the retroviral vector pRAV-Flag (36) (unpublished results). All T47D-YV-derived cell lines were routinely grown in modified Eagle's medium supplemented with 7% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin.
For the experiments, cells were plated in phenol red-free medium supplemented with 10% dextran-coated charcoal-treated FBS and, 24 h later, the medium was replaced by fresh serum-free medium. After 2 days under serum-free conditions, cells were treated with R5020 (10 nM) or ethanol for different times at 37°C. Prolactin was used at 500 ng/ml. When indicated, RU (1 μM) or ICI (10 μM) was added simultaneously, or PD (50 μM), WM (0.1 μM), or AG (50 μM) was added 1 h before hormone treatment.
Plasmids.
The pGL3-1778 11βLuc reporter vector was a gift from Lewis P. Rubin (1). The entire promoter construct pGL3-1778+117 11βLuc was obtained by adding 117 bp of exon 1, obtained by PCR amplification of human genomic DNA with primers −368 (5′-GTGTCCCGAACAAGCGTGAGTGGC-3′) and +608 (5′-TCTCACTTTCCCTCCAACACTCCC-3′), followed by digestion of the product with ApaI and NcoI. The unique restriction sites NcoI, AatII, and BssHII were used to obtain the deletion constructs starting at positions −1551, −839, and −571, respectively. The deletion construct at −1345 was generated by amplification from the pGL3-1778+117 11βLuc plasmid, with the oligonucleotide 5′-TTTGGTACCTCCCAGCCTCCCCTGAGATT-3′, corresponding to the nucleotides from −1345 to −1326, plus a KpnI restriction site, and the oligonucleotide 5′-CAGTGGAGGGTGGGGTGTCAG-3′, corresponding to the nucleotides from −748 to −769, as forward and reverse primers, respectively. The PCR product was cut with KpnI and AatII and then cloned into the KpnI and AatII sites of the pGL3-839+117 11βLuc construct.
The vector pGL3-1778-1345 11βLuc was generated by amplification from the pGL3-1778+117 11βLuc plasmid, with the oligonucleotide 5′-ATTTCTCTATCGATAGGTACC-3′, which includes the KpnI restriction sequence of the pGL3 reporter vector multiple cloning site, and the oligonucleotide 5′-AAAGGTACCGCCAGACCCACC-3′, corresponding to the nucleotides from −1345 to −1336, plus a KpnI restriction site added to perform the cloning, as forward and reverse primers, respectively. The PCR product was digested with KpnI and then cloned into the KpnI site of the pGL3 reporter vector. The vector pGL3-1778-1345/−368+117 11βLuc was generated by cloning the same PCR product into the KpnI site of the pGL3-368+117 11βLuc construct.
pSG5-PRB and pSG5-PRA were gifts from Pierre Chambon (34). pSTAT5A-WT and pSTAT5A*6 (STAT5A constitutively active [CA] mutant) were kindly provided by Toshio Kitamura (4). pSTAT5AΔ749 (STAT5A dominant-negative [DN] mutant) was a gift from Fabrice Gouilleux (43).
Immunofluorescence.
T47D cells were seeded onto coverslips in 35-mm dishes at 1.2 × 105 cells/cm2 as described above for serum-free hormone treatment experiments. After the treatment with ethanol or R5020, cells were washed, fixed by incubation in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, and permeabilized by incubation in 0.2% Triton X-100 in PBS for 10 min at room temperature. After three 5-min rinses in PBS, the coverslips were incubated for 1 h with 3% bovine serum albumin (BSA) in PBS at room temperature to reduce nonspecific staining. To detect 11β-HSD2 protein, cells were incubated with specific antibody against 11β-HSD2 diluted 1:1,000 in 3% BSA in PBS for 1 h at room temperature. After several washes in PBS, the coverslips were exposed to biotinylated-secondary anti-goat antibody (Molecular Probes) diluted 1:200 in 3% BSA in PBS for 1 h at room temperature. Then, the coverslips were washed in PBS and exposed to streptavidin-conjugated antibody Alexa 488 (Molecular Probes), diluted 1:1,000 in 3% BSA in PBS for 1 h at room temperature. The coverslips were mounted on slides with VectaShield DAPI (4′,6′-diamidino-2-phenylindole) mounting medium (Vector Laboratories) and subjected to a Leica DM IRBE inverted research microscope.
To detect phospho-JAK-2, cells were incubated with specific antibody diluted 1:500 in 3% BSA in PBS for 1 h at room temperature. After several washes in PBS, coverslips were incubated for 1 h with Alexa Fluor 555-conjugated goat anti-rabbit immunoglobulin G (IgG; Invitrogen), diluted 1:250 in 3% BSA in PBS for 1 h at room temperature. Nuclei were stained with DAPI, and the coverslips were mounted on slides with Mowiol mounting medium and subjected to a Leica SPE confocal inverted microscope.
Immunoblotting.
Cells treated as indicated above were lysed in Tris-HCl (pH 7.4; 25 mM), EDTA (1 mM), EGTA (1 mM), and sodium dodecyl sulfate (1%) plus protease and phosphatase inhibitors. Cell lysates were obtained by centrifugation, and protein concentration was determined by Micro bicinchoninic acid protein assays (Pierce). Lysates were adjusted to equivalent protein concentrations with lysis buffer, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western blotting using specific antibodies.
RNA extraction and RT-PCR.
Total RNA was prepared by using Trizol reagent (Invitrogen) as described in the manufacturer's instructions. The cDNA was generated from 100 ng of total RNA by using the Superscript first-strand synthesis system (Invitrogen). One microliter of cDNA was used as a template for PCR. Indicated gene products were analyzed by PCR with specific oligonucleotides, followed by visualization in agarose gel. When indicated, quantification of gene products was performed by real-time PCR using LightCycler 480 Sybr green I Master (Roche). Each value was corrected by the human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene and expressed as relative units. Gene-specific oligonucleotide sequences are available on request. Each experiment was performed in duplicate, as were the following real-time PCR quantifications (total n = 4). Results for one experiment representative of at least two experiments are shown.
Transfection assays.
For transient transfections, 2 × 105 cells plated in 35-mm plates were transfected with Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. A total amount of 3 μg of reporter and expression vectors was used per well. Six hours later, the medium was replaced by phenol red-free medium with antibiotics. After 2 days under serum-free conditions, cells were incubated with R5020 for 16 h. After incubation, cells were harvested in lysis buffer (Promega) and the protein amount was determined by a Micro bicinchoninic acid protein assay (Pierce). Lysates were adjusted to equivalent protein concentrations with lysis buffer, and Luc activity was determined with a Luc assay kit (Promega) according to the manufacturer's instructions. For deletion analysis of the 11β-HSD2 promoter, the vectors expressing Luc under the control of the different constructs of the promoter were transfected into T47D or T47D-YV cells, together with empty pSG5, pSG5-PRB, or pSG5-PRA expression vectors. For expression analysis of the 11β-HSD2 promoter under the control of WT STAT5A or mutant STAT5A forms, 11βLuc reporter constructs were cotransfected in T47D-YV cells with a pSG5-PRB expression vector and pSTAT5A-WT, pSTAT5A*6, or pSTAT5AΔ749 expression vectors. Each experiment was performed in duplicate, as were the following Luc measurements (total n = 4). Results for one experiment representative of at least two experiments are shown.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (56), using chromatin from TYML cells expressing Flag-tagged WT PRB or, when indicated, PRB DBD or AF2 mutants, cultured and treated as described previously. Chromatin was sonicated to an average fragment size of 400 to 500 bp, routinely, in a Branson Sonifier. For the experiment whose results are shown in Fig. 8C and D, fragments with an average size of 200 bp were obtained using a Diagenode Bioruptor. Rabbit or mouse IgG (Sigma) was used as a control for nonspecific interaction of DNA. In order to determine the linear range of the amplification, different numbers of cycles and dilution series of input DNA were used for PCR analysis of each amplicon. Input was prepared with 10% of the chromatin material used for an immunoprecipitation. Later, a 1:10 dilution of input material was performed before PCR amplification. The human β-globin gene amplicon was used as a loading control, detecting unspecific binding of genomic DNA to beads or IgGs. PCR products were resolved in agarose gel. Alternatively, quantification of gene products was performed by real-time PCR using LightCycler 480 Sybr green I Master (Roche). Each value was corrected by the level for the human actin gene and expressed as relative units. Primer sequences and specific PCR conditions are available on request. The numbers of PCR amplification cycles were as follows: 28 for amplicons A, B, D, and G; 30 for amplicons C, E, F, H, I, and NucB; and 32 for β-globin. Results for one experiment representative of at least two experiments are shown.
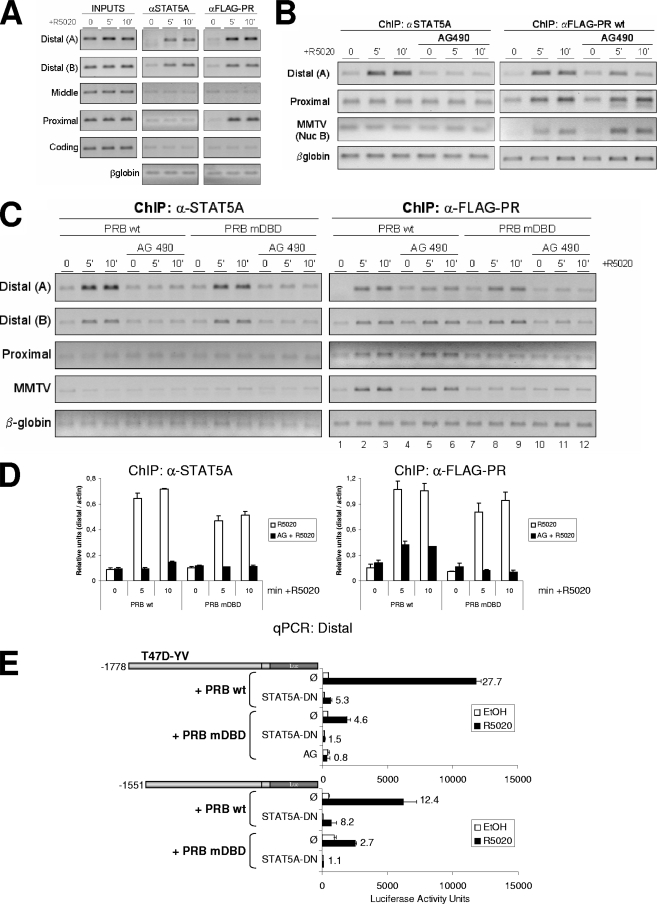
FIG. 8.
STAT5A and PR recruitment to the distal region depends on the JAK/STAT pathway. (A) TYML cells expressing FLAG-tagged WT PRB were cultured as described for Fig. 1A, untreated (0) or treated with R5020 (10 nM) for 5 or 10 min, harvested, and used for ChIP experiments using anti-STAT5A (αSTAT5A; middle panel) or anti-FLAG tag (right panel) antibodies. The precipitated DNA fragments were subjected to PCR analysis with primers corresponding to the indicated 11β-HSD2 promoter regions and the β-globin gene. Input material (1%) is shown for comparison. PCR products were run in a 1.2% agarose gel and visualized with ethidium bromide. (B) TYML cells expressing FLAG-tagged WT PRB, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 5 or 10 min, harvested, and used for ChIP experiments with anti-STAT5A or anti-FLAG-tag antibodies. When indicated, cells were treated with AG (50 μM) 1 h before hormone addition. The precipitated DNA fragments were subjected to PCR analysis with specific primers corresponding to the distal and proximal 11β-HSD2 promoter regions or MMTV nucleosome B and the β-globin gene as a control. PCR products were run in a 1.2% agarose gel and visualized with ethidium bromide. (C) ChIP experiment as in panel B from TYML cells expressing FLAG-tagged WT PRB or mutant DBD PRB (PRB-mDBD), combined with AG (50 μM). Here, the average chromatin fragment size was 200 bp (see Materials and Methods). (D) Real-time PCR quantification of ChIP samples from panel C for distal amplicon A, performed in duplicate and normalized to actin gene amplification levels. The values represent the means and data ranges. (E) T47D-YV cells cotransfected with 1 μg of the 11β-HSD2 −1778 or −1551 promoter construct, 1 μg of WT or PRB-mDBD, and 1 μg of DN STAT5A when indicated were treated with ethanol (EtOH) or R5020 (10 nM) for 16 h, and Luc activity was measured. Where indicated, 1 h of AG pretreatment was performed. Induction in response to hormone is shown for each construct. The values represent the means and ranges of a representative experiment performed in duplicate.
In silico analysis.
Screening for potential transcription factor binding sites was performed by Web-based prediction of regulatory elements, using ConSite (52) (http://www.phylofoot.org/consite) and Transfac (68) (http://www.gene-regulation.com/pub/databases.html#transfac). Data on RNA maps discussed by Kapranov et al. (32) were accessed through the National Center for Biotechnology Information's Gene Expression Omnibus (GEO; accession number GSE7576) (http://www.ncbi.nlm.nih.gov/geo/).
Microarray data accession number.
Microarray data are available at GEO under accession number GSE9286 (http://www.ncbi.nlm.nih.gov/geo/index.cgi).
RESULTS
11β-HSD2 transcription is induced by progestin in breast cancer cells and depends on PR.
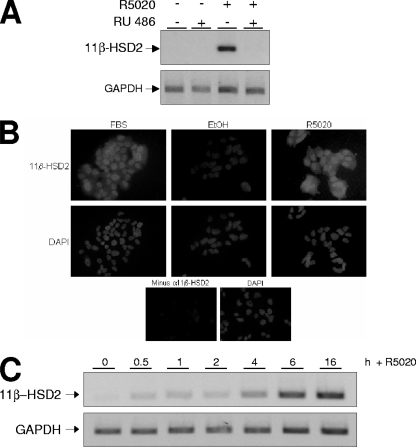
To check the hormone responsiveness of the 11β-HSD2 promoter in human breast cancer cells, serum-starved T47D cells were incubated with the progestin R5020, and mRNA expression was analyzed by RT-PCR (Fig. 1A). After 16 h of treatment, 11β-HSD2 expression was increased by R5020 treatment, and the induction was totally abolished by RU incubation (Fig. 1A). This result indicated that the promoter expression is specifically induced by progestin through the activation of PR in T47D cells. 11β-HSD2 protein accumulation in response to R5020 treatment was also detected with a specific antibody (Fig. 1B).
FIG. 1.
11β-HSD2 promoter expression in T47D cells is induced by progestin and depends on the classical PR. (A) T47D cells were cultured in phenol red-free medium under serum-free conditions for 48 h before the addition of R5020 (10 nM) and/or RU (1 μM) or vehicle for 16 h. Cells were harvested, and total RNA was extracted. The 11β-HSD2 mRNA expression was analyzed by RT-PCR with specific primers. GAPDH cDNA-specific primers were used as a control. PCR products were run on a 1.2% agarose gel and visualized with ethidium bromide. (B) Hormone-induced accumulation of 11β-HSD2 enzyme detected by immunofluorescence. T47D cells were cultured over coverslips in 10% FBS rich medium or in serum-free, steroid-deprived medium for 48 h before the addition of R5020 (10 nM) or ethanol (EtOH) for 24 h. Then, the cells were fixed, impermeabilized, and incubated with anti-11β-HSD2 antibody, followed by incubation with biotinylated secondary antibody and then streptavidin-conjugated Alexa 488 and, finally, DAPI, to stain nucleic acids (as described in Materials and Methods). (Lower panels) For a negative control, T47D cells cultured in rich medium were analyzed as previously described in the absence of the primary antibody. (C) T47D cells, cultured as described for panel A, were untreated (0) or treated with R5020 (10 nM) for the times indicated. 11β-HSD2 expression was analyzed as described for panel A, and GAPDH expression was used as a control.
To analyze the kinetics of promoter activation by progestin, 11β-HSD2 transcript accumulation was examined at different time points after R5020 addition. A gradual increase of 11β-HSD2 transcription was observed, already evident at 30 min and reaching nearly plateau levels after 6 h (Fig. 1C). Real-time PCR quantification of 11β-HSD2 cDNA repeatedly showed 8- to 12-fold induction after 6 h of R5020 treatment (see Fig. 4C and 5B as examples).
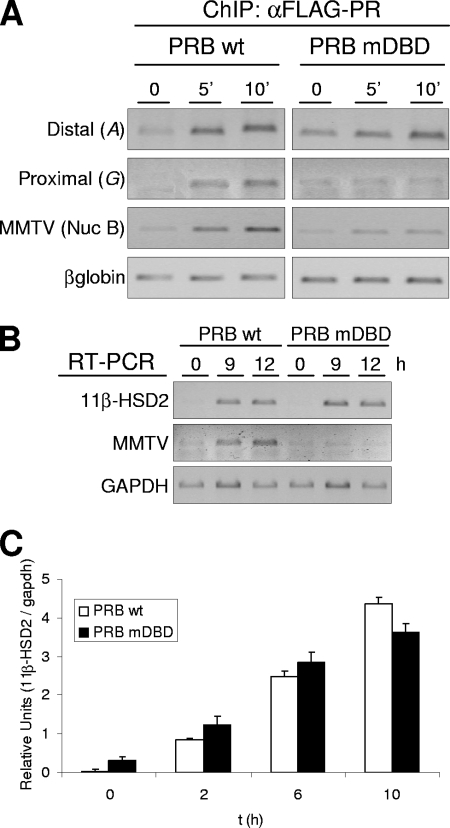
FIG. 4.
Effect of PR mutations at the DBD on progesterone-induced 11β-HSD2 promoter activity. (A) TYML cells expressing FLAG-tagged WT PRB or mutant DBD PRB (PRB-mDBD), cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 5 or 10 min, harvested, and used for ChIP experiments with anti-FLAG tag (αFLAG) antibody. The precipitated DNA fragments were subjected to PCR analysis with specific primers corresponding to the distal or proximal 11β-HSD2 promoter region, with MMTV nucleosome B and the β-globin gene as a control. (B) TYML cells expressing WT PRB or PRB-mDBD, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 9 or 12 h. Cells were harvested, and total RNA was extracted. The 11β-HSD2 and MMTV-Luc mRNA expression levels were analyzed by RT-PCR with specific primers. GAPDH cDNA-specific primers were used as a control. PCR products were run on a 1.2% agarose gel and visualized with ethidium bromide. (C) TYML cells expressing WT PRB or PRB-mDBD, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 2, 6, or 10 h. Cells were then harvested, and total RNA was extracted. 11β-HSD2 mRNA expression was analyzed by RT and real-time PCR with specific primers. GAPDH expression was measured for normalization. Data of a representative experiment performed in duplicate, with each sample analyzed in duplicate, are expressed as mean numbers of 11β-HSD2/GAPDH relative units, and error bars represent the data ranges.
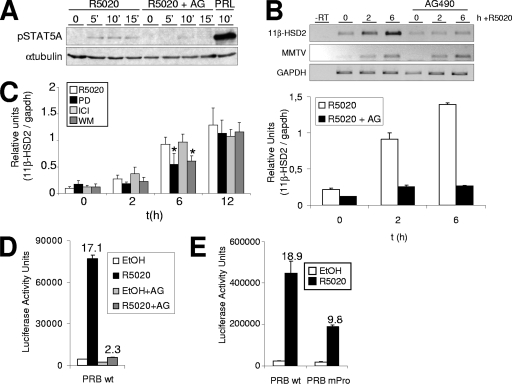
FIG. 5.
JAK/STAT pathway activation is involved in the hormonal induction of 11β-HSD2 gene expression. (A) T47D cells were cultured in phenol red-free medium under serum-free conditions for 72 h prior to the addition of 10 nM R5020 for 5, 10, or 15 min or 500 ng/ml prolactin (PRL) for 10 min. When indicated, 1 h before hormonal induction, cells were treated with 50 μM AG. Cell lysates were analyzed by Western blotting with antibodies against phosphorylated STAT5A (Tyr694/699) and tubulin. (B) T47D cells, cultured as described for Fig. 1A, were treated with R5020 (10 nM) for 0, 2, or 6 h. When indicated, AG (50 μM) was added 1 h before hormone addition. Cells were harvested, and total RNA was extracted. (Upper panel) 11β-HSD2 and MMTV-Luc mRNA expression levels were analyzed by RT-PCR with specific primers. GAPDH cDNA-specific primers were used as a control. PCR products were run on a 1.2% agarose gel and visualized with ethidium bromide. (Lower panel) 11β-HSD2 mRNA expression was analyzed by RT and real-time PCR with specific primers. GAPDH cDNA-specific primers were used as a control. The values represent the means and ranges of a representative experiment performed in duplicate, with each sample analyzed in duplicate, expressed as numbers of 11β-HSD2/GAPDH relative units. (C) Involvement of MAPK and PI3K pathways in hormone activation of 11β-HSD2. T47D cells, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 2, 6, or 12 h. When indicated, PD (50 μM), WM (0.1 μM), or ICI (10 μM) was added 1 h before hormone induction. Cells were harvested, total RNA was prepared, and cDNA was generated by RT. 11β-HSD2 expression was analyzed by real-time PCR with specific primers. To normalize the data, GAPDH expression was used as a control. The values represent the means and standard deviations of two experiments performed in duplicate, expressed as numbers of 11β-HSD2/GAPDH relative units. Asterisks denote significant differences (P < 0.05) between treated (PD or WM) and untreated (only R5020) data sets, as analyzed by Student's t test. (D) T47D-YV cells cotransfected with WT pSG5-PRB and reporter 11β-HSD2-Luc vectors were treated with R5020 (10 nM) for 16 h, and Luc activity was determined. When indicated, cells were treated with AG (50 μM) 1 h before hormone addition. For normalization, equal amounts of cellular extract of each sample were used. The values represent the mean numbers of arbitrary Luc units and ranges of a representative experiment performed in duplicate. EtOH, ethanol. (E) Expression of 11β-HSD2 reporter construct in the presence of a Pro cluster PR mutant. T47D-YV cells cotransfected with WT pSG5-PRB or a Pro cluster mutant and reporter 11β-HSD2-Luc vectors were treated with R5020 (10 nM) for 16 h, and Luc activity was determined. For normalization, equal amounts of cellular extract of each sample were used. The values represent the mean numbers of arbitrary Luc units and ranges of a representative experiment performed in duplicate.
The distal −1778/−1345 promoter region is required for progesterone induction.
In order to investigate the mechanism underlying hormonal activation of 11β-HSD2 transcription, we first focused on defining the minimal promoter region required for hormone response. Previous reports have defined the 11β-HSD2 promoter as the 1,778 bp preceding the TSS and part of the first exon (1, 44). In silico analysis revealed several potential HRE half-sites along this promoter sequence (Fig. 2A).
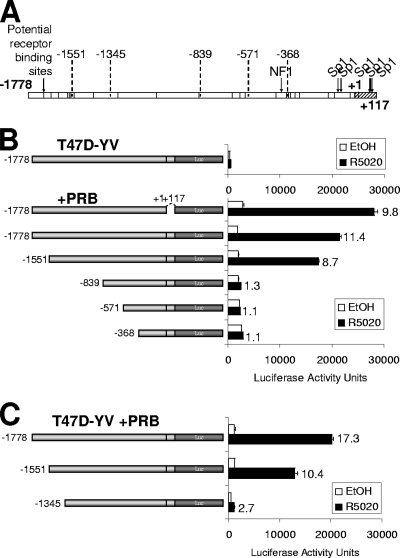
FIG. 2.
An 11β-HSD2 promoter region upstream of nucleotide −1345 is necessary for progesterone-induced activation. (A) Human 11β-HSD2 promoter region structure. Arrows indicate the positions of NF1 and Sp1 binding sites. The vertical bars along the 11β-HSD2 promoter region indicate the positions of potential progesterone receptor elements predicted by computational analysis with ConSite and Transfac software. The deletion endpoints used in the deletion analysis are indicated. (B, C) Analysis of promoter deletion constructs driving Luc reporter expression. Indicated constructs (−1778, −1551, −839, −571, and −368 in panel B; −1778, −1551, and −1345 in panel C) were transfected together with a PRB expression vector into T47D-YV cells. After transfection, cells were serum starved for 48 h and then treated with ethanol (EtOH) or R5020 (10 nM) for 16 h and Luc activity was measured. For normalization, equal amounts of extracted protein of each sample were used. Data of a representative experiment performed in duplicate, with each sample analyzed in duplicate, are expressed as mean numbers of Luc arbitrary units, and error bars represent the data ranges. Induction in response to hormone compared to the level for ethanol is shown for each construct.
To define the regions mediating induction by progestin, serial deletion constructs of the human 11β-HSD2 promoter were fused to the Luc reporter and tested in transient transfection experiments with the PR-negative clonal derivative T47D-YV (30, 53), with and without expression vectors for PRB or PR isoform A (PRA) (Fig. 2 and Table 1).
TABLE 1.
Summary of 11β-HSD2 promoter deletion analysis results obtained with T47D-YV and T47D cellsa
| Cell type and overexpressed isoform | 11β-HSD2 promoter region | Fold induction | Range |
|---|---|---|---|
| T47D-YV | −1778/+117 | 1.51 | ±0.10 |
| PRB | −1778/+117 | 11.41 | ±0.21 |
| −1778/+1 | 10.38 | ±0.34 | |
| −1551/+117 | 8.66 | ±0.02 | |
| −839/+117 | 1.26 | ±0.01 | |
| −571/+117 | 1.10 | ±0.01 | |
| −368/+117 | 1.12 | ±0.02 | |
| PRA | −1778/+117 | 8.07 | ±1.54 |
| −1778/+1 | 9.25 | ±0.44 | |
| −1551/+117 | 6.57 | ±0.03 | |
| −839/+117 | 1.35 | ±0.03 | |
| −571/+117 | 1.13 | ±0.03 | |
| −368/+117 | 1.07 | ±0.01 | |
| T47D | −1778/+117 | 3.15 | ±0.11 |
| −1778/+1 | 2.92 | ±0.07 | |
| −839/+117 | 1.66 | ±0.20 | |
| −571/+117 | 1.53 | ±0.10 | |
| −368/+117 | 1.53 | ±0.07 | |
| PRB | −1778/+117 | 12.24 | ±0.77 |
| −1778/+1 | 13.43 | ±0.51 | |
| −1551/+117 | 5.49 | ±0.29 | |
| −839/+117 | 2.70 | ±0.79 | |
| −571/+117 | 3.28 | ±0.17 | |
| −368/+117 | 3.05 | ±0.36 | |
| PRA | −1778/+117 | 8.68 | ±0.21 |
| −1778/+1 | 7.95 | ±0.51 | |
| −839/+117 | 1.38 | ±0.48 | |
| −571/+117 | 2.09 | ±0.06 | |
| −368/+117 | 1.59 | ±0.26 |
The average induction levels and ranges of Luc activity for hormone/vehicle from two independent experiments performed in duplicate are shown.
The entire 11β-HSD2 promoter (−1778/+117) transfected into T47D-YV cells was unsensitive to R5020 due to the lack of endogenous PR isoforms. When the promoter construct was cotransfected with a PRB expression vector, a robust response to R5020 (up to 11-fold induction) was obtained (Fig. 2B). Deletion of the +1/+117 region, which contains several previously identified Sp1 binding sites (1), did not significantly affect the hormone response. Deletion of the −1778/−1551 region led to a partial reduction in R5020 induction, while deletion of the −1778/−839 region (or further deletions) completely abolished 11β-HSD2 promoter activity (Fig. 2B). In a separate experiment, deletion at −1345 also showed a substantial reduction of transcriptional response to hormone (Fig. 2C). Similar results were obtained when PRA was used instead of PRB for cotransfection with promoter constructs into T47D-YV, although the induction was slightly lower, indicating that PRA is also transcriptionally active on this gene and acts through the same promoter region (Table 1).
Similar experiments were performed with T47D cells to investigate the hormone responses of some of the 11β-HSD2 promoter constructs in the context of endogenous PRB and PRA levels (Table 1). Full-length promoter constructs responded to R5020 with a threefold induction. Deletions to −839 or further downstream lost this response. When T47D cells were cotransfected with a PRB or PRA expression vector, a strong hormone response (up to 13-fold) was observed, similar to that of transfected T47D-YV (Table 1). In conclusion, overexpression of any PR isoform improves the response obtained by endogenous PR in transfection assays. Interestingly, shorter promoter constructs (down to −368/+117) retained a significant hormone-dependent activity (threefold) in PRB-overexpressing T47D cells, indicating that this proximal region may also contain a weak but independent element responding to hormone activation.
These results led us to conclude that the region between −1778 and −1345 encompasses the information required for most of the promoter responsiveness to progestin in the context of PR-expressing cells. Interestingly, deletion at −1551 retains a proportion of this capacity. Under receptor overexpression conditions, the proximal −368/+117 region also presents some hormone response in the context of reporter constructs.
PR associates with two different regions of 11β-HSD2 promoter after hormone activation.
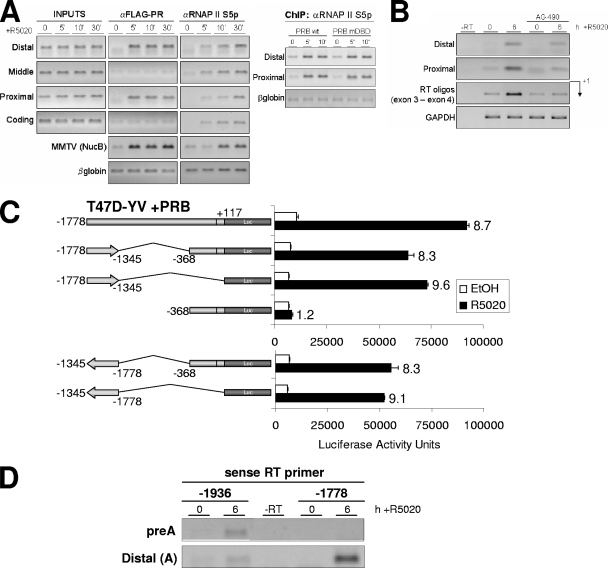
In order to further characterize the regulation of 11β-HSD2 gene expression by progesterone, PR recruitment to the endogenous promoter was investigated by ChIP. For this, we used a T47D-YV-derived cell line containing an integrated copy of the progesterone-responsive reporter MMTV-Luc (TYML cells), stably expressing a FLAG-tagged version of the WT PRB isoform (unpublished results). Expression of tagged PRB in this cell line was comparable to endogenous PRB expression in T47D cells (data not shown). PR recruitment in response to R5020 treatment was analyzed at 5, 10, and 30 min using an anti-FLAG-tag antibody, and the −1778/+117 promoter region was extensively covered with eight PCR amplicons (Fig. 3A).
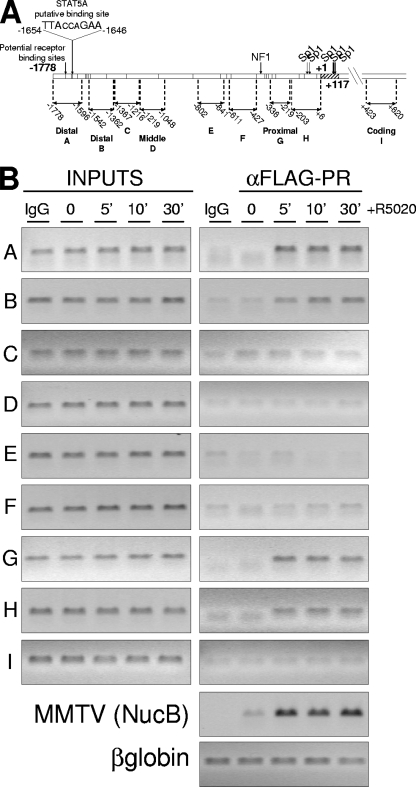
FIG. 3.
PR binds to two different regions of the 11β-HSD2 promoter after hormone activation. (A) Schematic representation of the 11β-HSD2 promoter region with the locations of amplicons for ChIP analysis as well as the location and sequence of the potential STAT5A binding site as predicted by ConSite and Transfac software. (B) TYML cells (T47D-YV cell line carrying an integrated copy of the progesterone reporter MMTV-Luc), expressing FLAG-tagged WT PRB, were cultured as described for Fig. 1A, untreated (0) or treated with R5020 (10 nM) for 5, 10, or 30 min, harvested, and used for ChIP experiments with anti-FLAG antibody. The precipitated DNA fragments were subjected to PCR with primers for the indicated 11β-HSD2 promoter regions or the β-globin gene as a control. The MMTV nucleosome B region was also amplified to detect PR recruitment as a control for the experiment. Amplification of input DNA (representing 1% of immunoprecipitated material) is shown for comparison. PCR products were run in a 1.2% agarose gel and visualized with ethidium bromide.
In accordance with the involvement of the −1778/−1345 region described above, ChIP analysis showed that, in response to hormone activation, PR was recruited to this distal region, represented by amplicons A and B (Fig. 3B). Similarly, upon hormone addition, PR was recruited to the proximal −368/+1 promoter region (amplicons G and H) (Fig. 3B). However, the promoter deletion analysis showed limited induction by R5020 in this region (Table 1). PR recruitment was not observed in the middle amplicons or in a coding region (+423/+620). As controls, in the same experiment we confirmed PR recruitment to the well-studied MMTV promoter after progestin treatment and no recruitment to the β-globin gene. These results demonstrate that R5020 treatment induces rapid and stable (up to 30 min) PR recruitment to the distal and the proximal regions of the 11β-HSD2 promoter. This finding was confirmed for T47D cells expressing endogenous PR by means of a PR-specific antibody (data not shown).
Mutations in the DBD of the receptor affect PR recruitment to the proximal promoter but not endogenous 11β-HSD2 expression.
We next investigated the mechanisms involved in hormone-dependent PR recruitment to the two 11β-HSD2 promoter regions. The classical mode of action of PR on gene expression involves direct binding of PR to the HREs in the target promoter. This is the case of the MMTV promoter which contains five HREs in a region covered by a regulatory nucleosome (16). The first zinc finger of the receptor DBD is responsible for the direct contact with DNA via its so-called P box (8). Point mutations in this zinc finger have been shown to interfere with PR binding to target promoters (9, 57). We have used a TYML-derived cell line expressing FLAG-tagged PRB mutated at residues G584, S585, and V589 (PRB-mDBD) of the P box of the zinc finger (unpublished results) to study whether PR recruitment to the 11β-HSD2 promoter involves direct DNA binding. As we expected, PRB-mDBD recruitment to the MMTV promoter at the integrated MMTV-Luc transgene was impaired (Fig. 4A; also see Fig. 8C). Accordingly, endogenous MMTV-Luc transcriptional activation was abrogated by the DBD mutation (Fig. 4B).
ChIP analysis of progestin-induced PR recruitment showed that mutation in the DBD of PRB did not affect its recruitment to the distal 11β-HSD2 promoter region upon hormone treatment but completely abrogated recruitment to the proximal region (Fig. 4A; also see Fig. 8C). These results indicated that direct contact of PRB with DNA is required for association with the proximal region only.
To explore whether the inability of PRB-mDBD to bind to the proximal 11β-HSD2 promoter affects promoter activation, we checked endogenous 11β-HSD2 mRNA expression after R5020 treatment in this cell line by RT-PCR. In contrast to MMTV expression, 11β-HSD2 expression in TYML cells expressing PRB-mDBD was activated similarly to that in WT-PRB-expressing cells (Fig. 4B and C). This is in accordance with the deletion analysis, indicating that PR binding to the proximal region is not essential for the hormonal induction of 11β-HSD2 gene expression, whereas the distal region plays an essential role.
Inhibition of JAK/STAT pathway activation compromises hormone induction of 11β-HSD2.
We next explored the mechanism by which PR is recruited to the distal promoter region and induces its activation. Steroid receptors can also be recruited to promoters indirectly, through interaction with other transcriptional factors. A sequence analysis of the distal region of the 11β-HSD2 promoter revealed the presence of a putative STAT5A binding site at nucleotides −1654/−1646 (Fig. 3A), close to the consensus STAT5A sequence TTC/ANNNG/TAA (18). STAT transcription factors bind to target promoters to alter gene expression upon phosphorylation by JAK-2, dimerization, and translocation to the nucleus (28, 29). JAK-2 is activated by a number of membrane-associated, phosphotyrosine-dependent receptors. A recent work with breast cancer cells has described that progestin also induces activation of the JAK/STAT pathway, STAT3 phosphorylation, association of PR with STAT3, and translocation to the nucleus (47). Other reports have shown progestin-induced nuclear translocation of STAT5A in T47D cells (49) or progestin-PR-dependent recruitment of STAT5A to the β-casein promoter (13). Nonetheless, these reports have failed to detect progestin-stimulated tyrosine phosphorylation of total cellular STAT5A and suggested that only a small proportion of total STAT5A may get phosphorylated and may intervene in specific promoter recruitment (13). In agreement with this, we have been able to slightly detect phosphorylation of STAT5A shortly after R5020 treatment of serum-starved T47D cells by immunoblotting with a specific antibody (Fig. 5A). Prolactin was a stronger inducer of STAT5A phosphorylation. R5020-induced phosphorylation was blocked by preincubation with the specific JAK-2 inhibitor AG. Similarly, JAK-2 phosphorylation was detected by immunofluorescence shortly after R5020 treatment of T47D cells and blocked by AG preincubation (data not shown).
In order to test the possibility that STAT5A could be involved in PR recruitment to the distal promoter region and hormone-dependent 11β-HSD2 expression, we explored whether the JAK/STAT pathway was involved in 11β-HSD2 activation by R5020. First, we investigated by RT-PCR the effect of the specific JAK-2 inhibitor (AG at 50 μM) on endogenous 11β-HSD2 mRNA accumulation upon hormone treatment (Fig. 5B). Normal induction of 11β-HSD2 expression was abrogated in the presence of the JAK inhibitor. The integrated MMTV-Luc reporter, analyzed for comparison, was normally activated after addition of R5020 to AG-treated cells (Fig. 5B). These results indicate that JAK/STAT pathway activation has an important and specific role in 11β-HSD2 hormone-induced expression.
Other signaling pathways have been reported to be rapidly activated by progestin and to mediate their effects, including cell proliferation of breast cancer cells (5, 11, 42, 51). This includes PI3K and mitogen-activated protein kinase (MAPK) pathways. Hormonal activation of the MMTV promoter depends not only on PR interaction with several HREs but also on the ERα/c-Src/Ras/Erk pathway (62). Inhibitors of ERα or MEK1 interfere with MMTV activation. In order to test whether these pathways are also essential for 11β-HSD2 activation by progestin, we measured 11β-HSD2 transcript accumulation after R5020 treatment alone or in the presence of an ERα antagonist (ICI), an MEK1 inhibitor (PD), or an inhibitor of the PI3K pathway (WM). We observed small and transient effects with the inhibitors of MEK1 and PI3K on 11β-HSD2 induction and no effect with ICI (Fig. 5C). This indicates that the activation of the ERα/c-Src/Ras/Erk and PI3K pathways is not by any means as important as the activation of JAK/STAT on 11β-HSD2 expression, in contrast to what has been reported for the MMTV promoter.
The effect of AG treatment on the hormone response of a transfected 11β-HSD2-Luc full-length construct was also tested. Induction of Luc activity by 16 h of R5020 treatment was abolished when cells were pretreated with AG for 1 h (Fig. 5D).
JAK/STAT pathway activation by progestin involves c-Src tyrosine kinase activation that then phosphorylates JAK-2 (47). PR directly contacts the c-Src SH3 domain through a proline (Pro) cluster located at the inhibition function domain of PR. A PR mutant on this Pro cluster abrogates this contact and c-Src activation by progestin (11). We tested the induction of the 11β-HSD2-Luc construct in the presence of WT and Pro cluster mutant (PR-mPro) PR-expressing vectors. The mutant showed reduced activation of the 11β-HSD2 promoter upon progestin treatment (Fig. 5E). In parallel, MMTV-Luc induction was unaffected by the Pro cluster defect (data not shown). Alternatively, in certain contexts c-Src is activated by ERα upon progestin treatment, through an interaction of PR with ERα (5). When we performed the experiment using a PR mutant unable to interact with ERα (PR-ΔERID I), progestin induction of 11β-HSD2-Luc was not affected (data not shown). Taken together, the data confirm the involvement of JAK/STAT pathway activation in progestin-induced 11β-HSD2 expression.
JAK-2 inhibition compromises hormone induction of other progesterone target genes.
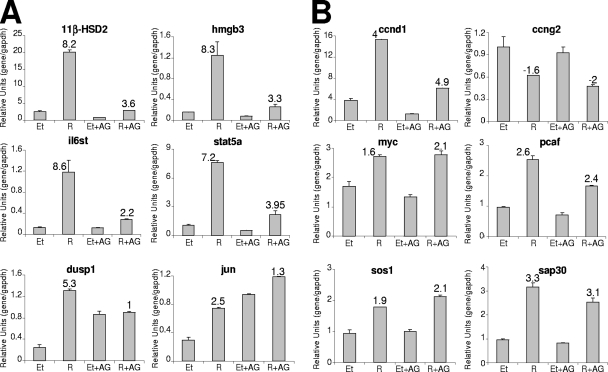
We next considered whether hormone induction of other target genes is dependent on activation of the JAK/STAT pathway. With this purpose, we used a breast cancer microarray platform containing 826 cDNA clones (C. Ballaré, B. Miñana, and M. Beato, unpublished results). Analysis of T47D cells treated with R5020 or vehicle for 6 h and pretreated or not with AG for 1 h pinpointed a number of genes regulated by progestin and affected by JAK/STAT pathway inhibition (see the supplemental material). Nineteen genes showed ≥2-fold inductions in response to R5020 compared to the vehicle level in the absence of AG. Among them, only two (Dusp1 and Il6st) showed more than 50% reduction in hormone response in the presence of the inhibitor, and six genes had lost 30 to 50% of the response (Dnah1, Jun, Hmgb3, Muc2L, Cyr61, and Atf3). Figure 6A and B show examples of progestin-regulated genes affected or not, respectively, by AG pretreatment, validated by RT/real-time PCR. Progestin activation of Dusp1, Il6st, Jun, Hmgb3, and Stat5A genes was affected by AG to an extent comparable to that of 11β-HSD2 inhibition. On the other hand, hormone responsiveness of Ccnd1, Ccng2, Myc, Pcaf, Sos1, and Sap30 was not significantly affected, despite the fact that basal expression levels are, in some cases, altered by this JAK/STAT inhibitor.
FIG. 6.
Involvement of JAK/STAT pathway activation in progestin-induced gene expression. T47D cells, cultured as described for Fig. 1A, were treated for 6 h with R5020 (10 nM) or ethanol (Et), and when indicated, AG was added 1 h before hormone. Cells were harvested, and total RNA was extracted. Expression of the genes indicated was analyzed by RT and real-time PCR with specific primers. GAPDH cDNA-specific primers were used as a control. The values represent the means and ranges of a representative experiment performed in duplicate, expressed as numbers of specific gene/GAPDH relative units. Inductions in response to hormone are indicated.
STAT5A is functionally important for hormone-dependent 11β-HSD2 expression and exerts its action through the distal promoter region.
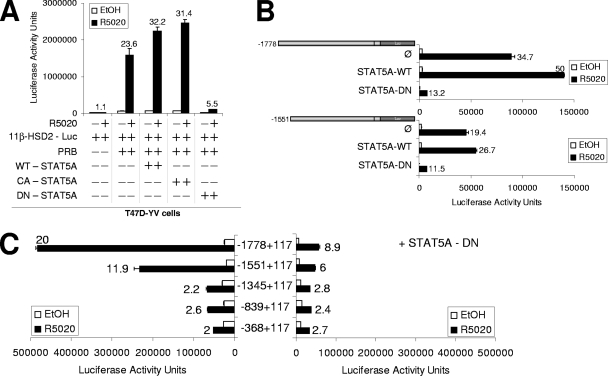
The involvement of the JAK/STAT pathway in 11β-HSD2 expression suggested that the predicted STAT5A site found at the distal region could be important. In order to analyze the functional involvement of STAT5A on progestin-induced 11β-HSD2 gene expression, we took advantage of existing DN or CA STAT5A mutants. The DN form consists of a deletion in the C-terminal transactivation domain that still binds to DNA upon activation but is unable to interact with many coregulators or to induce transcription (43). The CA form contains mutations that mimic stable Tyr phosphorylation, leading to nuclear accumulation, DNA binding, and transactivation activity (4). T47D-YV cells were cotransfected with the full-length 11β-HSD2-Luc reporter construct, PRB expression vector, and WT, CA, or DN STAT5A expression vectors, and the response to R5020 was measured (Fig. 7A). 11β-HSD2-driven Luc activity depended on PRB expression and hormone. Overexpression of WT and CA STAT5A modestly enhanced the hormone response of the promoter compared to that of endogenous STAT5A. Importantly, DN STAT5A impaired the response to the progestin, confirming that STAT5A plays an important role in the hormone activation of the 11β-HSD2 promoter. In addition, the DN form also reduced the basal promoter activity (Fig. 7A).
FIG. 7.
Expression of DN STAT5A affects the hormone activation of the 11β-HSD2 promoter. (A) T47D-YV cells were cotransfected when indicated with 1 μg of 11β-HSD2-Luc (−1778/+117) reporter vector, 1 μg of pSG5-PRB, and 1 μg of WT STAT5A, CA STAT5A, or DN STAT5A. Afterwards, cells were treated with ethanol (EtOH) or R5020 (10 nM) for 16 h and Luc activity was measured. For normalization, equal amounts of cellular extract of each sample were used. The values represent the mean numbers of arbitrary Luc units and ranges of a representative experiment performed in duplicate. Induction in response to hormone is shown for each construct. (B, C) T47D-YV (B) or T47D (C) cells cotransfected with 1 μg of the indicated 11β-HSD2 promoter constructs, 1 μg of pSG5-PRB, and 1 μg of plasmid expressing WT STAT5A or DN STAT5A were treated with ethanol or R5020 (10 nM) for 16 h, and Luc activity was measured. Induction in response to hormone is shown for each construct. The values represent the means and ranges of a representative experiment performed in duplicate.
In a different experiment, we compared the effects of WT and DN STAT5A on the full-length (−1778/+117) and −1551/+117 (lacking the identified well-conserved STAT5A binding site) 11β-HSD2-Luc constructs. Interestingly, both constructs were similarly affected by the STAT5A forms (Fig. 7B). The fact that activity of the −1551 deletion is also enhanced by WT STAT5A and abolished by DN STAT5A indicates that STAT5A exerts its function not only through the predicted STAT5A binding site but also through an unknown sequence located downstream.
To test whether the reduced activity of the proximal promoter constructs (down to −368/+117) identified in transient transfection experiments on T47D cells overexpressing PRB (Table 1) depended on STAT5A activation, we performed a similar experiment, cotransfecting or not cotransfecting DN STAT5A (Fig. 7C). As expected, DN STAT5A affected the hormone induction of the long constructs but not the shortest ones that show the same reduced hormone response irrespective of STAT5A. This indicates that residual activation mediated through the proximal promoter region is not dependent on STAT5A, in accordance with the finding that PRB binds there through its DBD (Fig. 4A).
STAT5A is recruited to the distal region of the 11β-HSD2 promoter in response to progestin.
We next investigated STAT5A recruitment to the 11β-HSD2 promoter in TYML cells expressing FLAG-tagged WT PRB, using ChIP experiments. Upon hormone addition, STAT5A rapidly associated to 11β-HSD2 distal promoter regions A and B (Fig. 8A). Irrespective of the hormone, no STAT5A bound the proximal or middle region of the 11β-HSD2 promoter. As a control, we confirmed in the same experiment that PR is recruited to both distal and proximal promoter regions (Fig. 8A). These results showed that STAT5A is recruited to the distal region of the 11β-HSD2 promoter in a hormone-dependent manner and with kinetics similar to that of PR. We expected recruitment to the predicted STAT5A binding site (amplicon A), but association was also found with amplicon B, in agreement with the previous data showing that DN STAT5A also affected the −1551 but not the −1345 deletion.
Hormone-dependent PR recruitment to the distal promoter depends on JAK/STAT pathway activation.
In order to test the possibility that STAT5A hormone-dependent recruitment to its binding site at the distal 11β-HSD2 promoter region could be involved in PR recruitment, we investigated the effect of blocking JAK/STAT pathway activation with AG on receptor recruitment. As expected, the inhibitor abrogated the observed STAT5A recruitment to the distal region (Fig. 8B). Significantly, in the presence of the JAK/STAT inhibitor, PR recruitment in response to R5020 to the distal 11β-HSD2 promoter region was diminished, while recruitment to the proximal region was unaltered (Fig. 8B). As a control, we tested if interfering with JAK/STAT activation would affect PR recruitment to another hormone-regulated promoter. ChIP experiments using MMTV nucleosome B-specific primers showed that PR is normally recruited in the presence of AG (Fig. 8B).
Because a residual fraction of PR associated with the distal promoter region in the presence of AG and consequently in the absence of STAT5A recruitment (Fig. 8B), we investigated whether this depended on direct interaction with DNA. When the AG effect in the cell line expressing PRB-mDBD was investigated, PR recruitment to the distal region (amplicons A and B) was found to be completely abrogated. This indicates that direct interaction of PR with DNA also plays some role in this region, although STAT5A-mediated recruitment seems to be the main mechanism (Fig. 8C, upper panel, compare lanes 4 to 6 with 10 to 12; real-time PCR quantification of amplicon A is shown in Fig. 8D). Interestingly, STAT5A recruitment was slightly reduced in the context of PRB-mDBD. All in all, this suggests a cross talk between the two mechanisms of PR recruitment converging into the distal region. STAT5A might be the driving force behind PR recruitment, but then, this might be stabilized by direct contacts of PR with DNA. A conformational change or chromatin remodeling might facilitate PR-DNA contacts.
To further investigate the potential involvement of PR binding to the distal 11β-HSD2 promoter region, evidenced in vivo only when the JAK/STAT pathway is blocked, we combined PRB-mDBD and DN STAT5A expression in Luc reporter transfection experiments (Fig. 8E). In this context, PRB-mDBD was not able to fully support the hormone response of −1778 and −1551 deletions, and combination of PRB-mDBD with DN STAT5A completely abrogated promoter expression. This suggests that both mechanisms, STAT5A-mediated and direct interaction of PRB with DNA, play roles in the context of transiently transfected 11β-HSD2 promoter constructs, this effect being mediated by a region located between −1551/−1345. The difference with the endogenous situation could be that progesterone response elements (PREs) are likely more exposed in the poorly chromatinized transfected templates. Later on, we noted that the negative effect of mDBD could also be observed when using the construct containing only the distal region (−1778/−1345) driving Luc expression (see below and Fig. 10C). Hormone induction was 2.7 times lower in PRB-mDBD-expressing cells than in WT PRB (data not shown). This experiment confirmed that functional putative HREs in transient reporter constructs are within the distal region, not provided by the proximal region.
FIG. 10.
The distal region works as a hormone-responsive RNAP II entry site from where RNA synthesis occurs and enhances 11β-HSD2 expression. (A) TYML cells expressing WT FLAG-tagged PRB, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 5 or 10 min, harvested, and used for ChIP experiments with anti-FLAG tag (αFLAG) and phospho(Ser5)-RNAP II (RNAP II S5p) antibodies. The precipitated DNA fragments were subjected to PCR analysis with specific primers corresponding to the indicated 11β-HSD2 promoter regions or MMTV nucleosome B and the β-globin gene as a control. On the right panel, anti-phospho-RNAP II ChIP was performed on chromatin extracted from TYML cells expressing FLAG-tagged WT PRB or PRB-mDBD, untreated or treated with R5020. (B) TYML cells expressing FLAG-tagged WT PRB, cultured as described for Fig. 1A, were treated with ethanol or R5020 (10 nM) for 6 h. When indicated, cells were pretreated with AG (50 μM). Cells were harvested, total RNA was prepared, and cDNA was generated by RT using oligo(dT). RNA synthesis was analyzed using primers that specifically amplified the 11β-HSD2 promoter distal region and the proximal region and primers located in exon 3 and exon 4 (RT oligos). GAPDH-specific primers were used as a control. (C) T47D-YV cells cotransfected with 1.5 μg of WT pSG5-PRB expression vector, 1.5 μg of the indicated 11β-HSD2 deletion constructs was treated with ethanol (EtOH) or R5020 (10 nM) for 16 h, and Luc activity was measured. For normalization, equal amounts of cellular extract of each sample were used. The values represent the mean numbers of arbitrary Luc units and ranges of a representative experiment performed in duplicate. Inductions in response to hormone are indicated. (D) TYML cells expressing FLAG-tagged WT PRB, cultured as described for Fig. 1A, were treated with EtOH or R5020 (10 nM) for 6 h. Then, cells were harvested, total RNA was prepared, and cDNA was generated by RT using gene-specific sense primers at positions −1936 and −1778 upstream of the TSS. The presence of antisense RNA upstream of the TSS was analyzed using PCR primers at positions −1936/−1779 (preA) and −1778/−1596 (distal A) that specifically amplified the promoter region.
PR/STAT5A cooperation for transcriptional activation of 11β-HSD2.
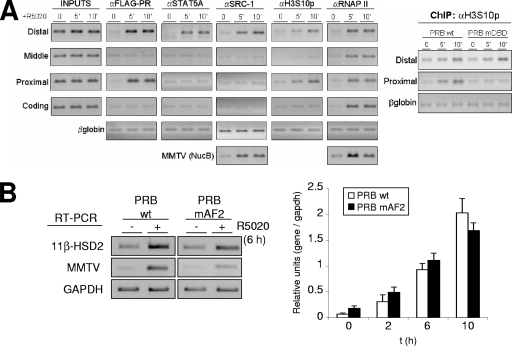
We have shown that 5 min after hormone addition, PR and STAT5A are recruited to the distal 11β-HSD2 promoter region in a process that depends on JAK/STAT pathway activation and that PR is also recruited to the proximal region, requiring an intact DBD and implying PR binding to HREs. We have further studied recruitment of transcription coregulators and the changes in posttranslational modifications at relevant histone residues. SRC-1 is a known coactivator of PR and STAT proteins with intrinsic histone acetyltransferase activity and the ability to recruit additional histone acetyltransferases. Ten minutes after hormone addition, we found SRC-1 only at the distal region (Fig. 9A).
FIG. 9.
Coactivator recruitment and histone modifications at the 11β-HSD2 promoter. (A) TYML cells expressing FLAG-tagged WT PRB, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 5 or 10 min, harvested, and used for ChIP experiments with anti-FLAG tag (αFLAG), STAT5A, SRC-1, H3S10p, and RNAP II antibodies. The precipitated DNA fragments were subjected to PCR analysis with specific primers corresponding to the indicated 11β-HSD2 promoter regions or MMTV nucleosome B and the β-globin gene as a control. For the right panel, ChIP with H3S10p antibody was performed on chromatin extracted from TYML cells expressing FLAG-tagged WT PRB or PRB-mDBD, untreated or treated with R5020. (B) TYML cells expressing WT PRB or PRB-mAF2, cultured as described for Fig. 1A, were untreated (0) or treated with R5020 (10 nM) for 2, 6, or 10 h. Cells were harvested, and total RNA was extracted. (Left) 11β-HSD2 and MMTV-Luc mRNA expression levels were analyzed by RT-PCR with specific primers. GAPDH cDNA-specific primers were used as a control. PCR products were run on a 1.2% agarose gel and visualized with ethidium bromide. (Right) 11β-HSD2 mRNA expression was analyzed by RT and real-time PCR with specific primers. GAPDH cDNA-specific primers were used as a control. The values represent the means and ranges of a representative experiment performed in duplicate, expressed as numbers of 11β-HSD2/GAPDH relative units.
In order to explore whether SRC-1 recruitment was mediated by PR or by STAT5A, we have used a TYML cell line stably expressing a mutant PRB with a single amino acid exchange, E911A, at activation function 2 (PRB-mAF2) (25), as AF2 is involved in coactivator recruitment (24, 38). The AF2 mutant impairs progestin response of MMTV in comparison to the WT, despite normal recruitment of the mutated receptor (data not shown). In contrast, the 11β-HSD2 promoter is normally activated by the AF2 mutant (Fig. 9B). These results suggest that coactivators such as SRC-1 are being recruited by STAT5A, and PR is not contributing with its transactivation functions.
Histone H3 phosphorylation at Ser10 (H3S10p) has been associated with immediate-early gene activation by diverse stimuli and with MMTV induction by R5020 (58, 62). We have also observed phosphorylation of H3S10 upon hormone addition, localized to the two regions where PR is recruited (Fig. 9A). Mutation at DBD that abrogates PR recruitment to the proximal region also abolished H3 Ser10 phosphorylation in this region without affecting the distal region (Fig. 9A, right panel).
We conclude that at the 11β-HSD2 promoter, whereas an H3S10 kinase is recruited by PR, coactivators such as SRC-1 are recruited solely by activated STAT5A.
RNAP II binds the distal region and tracks to the proximal promoter.
Binding of PR to two distant promoter regions of the 11β-HSD2 promoter could cause or reflect the formation of a chromatin loop, bringing factors recruited by the PR-STA5A association to the basal transcriptional machinery that may initiate mRNA synthesis at the TSS. Against this loop formation hypothesis is the fact that some factors (mainly STAT5A) are not detected at the proximal region. To further clarify the mechanism by which PR-STAT5A binding to the distal region activates transcription about 1.6 kb downstream, we performed ChIP to detect RNAP II. Surprisingly, RNAP II was found to associate after 5 min of hormone treatment not only to the proximal region but also to the distal and middle regions (Fig. 9A). In order to check whether RNAP II is in its active conformation along the 11β-HSD2 promoter upon hormone treatment, we used the H14 RNAP II antibody, specifically detecting the phosphorylated form at Ser5 of its carboxyl-terminal domain (45). This antibody indicated that RNAP II engaged in, at least, preinitiation or initiation is already present at the distal region and all along the promoter shortly after hormone addition (Fig. 10A). Moreover, cells expressing the PR DBD mutant show equivalent loadings of active RNAP II to the proximal region, where PR-mDBD cannot be recruited (Fig. 4A), indicating that PR is not involved in recruiting the polymerase to the proximal region (Fig. 10A, right panel).
These results indicate that a processing polymerase is loaded at the distal region, coinciding with PR-STAT5A recruitment upon hormone treatment, and probably tracks toward the proximal promoter and initiation site. In order to test the possibility that RNA is being synthesized upstream of the TSS of 11β-HSD2 identified in several tissues (2), we have performed RT-PCR with oligonucleotide pairs covering the distal and proximal promoter regions on T47D cells treated or not with hormone and AG. Hormone-dependent, AG-sensitive transcription is detected upstream of the reported TSS (Fig. 10B).
Our deletion analysis described above indicated that the distal region was required for most of the hormone response of the 11β-HSD2 promoter and the proximal region retained little responsiveness. Further constructs were prepared to explore the possibility that the distal region acted as a polymerase entry site and were cotransfected with PRB into T47D cells (Fig. 10C). First, an internal deletion, Δ−1345/−368, showed the same hormone response as the full-length promoter, indicating that the sequences between the distal and proximal regions are devoid of regulatory elements, and the distance between distal and proximal regions is not relevant. Noteworthy is the fact that the distal −1778/−1345 region alone also conserved full capacity to drive expression of the reporter gene and was normally induced by hormone (Fig. 10C, upper panel). This could indicate that this region, in fact, contained a promoter, but data could also fit with the presence of a transcriptional enhancer. Gene activation is a property of enhancers, which are defined by their ability to direct high-level expression of linked genes in transient transfection assays. Enhancer function includes not only long-distance but also orientation-independent transcriptional activation. In order to test whether the distal region acts as an enhancer or is a promoter by itself, we inverted this region in the two previously reported constructs and analyzed their hormone responses (Fig. 10C, lower panel). The inverted constructs were as active on Luc expression as the sense constructs, confirming that the distal region is not a promoter by itself but an entry site for the transcriptional machinery, probably at multiple, weakly defined sites, that then tracks in the two directions.
In order to test whether antisense transcription occurred from RNAP II entry sites at the endogenous promoter region, we performed RT reactions with specific sense oligonucleotides matching positions −1936 and −1778 upstream of the TSS, followed by PCR amplification with specific oligonucleotide pairs covering several promoter regions (Fig. 10D). RNA extracted from T47D cells treated or not with R5020 for 6 h was used. Amplification was obtained from the hormone-treated cells, indicating that, concomitant with STAT5A, PR, and RNAP II recruitment, transcripts covering the promoter region were synthesized not only from the positive strand (Fig. 10B and data not shown) but also from the negative strand (Fig. 10D).
In conclusion, our data show that the distal region works as an enhancer, where STAT5A and PR recruitment brings the transcriptional machinery that generates upstream RNAs coexpressed with the main 11β-HSD2 transcript. This high density of RNAP II tracks along the promoter and may initiate 11β-HSD2 expression at the reported start site.
DISCUSSION
The aim of this work was to investigate the mechanisms involved in gene expression activation by progesterone and whether the two modes of action of the hormone receptor (i.e., acting as a transcription factor and interacting with cytoplasmic signaling pathways) converge to regulate target promoters. The MMTV promoter contained in its 5′ long terminal repeat has been extensively studied and has become the model for progesterone- and glucocorticoid-induced gene expression in human breast cancer cell lines (17, 63). Using this model, we have recently contributed to connecting the rapid signaling activation by progesterone with its transcriptional effect (62). In this work, we have explored the activation of the endogenous 11β-HSD2 promoter, one of the strongest progestin and glucocorticoid-induced genes in breast cancer cells, in order to extend the understanding of the hormone receptors' function. In summary, our results show that PR is recruited to two different regions of the 11β-HSD2 promoter shortly after progestin treatment of serum-starved breast cancer cells. Recruitment to a distal region is essential for promoter response and involves JAK/STAT pathway activation and STAT5A promoter binding. PR association with a proximal region involves direct interaction with DNA, but this is not required for 11β-HSD2 gene expression upon hormone treatment.
JAK/STAT pathway activation by progestin is required for 11β-HSD2 gene expression.
We found that progestin activation of the JAK/STAT pathway plays an essential role in PR recruitment to the promoter, again suggesting that the direct transcriptional action of the receptor requires earlier events initiated by the ability of the ligand-bound receptor to interact with and activate cytoplasmic kinases engaged in intracellular signaling. We investigated the participation of the JAK/STAT pathway in hormone-induced 11β-HSD2 expression after identifying a putative STAT5A binding site in the distal promoter region. The JAK/STAT pathway induction by progestin leads to STAT5A activation and corecruitment together with PR to the distal enhancer region, which we have shown to be the relevant region of the promoter for hormone response. Interfering with JAK/STAT activation with AG blocks hormonal induction of 11β-HSD2 expression, STAT5A recruitment, and PR association with the enhancer. However, none of this occurred in the case of the MMTV promoter, indicating that JAK/STAT activation by progestin is involved in 11β-HSD2 but not in MMTV activation.
On the other hand, STAT5A activation is not sufficient, as CA STAT5A is unable to increase 11β-HSD2 expression in the absence of hormone. This fact indicates that PR also has a transcriptional role, presumably in the recruitment of histone-modifying enzymes or chromatin-remodeling complexes. In accordance with this, prolactin, a strong inducer of JAK/STAT activation and STA5A phosphorylation (as shown by phosphoimmunoblotting) does not activate the expression of 11β-HSD2 (data not shown).
Activation of the JAK/STAT pathway by progestin requires c-Src tyrosine kinase activation (47). It has been proposed that c-Src activation by progestin either occurs by direct contact between a Pro cluster at the PR inhibition function domain and the SH3 domain of c-Src (11) or is mediated by an interaction between the ER-interacting domains (ERIDs) of PR and the ligand-binding domain of ERα, which then interacts with the SH2 domain of c-Src (5, 42). Deletion of PR ERIDs abrogates progestin activation of Erk and induction of an integrated MMTV promoter in T47D cells (5, 62). Progestin induction of the transfected 11β-HSD2-Luc construct, shown to depend also on JAK/STAT pathway activation, was reduced when a PR mutant on the Pro cluster was coexpressed. In the presence of an ERID I-deleted PR, hormone induction was normal. This supports the involvement of direct c-Src activation by PR on JAK and STAT5A activation and 11β-HSD2 induction. Nonetheless, we cannot rule out a hypothetical involvement of the ERIDs and PR-ER interaction in another step of the induction process, if the promoter was immersed in chromatin. In this vein, an ERID I-deleted PR supports MMTV activation when the promoter is transiently transfected, but not in chromatin, due to the role of the receptor in the PR/ERα/c-Src/Ras/Erk/Msk pathway (5, 62).
We used the JAK inhibitor AG to test whether the JAK/STAT pathway activation was required for the hormone response of other progestin target genes and found that only a small proportion of R5020-responsive genes decreased their response. This indicates that, although 11β-HSD2 is not a unique case, this pathway is not generally involved in progestin-induced gene expression in breast cancer cells. Interestingly, the hormone responses of some genes relevant to growth control, such as Jun and Stat5A, are affected by the JAK/STAT inhibitor. Interestingly, the JAK/STAT pathway activation may be of relevance for breast cancer progression, as blockage of STAT3 activation by a DN form resulted in inhibition of in vivo breast tumor growth in an immunocompetent mouse model (47).
Although our data suggest the JAK/STAT pathway activation by progestin to be involved in the induction of specific promoters, detection of progestin-stimulated tyrosine phosphorylation of total cellular STAT5A by immunoblotting with different available Pho-STAT5 antibodies was challenging (Fig. 5A). This supports the hypothesis that progestin might stimulate the phosphorylation of a small fraction of cellular STAT5A and that phospho-STAT5A would be recruited to specific promoters, including β-casein (13) and 11β-HSD2.
Involvement of other cytoplasmatic signaling pathways in 11β-HSD2 expression.
Other signaling pathways have been reported to be rapidly activated by progestin and mediate their effects, including cell proliferation of breast cancer cells (6, 22). Hormonal activation of the MMTV promoter depends not only on PR interaction with several HREs but also on the hormone-activated ERα/c-Src/Ras/Erk pathway (62). Inhibitors of ERα, MEK1, or Msk1 interfere with MMTV activation. The PI3K pathway has also been reported to be activated by hormone treatment. In particular, ligand-bound ERα interacts with the regulatory subunit of PI3K (p85) and activates the PI3K/Akt/Ras/Erk pathway (15). By using specific inhibitors, we have described a minor participation of MAPK (MEK1) and PI3K in endogenous 11β-HSD2 induction. These or other pathways may participate in activating the kinases responsible for phosphorylation of PR or other transcription factors involved. For the MMTV, MAPK activation was involved in PR phosphorylation, Msk1 activation, and H3 Ser10 phosphorylation at the promoter, and this was essential for promoter induction (62). In 11β-HSD2, this process seems to be unessential attending at the low effect of MEK1 inhibition. Nonetheless, we have also observed histone H3 S10 phosphorylation concomitant to PR recruitment at the 11β-HSD2 promoter upon hormone addition. We therefore cannot discard the possibility that Msk1 and H3 Ser10 phosphorylation could also play an important role in 11β-HSD2 expression, via a signaling pathway other than MAPK. Further experiments are required to fully understand the involvement of these progestin-activated signaling pathways.
PR binds to two separate regions of the 11β-HSD2 promoter, but only binding to the distal enhancer depends on STAT5A and is functionally relevant.
We have found that PR is recruited to two different regions of the 11β-HSD2 promoter as early as 5 min after hormone addition. PR recruitment to the distal region was monitored with PCR amplicons covering nucleotide positions −1778 to −1362, whereas PR association with the proximal region was detected with amplicons covering positions −336 to +6. PR recruitment to the distal region occurred in parallel to STAT5A recruitment. Interfering with JAK/STAT pathway activation impaired PR recruitment, indicating that STAT5A binding to its binding site may bring PR to the distal region. Direct association of PR with DNA at this region was discarded for two reasons. First, a DBD mutant of PR associated with the distal region similarly to WT PR upon hormone treatment. Second, cells expressing this DBD mutant induced endogenous 11β-HSD2 gene expression normally. On the other hand, PR recruitment to the proximal promoter region depended on an intact DBD and was unaffected by inhibition of the JAK/STAT pathway, suggesting that this region does not contribute significantly to the hormone response of endogenous 11β-HSD2.
In agreement with this, the deletion analysis of the promoter with Luc reporter constructs showed that the proximal region is hormone unresponsive. It retains minor hormone responsiveness only when PRB is transiently overexpressed in PRA/PRB-expressing T47D cells. In contrast, the distal region is strictly required for strong hormone response. Interestingly, deletion at the intermediate −1551 position partially affected induction, indicating that regulatory elements at the two sides of this position may contribute to the transcriptional hormone response. Coexpression of a DN form of STAT5A greatly impaired reporter gene activity not only from the full-length promoter but also from the −1551 construct, indicating that STAT5A binding may not be limited only to the −1654/−1646 site, where a canonical STAT5A binding site was predicted. Additional cryptic STAT5A binding sites may exist after position −1551. This is in accordance with our ChIP data showing the STAT5A recruitment observed when an amplicon covering positions −1542 to −1362 was used. We discard the notion that this is due to the sizes of sonicated chromatin fragments since, for the experiment shown in Fig. 8C, fragments had an average size of 200 bp (data not shown). It also explains why, with JAK/STAT activation being so crucial for 11β-HSD2 expression, the −1551 construct retains considerable hormone responsiveness.
Expression of DN STAT5A also showed that residual activation mediated through the proximal promoter region is not dependent on STAT5A. This is in agreement with the findings that PRB binds there through its DBD, that no STAT5A was detected, and that PRB binding is insensitive to AG addition.
Further analysis of promoter reporter constructs revealed that the distal region is not only required for progestin-induced promoter expression but also sufficient in the absence of the proximal region in the context of transiently transfected templates. This was unexpected, as we assumed that this distal region worked as an enhancer of the basal promoter located at the proximal region and could indicate that the distal region may have full promoter activity by itself. Alternatively, it has been previously reported that enhancers may direct high-level expression of linked reporter genes in transient transfection assays. A prostate-specific antigen (PSA) reporter construct containing only the enhancer was as active to androgens as the full-length promoter (66). Also, the HS2 enhancer of the β-globin locus control region initiates synthesis of noncoding RNAs (nc-RNAs) autonomously, independent of a cis-linked promoter (40). The human growth hormone promoter is activated by a distal locus control region which creates a domain of bidirectional noncoding transcription (26).
Given that enhancers may drive orientation-independent transcriptional activation by loading RNA polymerases at multiple sites that then track in the two directions and that promoters work in only one orientation, we constructed reporter constructs containing the 11β-HSD2 distal region in the antisense direction with respect to the Luc gene. The results showed that the distal region was equally active to drive hormone-dependent expression of the reporter when placed in the antisense direction, suggesting that it was acting as an enhancer instead of as a promoter.
In the absence of STAT5A, residual PR binds through DBD to the distal region.
As already mentioned, expression of a DBD mutant of PR did not impair receptor and STAT5A recruitment to the distal region. In the presence of the JAK/STAT inhibitor AG, STAT5A was not recruited, but a minor proportion of PR still associated to the distal region. Combining AG with the DBD mutant showed that in the absence of STAT5A recruitment, there was some DBD-dependent association of PR with the distal promoter. This indicates that the distal promoter region may contain potential PR binding sites (HREs) that are not used when STAT5A can be activated to recruit PR. Again, normal induction of endogenous 11β-HSD2 in the presence of the PR DBD mutant supported this model.
On the other hand, analysis of transiently transfected 11β-HSD2 reporter constructs demonstrated that hormone response was lower when PR-mDBD was coexpressed than with WT PR. We speculate that this might be due to the poor chromatinization of the transfected promoter, which might leave potential HREs exposed, allowing direct binding of PR to DNA and contributing to the hormonal response observed. The remaining activity of the constructs seems to be due to STAT5A-mediated PR recruitment, as combining PR-mDBD with DN STAT5A completely abrogates their activation. This effect was also seen with the −1551 deletion, indicating that PR and STAT5A may contact the −1551/−1345 region. Involvement of the putative STAT5A binding site identified in −1778/−1551 (amplicon A) comes only from ChIP data at this point.
Given these data, we can talk about convergence of two mechanisms for PR recruitment to the 11β-HSD2 promoter distal region, mediated by STAT5A-contacting DNA and direct association of PR with HREs. The first one seems to be the principal driving force behind PR transcriptional activity, and the second is apparent only when STAT5A activation is impaired or in transiently transfected templates. We cannot discard the possibility of a cross talk between the two mechanisms of PR recruitment converging to the distal region. PR might be recruited principally by STAT5A, but then, this might be stabilized by direct contacts of PR with DNA. A conformational change or a chromatin remodeling may also facilitate the PR-DNA contacts. Although we have proven that PR is also recruited to HREs at the proximal promoter, we have not found this to be functionally important. Further identification and mutagenesis of participating HREs will help clarify whether PR binding to DNA plays any as-yet-unrecognized role.
RNAP II is recruited to the enhancer region and tracks across the promoter, producing upstream RNAs.
As early as 5 min after hormone addition to serum-free cultured cells, both PR and STAT5A got recruited to the distal enhancer region of the 11β-HSD2 promoter and only PR to the proximal region. Concomitantly, RNAP II showed association with all tested regions of the promoter and the coding region. ChIP using an antibody against phosphorylated RNAP II indicated that the initially recruited enzyme was activated at Ser5 a few minutes later, coinciding with other changes occurring at the promoter. Histone H4 became acetylated, and H3 methylated at Lys4 along the promoter (data not shown). In contrast, H3 was phosphorylated at Ser10 only in the regions where PR was recruited. This resembles the recently described situation with the MMTV promoter where a PR/Erk/Msk complex is recruited to the regulatory nucleosome upon hormone stimulation and phosphorylates H3S10 locally to start a chromatin-remodeling cascade that facilitates transcription factor loading and leads to transcription initiation (62). Accordingly, a mutation at DBD that abrogated PR recruitment to the proximal region showed no H3 Ser10 phosphorylation in this region without affection of the distal region. SRC-1, a known STAT and PR coactivator, is recruited only to the distal region, probably reflecting the fact that it is the PR-associated complex recruited to the distal region that is functionally important.
The inability of PR binding to the proximal region to recruit SRC-1 may explain why this PR interaction is not required for functional response. It might also reflect an inappropriate receptor conformation due to promoter context effects, allosteric effects of DNA sequence and architecture (39), or the reported inability of PR binding to a single PRE to efficiently recruit SRC-1 or a full complement of transcription factors (60). No palindromic PRE and only few PRE half-sites are predicted at the proximal 11β-HSD2 promoter region (data not shown).
Several models for explaining how a distal enhancer communicates with a proximal promoter have been proposed: looping, scanning, tracking, and linking (10, 12, 35, 46). Several observations lead us to discard a pure looping mechanism. Active RNAP II is observed not only in the distal and proximal regions but also in the middle region, indicating that some sort of scanning or tracking may take place. In addition, some factors are only found at the distal region (STAT5A and SRC-1). PR would be the hypothetical candidate for the establishment of a loop, as it has contacts with the two distant regions, but in that case, PR-mDBD would interfere with loop formation and affect gene activation, something we do not observe. Moreover, cells expressing PR-mDBD show equivalent loading of active RNAP II to the proximal region, indicating that the enzyme may be tracking from the entry site in the distal region.
From our data, we can speculate that RNAP II and some chromatin-modifying enzymes (responsible for H4 acetylation and H3K4 methylation) track (or scan) along the promoter without carrying PR, STAT5A, or SRC-1. We have also tested whether active RNAP II tracking involves RNA synthesis. We have detected by RT-PCR the synthesis of hormone-dependent, AG-sensitive, polyadenylated upstream transcripts covering the distal and proximal regions. Moreover, by using specific sense and antisense RT oligonucleotides, we have detected bidirectional, hormone-dependent transcripts covering the promoter region. This is in accordance with the observed capacity of the distal promoter region to direct expression of a reporter gene in response to hormones, independently of directionality, in transfection experiments. It will be interesting to further explore the length and coverage of the upstream RNA molecules generated as well as whether they play a specific role in transcription initiation from the promoter or if they are a by-product of RNAP II activity. Based on previous reports, we speculate that RNA polymerase tracking may involve upstream nc-RNA synthesis (in one or the two directions), maybe from poorly defined start sites at the enhancer, to deliver the machinery to the proximal promoter, where transcription starts consistently at a defined +1 site.
Many enhancers and promoters are being transcribed. nc-RNA transcription is a major regulatory mechanism of long-range control elements (33). Recently, novel classes of nc-RNAs that offer cis- and trans-regulatory potential have been identified, some of which correlate with the expression state of protein-coding genes they associate with (32). Analysis of functional elements in 1% of the human genome by the ENCODE consortium has concluded that the genome is massively transcribed, probably from the multitude of new TSSs identified, and many new nc-RNAs exist (23). Nevertheless, these studies do not distinguish between the importance of transcription per se and the production of functional RNA products. One consequence could be a better binding of transcription factors, as zinc finger proteins show better binding involving RNA. Interestingly, analysis of publically accessible RNA maps reported by Kapranov et al. (32). has revealed the existence of a high density of a new class of nc-RNAs (≥200 nucleotides, nuclear and cytosolic polyadenylated) originating from a region extending between the 11β-HSD2 TSS and approximately 2.5 kb upstream, expressed in all cell lines analyzed (data not shown). Additionally, the existence of bidirectional expressed sequence tags overlapping this promoter region can also be found in databases. Taken together, these data support the observed capability of this promoter region to generate new nc-RNAs coexpressed with the 11β-HSD2 coding region with potential, still-not-understood regulatory functions. Detailed analysis of this region would probably uncover previously unrecognized TSSs, as has been reported in the ENCODE project (21, 23). In this vein, several examples of noncoding transcription near to or overlapping a promoter with major or minor influence on its activity have been recently reported (26, 35, 40, 61).
RNAP II tracking has been also proposed as a requirement for activation of the PSA promoter by androgens (66). An androgen receptor-associated coactivator complex is predominantly recruited to the PSA enhancer, which communicates with the androgen receptor transcription complex weakly associated with the PSA promoter through the 4 kb of intervening DNA through which RNAP II tracks. Similarly, integration of prolactin and glucocorticoid signaling at the β-casein promoter involves STAT5A and GR recruitment both to the proximal promoter and to a distal enhancer and RNAP II tracking (31, 55). Interestingly, STAT5A is recruited to a STAT binding site and GR acts as a STAT coactivator. Whether RNAP II tracking generated nc-RNAs was not analyzed. These examples share striking similarities with the 11β-HSD2 model proposed here.
In summary, we describe a mechanism of PR-mediated gene activation, using the human 11β-HSD2 promoter as a model. Progestin activation of the JAK/STAT pathway and STAT5A-mediated recruitment of the receptor to a distal enhancer are essential to recruit active RNAP II that then may track toward the proximal promoter. The nature and role of the upstream synthesized nc-RNAs remain to be further investigated.
Supplementary Material
Acknowledgments
We thank K. Horwitz and B. Jacobsen (University of Colorado HSC at Fitzsimons, CO) for providing T47D-YV cells; L. P. Rubin (Women and Infants Hospital, Providence, RI) for the pGL3-1778 11bLuc reporter vector; Fabrice Gouilleux (INSERM, France) and Toshio Kitamura (University of Tokyo, Japan) for Stat5a expression vectors; H. Tilgner and R. Guigó (CRG) for the analysis of tiling array-generated RNA maps from the publicly available NCBI GEO data; B. Miñana and L. Sumoy (CRG) for the microarray experiments and data submission to GEO; E. Diani (CRG) for help with immunofluorescence experiments; I. Falero (CRG) and O. Millan (IAT-PRBB) for help with manuscript editing; G. Vicent, F. Le Dily, M. Sancho, and E. Gallastegui (CRG) for helpful discussion; and N. Spinedi for technical assistance.
This work was supported by grants from the Departament d'Universitats, Recerca i Societat de la Informacio (DURSI), and Ministerio de Ciencia y Tecnología (MCYT) and FEDER (SAF2002-03320). A.J. was a recipient of a Ramón y Cajal appointment from the MCYT. I.Q. was a recipient of an FPU predoctoral fellowship from the Ministerio de Educación.
Footnotes
Published ahead of print on 31 March 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agarwal, A. K., and P. C. White. 1996. Analysis of the promoter of the NAD+ dependent 11 beta-hydroxysteroid dehydrogenase (HSD11K) gene in JEG-3 human choriocarcinoma cells. Mol. Cell. Endocrinol. 12193-99. [DOI] [PubMed] [Google Scholar]
- 2.Albiston, A. L., V. R. Obeyesekere, R. E. Smith, and Z. S. Krozowski. 1994. Cloning and tissue distribution of the human 11 beta-hydroxysteroid dehydrogenase type 2 enzyme. Mol. Cell. Endocrinol. 105R11-R17. [DOI] [PubMed] [Google Scholar]
- 3.Alikhani-Koopaei, R., F. Fouladkou, F. J. Frey, and B. M. Frey. 2004. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Investig. 1141146-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariyoshi, K., T. Nosaka, K. Yamada, M. Onishi, Y. Oka, A. Miyajima, and T. Kitamura. 2000. Constitutive activation of STAT5 by a point mutation in the SH2 domain. J. Biol. Chem. 27524407-24413. [DOI] [PubMed] [Google Scholar]
- 5.Ballaré, C., M. Uhrig, T. Bechtold, E. Sancho, M. Di Domenico, A. Migliaccio, F. Auricchio, and M. Beato. 2003. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol. Cell. Biol. 231994-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballare, C., G. Vallejo, G. P. Vicent, P. Saragueta, and M. Beato. 2006. Progesterone signaling in breast and endometrium. J. Steroid Biochem. Mol. Biol. 1022-10. [DOI] [PubMed] [Google Scholar]
- 7.Beato, M., P. Herrlich, and G. Schutz. 1995. Steroid hormone receptors: many actors in search of a plot. Cell 83851-857. [DOI] [PubMed] [Google Scholar]
- 8.Beato, M., and J. Klug. 2000. Steroid hormone receptors: an update. Hum. Reprod. Update 6225-236. [DOI] [PubMed] [Google Scholar]
- 9.Berglund, H., M. Wolf-Watz, T. Lundback, S. van den Berg, and T. Hard. 1997. Structure and dynamics of the glucocorticoid receptor DNA-binding domain: comparison of wild type and a mutant with altered specificity. Biochemistry 3611188-11197. [DOI] [PubMed] [Google Scholar]
- 10.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 28160-63. [DOI] [PubMed] [Google Scholar]
- 11.Boonyaratanakornkit, V., M. P. Scott, V. Ribon, L. Sherman, S. M. Anderson, J. L. Maller, W. T. Miller, and D. P. Edwards. 2001. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol. Cell 8269-280. [DOI] [PubMed] [Google Scholar]
- 12.Bulger, M., and M. Groudine. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 132465-2477. [DOI] [PubMed] [Google Scholar]
- 13.Buser, A. C., E. K. Gass-Handel, S. L. Wyszomierski, W. Doppler, S. A. Leonhardt, J. Schaack, J. M. Rosen, H. Watkin, S. M. Anderson, and D. P. Edwards. 2007. Progesterone receptor repression of prolactin/signal transducer and activator of transcription 5-mediated transcription of the beta-casein gene in mammary epithelial cells. Mol. Endocrinol. 21106-125. [DOI] [PubMed] [Google Scholar]
- 14.Carsol, J. L., S. Gingras, and J. Simard. 2002. Synergistic action of prolactin (PRL) and androgen on PRL-inducible protein gene expression in human breast cancer cells: a unique model for functional cooperation between signal transducer and activator of transcription-5 and androgen receptor. Mol. Endocrinol. 161696-1710. [DOI] [PubMed] [Google Scholar]
- 15.Castoria, G., A. Migliaccio, A. Bilancio, M. Di Domenico, A. de Falco, M. Lombardi, R. Fiorentino, L. Varricchio, M. V. Barone, and F. Auricchio. 2001. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 206050-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavez, S., and M. Beato. 1997. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc. Natl. Acad. Sci. USA 942885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, J., H. K. Kinyamu, and T. K. Archer. 2006. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol. Endocrinol. 201-13. [DOI] [PubMed] [Google Scholar]
- 18.Clevenger, C. V. 2004. Roles and regulation of stat family transcription factors in human breast cancer. Am. J. Pathol. 1651449-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulter, C. L., R. E. Smith, M. Stowasser, H. Sasano, Z. S. Krozowski, and R. D. Gordon. 1999. Expression of 11beta-hydroxysteroid dehydrogenase type 2 (11betaHSD-2) in the developing human adrenal gland and human adrenal cortical carcinoma and adenoma. Mol. Cell. Endocrinol. 15471-77. [DOI] [PubMed] [Google Scholar]
- 20.Darnell, J. E., Jr. 1996. The JAK-STAT pathway: summary of initial studies and recent advances. Recent Prog. Horm. Res. 51391-403. [PubMed] [Google Scholar]
- 21.Denoeud, F., P. Kapranov, C. Ucla, A. Frankish, R. Castelo, J. Drenkow, J. Lagarde, T. Alioto, C. Manzano, J. Chrast, S. Dike, C. Wyss, C. N. Henrichsen, N. Holroyd, M. C. Dickson, R. Taylor, Z. Hance, S. Foissac, R. M. Myers, J. Rogers, T. Hubbard, J. Harrow, R. Guigo, T. R. Gingeras, S. E. Antonarakis, and A. Reymond. 2007. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 17746-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards, D. P. 2005. Regulation of signal transduction pathways by estrogen and progesterone. Annu. Rev. Physiol. 67335-376. [DOI] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447799-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng, W., R. C. Ribeiro, R. L. Wagner, H. Nguyen, J. W. Apriletti, R. J. Fletterick, J. D. Baxter, P. J. Kushner, and B. L. West. 1998. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 2801747-1749. [DOI] [PubMed] [Google Scholar]
- 25.Gong, W., S. Chavez, and M. Beato. 1997. Point mutation in the ligand-binding domain of the progesterone receptor generates a transdominant negative phenotype. Mol. Endocrinol. 111476-1485. [DOI] [PubMed] [Google Scholar]
- 26.Ho, Y., F. Elefant, S. A. Liebhaber, and N. E. Cooke. 2006. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23365-375. [DOI] [PubMed] [Google Scholar]
- 27.Hundertmark, S., H. Buhler, M. Rudolf, H. K. Weitzel, and V. Ragosch. 1997. Inhibition of 11 beta-hydroxysteroid dehydrogenase activity enhances the antiproliferative effect of glucocorticosteroids on MCF-7 and ZR-75-1 breast cancer cells. J. Endocrinol. 155171-180. [DOI] [PubMed] [Google Scholar]
- 28.Ihle, J. N. 1996. STATs: signal transducers and activators of transcription. Cell 84331-334. [DOI] [PubMed] [Google Scholar]
- 29.Ihle, J. N., B. A. Witthuhn, F. W. Quelle, K. Yamamoto, and O. Silvennoinen. 1995. Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol. 13369-398. [DOI] [PubMed] [Google Scholar]
- 30.Jacobsen, B. M., J. K. Richer, S. A. Schittone, and K. B. Horwitz. 2002. New human breast cancer cells to study progesterone receptor isoform ratio effects and ligand-independent gene regulation. J. Biol. Chem. 27727793-27800. [DOI] [PubMed] [Google Scholar]
- 31.Kabotyanski, E. B., M. Huetter, W. Xian, M. Rijnkels, and J. M. Rosen. 2006. Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers. Mol. Endocrinol. 202355-2368. [DOI] [PubMed] [Google Scholar]
- 32.Kapranov, P., J. Cheng, S. Dike, D. A. Nix, R. Duttagupta, A. T. Willingham, P. F. Stadler, J. Hertel, J. Hackermuller, I. L. Hofacker, I. Bell, E. Cheung, J. Drenkow, E. Dumais, S. Patel, G. Helt, M. Ganesh, S. Ghosh, A. Piccolboni, V. Sementchenko, H. Tammana, and T. R. Gingeras. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 3161484-1488. [DOI] [PubMed] [Google Scholar]
- 33.Kapranov, P., A. T. Willingham, and T. R. Gingeras. 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8413-423. [DOI] [PubMed] [Google Scholar]
- 34.Kastner, P., A. Krust, B. Turcotte, U. Stropp, L. Tora, H. Gronemeyer, and P. Chambon. 1990. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 91603-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, A., H. Zhao, I. Ifrim, and A. Dean. 2007. β-Globin intergenic transcription and histone acetylation dependent on an enhancer. Mol. Cell. Biol. 272980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knuesel, M., Y. Wan, Z. Xiao, E. Holinger, N. Lowe, W. Wang, and X. Liu. 2003. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell. Proteomics 21225-1233. [DOI] [PubMed] [Google Scholar]
- 37.Kostadinova, R. M., A. R. Nawrocki, F. J. Frey, and B. M. Frey. 2005. Tumor necrosis factor alpha and phorbol 12-myristate-13-acetate down-regulate human 11beta-hydroxysteroid dehydrogenase type 2 through p50/p50 NF-kappaB homodimers and Egr-1. FASEB J. 19650-652. [DOI] [PubMed] [Google Scholar]
- 38.Kucera, T., M. Waltner-Law, D. K. Scott, R. Prasad, and D. K. Granner. 2002. A point mutation of the AF2 transactivation domain of the glucocorticoid receptor disrupts its interaction with steroid receptor coactivator 1. J. Biol. Chem. 27726098-26102. [DOI] [PubMed] [Google Scholar]
- 39.Lefstin, J. A., and K. R. Yamamoto. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392885-888. [DOI] [PubMed] [Google Scholar]
- 40.Ling, J., B. Baibakov, W. Pi, B. M. Emerson, and D. Tuan. 2005. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Biol. 350883-896. [DOI] [PubMed] [Google Scholar]
- 41.Migliaccio, A., M. Di Domenico, G. Castoria, A. de Falco, P. Bontempo, E. Nola, and F. Auricchio. 1996. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J. 151292-1300. [PMC free article] [PubMed] [Google Scholar]
- 42.Migliaccio, A., D. Piccolo, G. Castoria, M. Di Domenico, A. Bilancio, M. Lombardi, W. Gong, M. Beato, and F. Auricchio. 1998. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 172008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moriggl, R., V. Gouilleux-Gruart, R. Jahne, S. Berchtold, C. Gartmann, X. Liu, L. Hennighausen, A. Sotiropoulos, B. Groner, and F. Gouilleux. 1996. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol. Cell. Biol. 165691-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nawrocki, A. R., C. E. Goldring, R. M. Kostadinova, F. J. Frey, and B. M. Frey. 2002. In vivo footprinting of the human 11beta-hydroxysteroid dehydrogenase type 2 promoter: evidence for cell-specific regulation by Sp1 and Sp3. J. Biol. Chem. 27714647-14656. [DOI] [PubMed] [Google Scholar]
- 45.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2703859-3870. [DOI] [PubMed] [Google Scholar]
- 46.Petrascheck, M., D. Escher, T. Mahmoudi, C. P. Verrijzer, W. Schaffner, and A. Barberis. 2005. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 333743-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proietti, C., M. Salatino, C. Rosemblit, R. Carnevale, A. Pecci, A. R. Kornblihtt, A. A. Molinolo, I. Frahm, E. H. Charreau, R. Schillaci, and P. V. Elizalde. 2005. Progestins induce transcriptional activation of signal transducer and activator of transcription 3 (Stat3) via a Jak- and Src-dependent mechanism in breast cancer cells. Mol. Cell. Biol. 254826-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabbitt, E. H., N. J. Gittoes, P. M. Stewart, and M. Hewison. 2003. 11beta-hydroxysteroid dehydrogenases, cell proliferation and malignancy. J. Steroid Biochem. Mol. Biol. 85415-421. [DOI] [PubMed] [Google Scholar]
- 49.Richer, J. K., C. A. Lange, N. G. Manning, G. Owen, R. Powell, and K. B. Horwitz. 1998. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J. Biol. Chem. 27331317-31326. [DOI] [PubMed] [Google Scholar]
- 50.Rocha-Viegas, L., G. P. Vicent, J. L. Baranao, M. Beato, and A. Pecci. 2006. Glucocorticoids repress bcl-X expression in lymphoid cells by recruiting STAT5B to the P4 promoter. J. Biol. Chem. 28133959-33970. [DOI] [PubMed] [Google Scholar]
- 51.Saitoh, M., M. Ohmichi, K. Takahashi, J. Kawagoe, T. Ohta, M. Doshida, T. Takahashi, H. Igarashi, A. Mori-Abe, B. Du, S. Tsutsumi, and H. Kurachi. 2005. Medroxyprogesterone acetate induces cell proliferation through up-regulation of cyclin D1 expression via phosphatidylinositol 3-kinase/Akt/nuclear factor-kappaB cascade in human breast cancer cells. Endocrinology 1464917-4925. [DOI] [PubMed] [Google Scholar]
- 52.Sandelin, A., W. W. Wasserman, and B. Lenhard. 2004. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 32W249-W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sartorius, C. A., S. D. Groshong, L. A. Miller, R. L. Powell, L. Tung, G. S. Takimoto, and K. B. Horwitz. 1994. New T47D breast cancer cell lines for the independent study of progesterone B- and A-receptors: only antiprogestin-occupied B-receptors are switched to transcriptional agonists by cAMP. Cancer Res. 543868-3877. [PubMed] [Google Scholar]
- 54.Stocklin, E., M. Wissler, F. Gouilleux, and B. Groner. 1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383726-728. [DOI] [PubMed] [Google Scholar]
- 55.Stoecklin, E., M. Wissler, R. Moriggl, and B. Groner. 1997. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 176708-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strutt, H., and R. Paro. 1999. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol. Biol. 119455-467. [DOI] [PubMed] [Google Scholar]
- 57.Takimoto, G. S., D. M. Tasset, A. C. Eppert, and K. B. Horwitz. 1992. Hormone-induced progesterone receptor phosphorylation consists of sequential DNA-independent and DNA-dependent stages: analysis with zinc finger mutants and the progesterone antagonist ZK98299. Proc. Natl. Acad. Sci. USA 893050-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomson, S., A. L. Clayton, C. A. Hazzalin, S. Rose, M. J. Barratt, and L. C. Mahadevan. 1999. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 184779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Truss, M., J. Bartsch, A. Schelbert, R. J. Hache, and M. Beato. 1995. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 141737-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tung, L., H. Abdel-Hafiz, T. Shen, D. M. Harvell, L. K. Nitao, J. K. Richer, C. A. Sartorius, G. S. Takimoto, and K. B. Horwitz. 2006. Progesterone receptors (PR)-B and -A regulate transcription by different mechanisms: AF-3 exerts regulatory control over coactivator binding to PR-B. Mol. Endocrinol. 202656-2670. [DOI] [PubMed] [Google Scholar]
- 61.Uhler, J. P., C. Hertel, and J. Q. Svejstrup. 2007. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. USA 1048011-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vicent, G. P., C. Ballare, A. S. Nacht, J. Clausell, A. Subtil-Rodriguez, I. Quiles, A. Jordan, and M. Beato. 2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell 24367-381. [DOI] [PubMed] [Google Scholar]
- 63.Vicent, G. P., C. Ballare, R. Zaurin, P. Saragueta, and M. Beato. 2006. Chromatin remodeling and control of cell proliferation by progestins via cross talk of progesterone receptor with the estrogen receptors and kinase signaling pathways. Ann. N. Y. Acad. Sci. 108959-72. [DOI] [PubMed] [Google Scholar]
- 64.Viegas, L. R., G. P. Vicent, J. L. Baranao, M. Beato, and A. Pecci. 2004. Steroid hormones induce bcl-X gene expression through direct activation of distal promoter P4. J. Biol. Chem. 2799831-9839. [DOI] [PubMed] [Google Scholar]
- 65.Wan, Y., and S. K. Nordeen. 2002. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol. Endocrinol. 161204-1214. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Q., J. S. Carroll, and M. Brown. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19631-642. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y., and C. H. Cheng. 2004. ERalpha and STAT5a cross-talk: interaction through C-terminal portions of the proteins decreases STAT5a phosphorylation, nuclear translocation and DNA-binding. FEBS Lett. 572238-244. [DOI] [PubMed] [Google Scholar]
- 68.Wingender, E., P. Dietze, H. Karas, and R. Knuppel. 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.