Abstract
Spindle integrity is critical for efficient mitotic progression and accurate chromosome segregation. Deregulation of spindles often leads to structural and functional aberrations, ultimately promoting segregation errors and aneuploidy, a hallmark of most human cancers. Here we report the characterization of a previously identified human sarcoma antigen (gene located at 19p13.11), Hice1, an evolutionarily nonconserved 46-kDa coiled-coil protein. Hice1 shows distinct cytoplasmic localization and associates with interphase centrosomes and mitotic spindles, preferentially at the spindle pole vicinity. Depletion of Hice1 by RNA interference resulted in abnormal and unstable spindle configurations, mitotic delay at prometaphase and metaphase, and elevated aneuploidy. Conversely, loss of Hice1 had minimal effects on interphase centrosome duplication. We also found that both full-length Hice1 and Hice1-N1, which is composed of 149 amino acids of the N-terminal region, but not the mutant lacking the N-terminal region, exhibited activities of microtubule bundling and stabilization at a near-physiological concentration. Consistently, overexpression of Hice1 rendered microtubule bundles in cells resistant to nocodazole- or cold-treatment-induced depolymerization. These results demonstrate that Hice1 is a novel microtubule-associated protein important for maintaining spindle integrity and chromosomal stability, in part by virtue of its ability to bind, bundle, and stabilize microtubules.
Mitosis is a dynamic process associated with dramatic physiological and morphological changes. For precise mitotic control, cells command a highly coordinated interplay among cell cycle regulators, spindle assembly molecules, and a multitude of cofactors. For instance, mitotic kinases, including Cdc2, Polo-like kinase Plk1, Aurora, and Nek2, are known for their involvement in mitotic regulation (reviewed in references 12, 23, 24, and 34). Upon centrosome maturation and mitotic entry, spindle assembly factors, such as Eg5, TPX2, and NUMA, are recruited for the assembly of a bipolar structure in a concerted manner (8, 9, 13, 21, 31, 36). The spindle undergoes progressive but dynamic changes concurrently with the ongoing mitosis (11). The integrity of a dynamic spindle is critical for proper chromosome alignment at metaphase and sister chromosome segregation at anaphase. Functional disturbance of the microtubule (MT)/spindle regulatory mechanisms often gives rise to altered dynamics and defective structures, which in turn lead to increased segregation errors and consequently aneuploidy (2, 27). Such phenotypes can be exacerbated when the spindle checkpoint is simultaneously inactivated (16). Thus, the maintenance of spindle integrity is of central importance for cells to undergo successful cellular division while maintaining genomic stability.
Among the cellular factors required for spindle regulation are those involved in MT binding and those involved in enhancing or reducing its activities, represented by the kinesin family members (e.g., Eg5 and MCAK) and the MT-associated proteins (MAPs) (reviewed in references 1 and 14), respectively. MAPs have intrinsic MT binding activity, at either the plus or minus end of MTs and/or along the longitudinal lattice. Different MAPs have both unique and shared functionalities in modulating MT structure and dynamics. They serve to regulate both global and local MT function, such as MT plus-end dynamics and attachment at kinetochores (e.g., TOG/XMAP215, EB1, or CLASPs), as well as to focus and stabilize MTs at the spindle poles (TPX2 and NUMA) (reviewed in references 1 and 14). Disruption of MAPs frequently generates aneuploidy in cells resulting from inaccurate mitotic segregation. Thus, it is of great interest to investigate the biochemical activity and cellular roles of novel MAPs in human cells.
Through a previous yeast two-hybrid screen, we have identified a novel Hec1-interacting partner, in this study designated Hice1 (Hec1-interacting and centrosome-associated 1) (4). The human Hice1 gene is located at 19p13.11 and encodes a 46-kDa protein containing two distinct coiled-coil domains. Hice1 is identical to the previously identified human sarcoma antigen (18), but its biological function remains elusive. The homologous counterparts of mammalian Hice1 can be found in vertebrates but not in lower eukaryotes such as worm, fly, or yeast. In this communication, we show that Hice1 is a novel MAP with MT-bundling and stabilization activities, which is important for the maintenance of spindle integrity and faithful mitotic division.
MATERIALS AND METHODS
Cloning and antibody production.
Human Hice1 cDNA (NY-SAR-48; GenBank accession no. NM_001011699) was cloned from a human lymphocyte cDNA library. Glutathione S-transferase (GST)-tagged full-length Hice1 (Hice1-FL) was purified from Escherichia coli and used to immunize mice. Crude and affinity-purified mouse antiserum was used in this study.
Cell lines and RNA interference (RNAi).
Human cancer cell lines HeLa, U2OS, and HCT116 from ATCC were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. U2OS cells stably expressing green fluorescent protein (GFP)-tagged Hice1 and its deletion mutants were established by retroviral infection using the pQCXIP system produced in the GP2-293 packaging cell line. Two small interfering RNAs (siRNAs) were synthesized (IDT, Coralville, IA) to target Hice1 (a, 5′GGGAGAACTTGATGTTGGTGATTCG3′; b, 5′CAAAGAGGCAGCCTTGGCAAACCAG3′). A third one was designed for DNA vector-based short-hairpin RNA (shRNA) expression (5′GAGTGATTGAGTCCCGGTA3′). Four tandem repeats of Hice1 shRNA expression cassettes derived from pSuperior were inserted into an adenoviral system (Quantum Technology, Montreal, Canada) for the production of adenovirus expressing Hice1 shRNA (7).
Microscopy and time-lapse study.
The immunostaining procedure was adapted as previously described (38). Briefly, cells were grown on polylysine-coated coverslips. Cells were gently washed with BRB80 buffer [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 5 mM EGTA, 1 mM MgCl2] before fixation with cold 100% methanol or 4% paraformaldehyde in BRB80 or phosphate-buffered saline (PBS). Following permeabilization with 0.4% Triton X-100, cells were blocked with 2% bovine serum albumin (BSA) in PBS and then incubated with primary antibodies diluted in 2% BSA in PBS. Secondary antibodies were conjugated with Alexa 488 or 594 (Molecular Probes, Eugene, OR). DAPI (4′,6′-diamidino-2-phenylindole) staining was applied after secondary antibody incubation, and cells were mounted on cover slides with Prolong gold antifade reagent (Invitrogen). Images were captured with a Zeiss Axioplan 2 microscope (equipped with a deconvolution module) or with a Zeiss LSM-510 META laser scanning confocal microscope. For live-cell time-lapse study, cells were maintained at 37°C in CO2-independent medium (Invitrogen) and monitored with a Zeiss Axiovert-200 M microscope equipped with a motorized stage controller and a temperature-controlled apparatus. Images were acquired with a Hamamatsu charge-coupled device camera controlled by the Axiovision software (Zeiss). Deconvolution was performed with the Autodeblur/Autovisualize software, and the obtained maximum projections of the Z-stack images were shown. Further image analysis or quantification was performed with ImageJ (NIH) or Adobe Photoshop (San Jose, CA). Commercial α-tubulin and γ-tubulin antibodies were purchased from Sigma-Aldrich (St. Louis, MO).
For electron microscopy, MTs were assembled with GMPCPP instead of GTP and then incubated with purified Hice1 or control proteins. The mixtures were then purified through a 50% glycerol cushion buffer by ultracentrifugation (see “MT cosedimentation and bundling assay” below). The pellet was resuspended and placed on Formvar-coated grids, negatively stained with uranyl acetate to enhance contrast. Samples were examined with a Philips CM-10 transmission electron microscope, and images were captured with a Gatan digital camera.
Mitotic chromosome spreads and microscopic counting for HCT116 cells were performed by following the standard cytogenetic procedure. In brief, after siRNA transfection (96 h), cells were treated with 100 ng/ml nocodazole for 1 hour and then harvested for mitotic spreading.
Protein expression and purification.
GST-tagged Hice1-FL was expressed in E. coli (BL21-pLys) and prepared by using glutathione-Sepharose beads as described in the GE Health Sciences manual. His6-tagged Hice1-FL and mutants were engineered in the pQE30 or pQE31 vector (Qiagen) and expressed in the Rosetta or M15 strain of E. coli. The proteins were prepared with Bio-Rad IMAC affinity resin by following the protocol provided in the kit.
MT cosedimentation and bundling assay.
The MT copelleting assay was performed according to manufacturer's instructions (Cytoskeleton Inc., Denver, CO). MTs were assembled with 5% glycerol and 1 mM GTP in BRB80 buffer (80 mM PIPES [pH 7.0], 1 mM EGTA, 1 mM MgCl2) at 35°C (30 min) and later stabilized with 20 μM Taxol. Hice1 was preclarified by ultracentrifugation (100,000 × g, 4°C, 20 min) to remove potential aggregates and then incubated with preassembled MTs at room temperature (RT) (20 min). The mixtures were then placed on a cushion buffer (50% glycerol, 1× BRB80, and 20 μM Taxol) for ultracentrifugation (100,000 × g, RT, 40 min) to allow the pelleting of the Hice1-MT complex. The MT bundling assay was performed as described previously (22). Rhodamine- or fluorescein isothiocyanate-labeled tubulin was purchased from Cytoskeleton Inc.
Statistics.
Statistical analysis or data fitting was performed by using the SigmaStat software.
RESULTS
Identification and characterization of Hice1.
Hice1 was originally identified as one of the Hec1-interacting proteins in a yeast two-hybrid screen using Hec1 as a bait (4). While the interacting significance of Hice1-Hec1 remains to be addressed, we first sought to explore the cellular role of Hice1. Hice1 encodes a 410-amino-acid protein identical to an uncharacterized human sarcoma antigen (18). Sequence analysis revealed two novel coiled-coil domains in the middle of Hice1 (Fig. 1A). To characterize the Hice1 gene product, we generated a mouse polyclonal antibody by using purified GST-tagged Hice1-FL as an immunogen. Both crude and purified mouse polyclonal antibodies recognize a 46-kDa species from human cells, which comigrates with the in vitro-translated 35S-labeled Hice1 (Fig. 1B).
FIG. 1.
Characterization of Hice1 protein. (A) Amino acid sequence and two coiled-coil domains of human Hice1. (B) Identification of Hice1 protein. Left panel (autoradiography), preimmune (pre I) or crude anti-Hice1 mouse serum (I, crude) was used to immunoprecipitate in vitro-translated 35S-labeled Hice1. IVT, loading control, Right panel (immunoprecipitation-Western blot), HeLa cell lysate was used for immunoprecipitation with control mouse immunoglobulin G (IgG) or purified anti-Hice1. (C) Hice1 distribution at centrosomes. Cells stably expressing Hice1-GFP were stained with an antibody against the centrosome marker γ-tubulin. (D and E) Costaining with an antibody against α-tubulin with Hice1-GFP in interphase and mitotic cells. Hice1 associates with MTs in a subset of cells expressing Hice1-GFP (D) and the spindles in mitotic cells (E). Bars, 10 μm.
Next, we examined the cellular distribution of Hice1 by establishing several stable clones of human U2OS cells expressing Hice1-GFP mediated by retroviral infection. Coimmunostaining of these cells with γ-tubulin revealed that Hice1 is distributed primarily in the cytoplasm with specific enrichment at centrosomes during interphase of the cell cycle (Fig. 1C). In about 10 to 15% of cells with an apparently higher expression level, Hice1 exhibited filamentous structures that colocalize with MTs (Fig. 1D). At each stage of mitosis, Hice1 is distributed to the spindle apparatus (preferentially at the vicinity of poles) as indicated by its colocalization with α-tubulin (Fig. 1E). During anaphase and telophase, Hice1 additionally associates with the midzone and midbody, respectively (Fig. 1E). The association of Hice1 with spindles is not an immunofluorescent-staining artifact, as Hice1-GFP alone shows spindle-like structures without costaining with any antibody (see Fig. S1A in the supplemental material). Furthermore, the distribution of endogenous Hice1 is similar to that of Hice1-GFP except during interphase; the endogenous Hice1 immunostaining signal appears to be less distinct on MTs (see Fig. S1B and C in the supplemental material). Hice1 localization at centrosomes and spindles was further revealed by a live-cell time-lapse study (see Fig. S2 in the supplemental material). Taken together, these results indicate that Hice1 is a centrosome- and spindle-associated factor in mammalian cells (hence named Hice1 for Hec-1-interacting and centrosome-associated 1).
Hice1 is required for efficient and accurate mitotic progression.
Functional defects of MAPs frequently result in spindle abnormalities and mitotic failure (13, 14). Given its specific cellular localization, Hice1 may play an important role in mitosis. To test this possibility, we have used two types of RNAi derived from either chemically synthesized siRNA or transcription-directed shRNA to efficiently deplete the endogenous Hice1 (Fig. 2A). Western blot analysis with serially diluted input showed greater than 90% reduction of endogenous Hice1 by siRNA, indicating the high efficiency of RNAi-mediated depletion (Fig. 2B). Immunostaining by anti-Hice1 antibodies further confirmed that the cellular Hice1 was largely depleted and barely detectable at the centrosome (Fig. 2C).
FIG. 2.
Depletion of Hice1 by siRNA hampers proper mitotic progression. (A) Depletion of Hice1 expression by siRNA and adenovirally expressed shRNA, targeting two different regions of Hice1 mRNA. Endogenous cellular Hice1 protein was measured by Western blotting probed with anti-Hice1 mouse serum. Hours after RNAi treatment are indicated. Luciferase RNAi (Luc) served as a control. (B) Western blot with serially diluted inputs to determine the extent of Hice1 depletion. (C) Reduction of Hice1 expression in cells treated with the Hice1 siRNA versus the control by immunostaining with purified anti-Hice1 antibodies. Cells were counterstained with DAPI to show the nucleus. (D) Increased mitotic index in Hice1-depleted cells. Mitotic cells were enumerated based on the mitotic patterns of H2B-GFP stably expressed in HeLa or U2OS cells. Both siRNA and adenovirally expressed shRNA were employed. (E) Fluorescence-activated cell sorter analysis of luciferase or Hice1 siRNA-treated cells to show cell cycle profile after 4 days of treatment. Percentages of each stage or cells with >4N DNA content are indicated. (F) Distribution of cell populations at each stage of mitosis. (G) Quantification of mitotic period from nuclear envelope breakdown to anaphase onset, which was monitored by time-lapse microscopy. The P value was obtained from a paired t test. Error bars indicate standard deviations. (H) DAPI staining of interphase or mitotic cells treated with control or Hice1 siRNA. Arrows, misaligned chromosomes; arrowheads, micronuclei or multinuclei. U2OS cells were used in all experiments in panels A to H except as otherwise indicated. Cells were treated with RNAi for 96 h. (I) Mitotic spread of Hice1 or luciferase control siRNA-treated HCT116 cells (a near-diploid human colon cancer line). Chromosomes in each spread were counted under a microscope, and the resulting number profile is presented in a dot plot (each dot representing an individual spread; random distribution on y axis). For each group, 100 spreads were enumerated. The distribution difference is significant (χ2 = 14.5; P = 0.006).
We then performed a systematic examination of the mitotic profiles of Hice1-depleted cells. Interestingly, we observed a moderate increase in the mitotic index (up to 15%, versus 3 to 6% in the control) in both HeLa and U2OS cells upon Hice1 depletion (Fig. 2D). Consistently, fluorescence-activated cell sorter analysis confirmed an increase in the G2/M population of ∼8%, while the G1 and S populations were slightly reduced (Fig. 2E). Further examination of Hice1-depleted mitotic cells showed a skewed mitotic distribution, in which the combined prometaphase and metaphase populations were expanded from ∼60% to ∼81%, suggesting a delay in mitosis (Fig. 2F). To support this observation, we performed time-lapse imaging of U2OS cells stably expressing H2B-GFP and observed that Hice1-depleted cells were significantly delayed in the mitotic periods from nuclear envelope breakdown to anaphase onset (∼3.5 h, versus 0.9 h in the control cells) (Fig. 2G). Consistently, the mitotic delay was associated with an activated spindle assembly checkpoint, since both Mad2 and BubR1 signals were retained on the kinetochores of delayed mitotic cells (data not shown). Taken together, these results suggest that Hice1 is important for cells to properly progress through prometaphase and metaphase.
The delay in progression through mitosis may result from improper mitotic division, which may increase chromosome segregation errors leading to aneuploidy. Chromosome misalignment at metaphase and formation of micro- and multinuclei during interphase were frequently observed in Hice1-depleted U2OS cells (Fig. 2H). After 4 days of Hice1-siRNA treatment, the populations of cells with micro- and multinuclei were increased from 5% to almost 40% and from 2 to 5%, respectively, compared with the control cells. These results suggest that depletion of Hice1 may affect cytokinesis, resulting in production of micro- and multinuclei in interphase cells. To directly assess whether Hice1 is required for maintaining chromosomal stability, we performed mitotic chromosome spreading using HCT116 cells, a near-diploid human colon cancer line (19). Consistently, we found that the number of mitotic cells with fewer than 42 or greater than 50 chromosomes was increased from 9% to 27% in the Hice1 siRNA-treated population compared with the control (n = 100 for each population) (Fig. 2I). Moreover, 16% of Hice1 siRNA-treated mitotic cells were near tetraploid but only 6% in the control siRNA-treated group were, thus suggesting a failure in cytokinesis. Although cell death was not obvious on the fourth day after RNAi treatment, it became more evident after 6 to 7 days (data not shown). Taken together, these results suggest that Hice1 is important for efficient and accurate mitotic division.
Hice1 is required for the maintenance of spindle integrity.
Mitotic delay and increased aneuploidy are frequent results of a functional defect(s) in mitotic regulation. To test this possibility, we specifically examined the potential effects of Hice1 siRNA on centrosome and spindle integrity, since Hice1 is present predominantly in these two locations. First, we enumerated the centrosomes and found that centrosome duplication at interphase is minimally affected (see Fig. S3A in the supplemental material). Centrosomes in Hice1-depleted cells also matured normally as indicated by the timely expansion of pericentriolar area when the G2 population was examined (25) (see Fig. S3B in the supplemental material). Thus, Hice1 may not be essential for centrosome duplication during interphase.
In contrast, we detected severe spindle abnormalities in mitotic cells when Hice1 was depleted. We divided the spindle abnormalities into two major categories, as shown in Fig. 3A: multipolar spindles, which have at least three half-spindles and/or astral structures (Fig. 3A, rows c to e), and split spindles, which have at least one pair of centrioles separated by ≥2 μm and still maintain bipolarity (Fig. 3A, rows f and g). Remarkably, multipolar spindles were readily detected in 22 to 27% of Hice1-depleted mitotic cells (four- to fivefold increase versus the control), whereas split spindles were detected in 9 to 13% of Hice1-depleted mitotic cells (3.5- to 5-fold increase versus the control) (Fig. 3A and B). Altogether, ∼35% of all spindles in Hice1-depleted cells exhibited one or more types of aberrant configurations 4 days after siRNA treatment, as opposed to 5 to 8% in the control cells. Similar abnormalities were recapitulated in HeLa cells, suggesting that the phenotype is not cell type specific (Fig. 3C). Interestingly, Hice1 siRNA-treated cells exhibited fragmented centrosomes as shown by dispersed γ-tubulin staining (see Fig. S4 in the supplemental material), indicating the instability of the spindle pole during mitosis in the absence of Hice1. Although the interphase centrosome appears normal in Hice1 knockdown cells, the mitotic spindle integrity is compromised in multiple aspects. Taken together, these results suggested that Hice1 is critical for the maintenance of a dynamic bipolar spindle in mitosis.
FIG. 3.
Spindle configurations in Hice1-depleted cells by immunofluorescent staining with γ-tubulin and α-tubulin antibodies upon siRNA treatment. (A) Maximum projection of deconvoluted images for normal spindles (rows a and b; control siRNA treated) and multipolar spindles (rows c to e; Hice1 siRNA treated). Row, f and g, split spindles (images not deconvoluted). Arrows indicate centrosomal material of split or multipolar spindles; arrowheads indicate misaligned chromosomes. DAPI staining shows the chromosomes. Bars, 10 μm. (B and C) Quantification of abnormal spindle configurations in U2OS (B) and HeLa (C) cells. Error bars indicate standard deviations.
To corroborate the observed spindle phenotypes, we further examined the dynamics of the aforementioned aberrant spindles by using time-lapse imaging of U2OS cells stably expressing GFP-α-tubulin. Importantly, about 22 to 35% of Hice1 RNAi-treated cells with abnormal spindle structures were visible early in mitosis (<20 min after entering M phase) or after a prolonged mitotic delay (∼3 h) (Fig. 4A and B), while fewer (10 to 15%) luciferase RNAi-treated cells with aberrant spindles were observed. Together, these results suggest that the spindle abnormality was at least in part (if not all) directly attributable to the loss of Hice1, rather than the side effects of prolonged M-phase progression. Moreover, the configuration of aberrant spindles was unstable in a given mitotic cell. For instance, a small MT aster arising proximally to the bipolar structure could gradually grow over time and eventually lead to the formation of a tripolar structure (Fig. 4B), suggesting that MT growth could occur at the newly formed aster to give rise to a new half-spindle. On the other hand, the spindle poles appeared to split and remerge in an erratic manner (Fig. 4C). These observations obtained from Hice1-depleted living cells indicate aberrant MT dynamics associated with quick shrinkage and changes of the spindle configurations, suggesting that Hice1 has an important role in maintaining spindle integrity.
FIG. 4.
Live-cell time-lapse microscopy of Hice1-depleted cells. (A) Top row (control siLuc treated), a cell with normal mitotic progression; bottom row (siHice1 treated), a representative cell that exhibited early tripolar spindle formation (<20 min after entering mitosis). (B) A representative cell (siHice1 treated) displaying an additional small half-spindle (MT aster) at ∼3 h after entering mitosis. The aster underwent gradual but progressive growth over time and eventually gave rise to a tripolar structure. (C) An Hice1 siRNA-treated cell with a dynamically unstable and abnormal spindle (arrowheads), which appeared as a near-bipolar spindle with fragmented poles (time points 0:40 to 1:20), later became a near-bipolar structure with a single extra half-spindle (time points 1:40 to 2:20), and then returned to a near-bipolar one with fragmented poles (time point 3:20). Bars, 10 μm.
Hice1 is a novel MAP with MT-bundling and -stabilizing activities.
To explore the biochemical basis for the role of Hice1 in regulating mitotic spindles and to establish Hice1 as a MAP, we performed MT cosedimentation and bundling assays. A series of Hice1 truncated mutants were generated, expressed in E. coli, and then used in the MT cosedimentation assays (Fig. 5A and B). As shown in Fig. 5D, Hice1-FL is capable of binding to MTs. Hyperbolic data fitting yielded a dissociation constant of ∼0.9 μM (tubulin concentration required to precipitate 50% of Hice1) (Fig. 5E), indicating a moderate affinity toward MTs. Similar binding activity was also detected for Hice1-N1 (the N-terminal region, amino acids 1 to 149), Hice1ΔCoil1 (coiled-coil domain 1 deleted), and Hice1ΔCoil2 (coiled-coil domain 2 deleted) but not Hice1ΔN (first 130 amino acids deleted) (Fig. 5F). These results indicated that Hice1 is a MAP and that its MT binding activity resides at the N-terminal domain (Hice1-N1). Notably, Hice1-N1 or its corresponding region in Hice1 homologues from other vertebrate species has a highly basic amino acid composition (the pI of human Hice1-N1 is 10.04) (Fig. 5G), consistent with known basic MT-binding domains of Tau, ASAP, and NuSAP (17, 28, 30). Further sequence alignment revealed about 50% identity between human and mouse Hice1 (Fig. 5G). However, BLAST search against the NCBI database failed to reveal any conserved motif between Hice1 and other known MAPs, suggesting that Hice1 represents a new class of MAPs emerging at a later stage in evolution.
FIG. 5.
Hice1 harbors an MT-binding domain at its N terminus. (A) Diagram of Hice1 expression constructs. (B and C) His-tagged Hice1 proteins, corresponding to the constructs in panel A, were purified by using the Bio-Rad IMAC resins and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and with Coomassie blue staining. (D and E) MT/Hice1 cosedimentation assay. Each reaction mixture contained ∼150 nM of Hice1 and various concentrations of MTs and then was analyzed by Western blotting. The amount of each protein measured from the blot was plotted using SigmaStat software. (F) MT/Hice1 cosedimentation assay for Hice1 mutants. The concentration of the input Hice1 was ∼150 nM. s, supernatant; p, precipitate. (G) Sequence alignment of Hice1 N-terminal domains across the vertebrates. Sequences were aligned by the Clustal W method with JalView software. Numbers denote amino acid positions. Asterisks indicate positively charged residues. The pI value of human Hice1-N1 (amino acids 1 to 149) is 10.04.
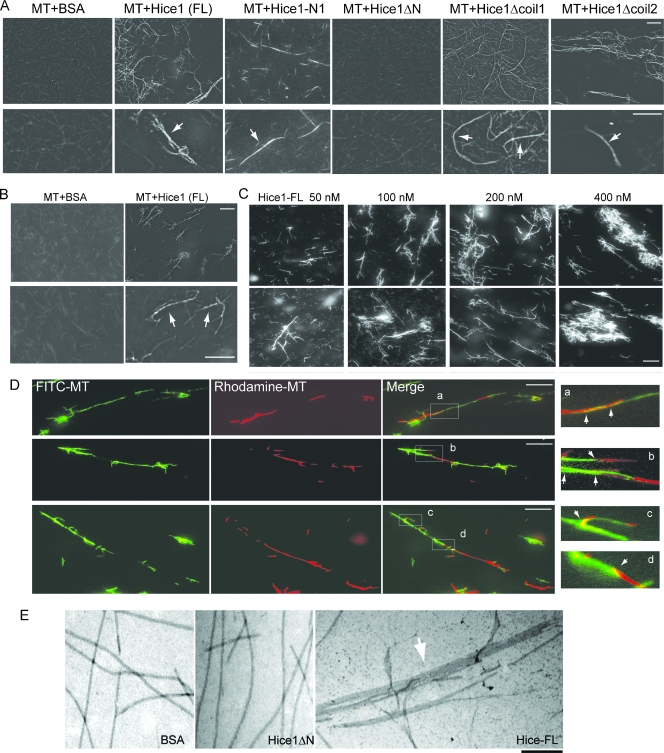
Next, we tested whether Hice1 may affect preassembled MTs stabilized by Taxol. Upon addition of Hice1-FL or Hice1-N1, these preassembled MTs quickly bundled into long (50 to 80 μm) and thick fibers within minutes (Fig. 6A), suggesting that the N-terminal region of Hice1 is responsible for its MT-bundling function. Furthermore, the bundled fibers were stable even after an 18-h incubation at 4°C (Fig. 6B). In contrast, when BSA or Hice1ΔN (MT binding deficient) was added, the preassembled MTs remained as short and thin filaments (usually 2 to 10 μm long). Hice1ΔCoil2 had MT-bundling activity similar to that of Hice1-FL, whereas Hice1ΔCoil1 had slightly reduced activity in the same assays, implying that the first coiled-coil domain of Hice1 may be also required for optimal MT bundling.
FIG. 6.
Hice1 bundles and stabilizes MTs. (A) Hice1 bundles preassembled MTs (rhodamine labeled), with representative MT fluorescent images. Assays were performed with 2 μM MTs and ∼100 nM Hice1 (full length or mutants) at RT for 10 min. BSA served as a negative control. (B) Preassembled MTs were incubated with BSA or Hice1-FL for 10 min at RT and then incubated at 4°C for 18 h. Representative fluorescent images of MTs are shown. (C) Dosage-dependent MT-bundling reaction by Hice1. Representative fluorescent images of rhodamine-labeled MTs are shown. Assays were performed with 2 μM MTs and increasing levels of Hice1 (full length) at RT for 10 min. (D) Fluorescent images to show miscellaneous fiber junctions of bundled MTs. Fluorescein isothiocyanate- and rhodamine-labeled MTs were individually preassembled and then used for bundling assay with Hice1 (100 nM). Note the long junction of two MT fibers (>10-μm overlap) (inset a, arrowhead), short junctions (<2-μm overlap) between two MT fibers (inset b, top arrowhead; also inset d), junction with at least four MT fibers (inset b, arrows showing two green and two red fibers); and fork-shape junction (inset c). Bars, 10 μm. (E) Electron microscopic images of free MTs incubated with BSA, Hice1ΔN, or MT bundles induced by Hice1-FL. Bar, 200 nm.
Importantly, the bundling activity of Hice1 is substoichiometric to that of tubulin dimer. The endogenous Hice1 concentration is estimated to be 50 to 150 nM in U2OS cells and 2 to 3 times lower in HeLa cells (data not shown). The bundling activity was already evident with only 100 nM of Hice1 (1:20 Hice1/tubulin ratio) and was gradually enhanced with increasing Hice1 concentration (100 to 400 nM) (Fig. 6C). Moreover, using mixtures of MTs labeled with rhodamine and fluorescein individually, we observed that the bundles were made of overlapping fibers (Fig. 6D). Most of these overlapping regions consisted of at least two to four bound MTs, with either short (<2 μm) or long (>10 μm) overlap (Fig. 6D, panels a, b, and d). Some of them are fork-like structures (Fig. 6D, panel c), suggesting that Hice1 may promote both end-to-end and end-to-lattice association between MTs. Finally, high-resolution imaging by electron microscopy further confirmed the MT bundle structure induced by Hice1-FL, but not by BSA or Hice1ΔN (Fig. 6E). The Hice1-induced bundles were similar but otherwise more compact than those formed by several previously documented MAPs, including CDC14B, HURP, Hec1/Nuf2, and Cep55 (3, 5, 15, 33, 39).
Hice1 stabilizes MTs in cells.
To verify the significance of the biochemical activity of Hice1 in vivo, we generated and expressed a series of GFP-tagged Hice1 mutants in HeLa cells (Fig. 7A). Remarkably, a subset of interphase cells overexpressing Hice1-FL and Hice1ΔCoil2 showed bright cytoplasmic fibers that colocalized with unusually thickened and elongated MTs (Fig. 7B and C), reminiscent of the MT bundles induced by Hice1 in vitro (Fig. 6). It is noteworthy that a portion of these unusual MTs (20 to 30%) are curly and intertwined, suggesting disorganized MT radial structures (Fig. 7B and C). Interestingly, GFP-Hice1-N1 or GFP-Hice1ΔCoil1 failed to display as MT fibers, suggesting that both the N-terminal MT-binding domain and the coiled-coil domain 1 are essential for proper MT association. Consistently, Hice1NC, a mutant containing the MT-binding domain and coiled-coil domain 1 only, was able to associate with MTs and trigger the MT thickening and curling. Thus, it is likely that the coiled-coil domain 1 may be important for protein-protein interaction to allow proper MT targeting of Hice1 and that the N-terminal region is responsible for direct binding to the MT subunits.
FIG. 7.
Hice1 promotes stabilization of MTs in cells. (A) Western blot (WB) of GFP or GFP-tagged Hice1 versions in HeLa cells. The mutants were illustrated in Fig. 5A, except for NC (N terminus and coiled-coil1 only; amino acids 1 to 226 of Hice1). (B) Fluorescent images of various Hice1 versions expressed in HeLa cells. Note the MT-like structures with the Hice1-FL, NC, and ΔCoil2 versions but not the others. (C) Costaining of Hice1-FL, NC, and ΔCoil2 with α-tubulin. (D) Hice1-FL-expressing cells were mock treated (dimethyl sulfoxide) or treated with nocodazole (100 ng/ml, 5 h) or cold shock on ice (20 min). Fluorescent images show retention of fiber structures (MTs) after treatment. Bars, 10 μm.
To further substantiate that Hice1 stabilizes the MT fibers in vivo, we treated cells overexpressing Hice1-FL with nocodazole or cold shock on ice, which are two independent methods to induce MT depolymerization. We found that even after 5 hours of nocodazole treatment (100 ng/ml) or after cold shock for 20 min, the fiber-like structures (presumably MTs) were still present in 50% of the original GFP-Hice1-FL-positive cells. In contrast, <1% cells expressing GFP alone retained an intact MT structure after similar treatment (data not shown). Taken together, these results suggest that Hice1 binds and stabilizes the MT fibers in vivo, consistent with its in vitro activity (Fig. 6).
DISCUSSION
In this work, we have studied the function of Hice1, a previously uncharacterized coiled-coil protein. In cells, Hice1 associates with interphase centrosomes and mitotic spindles. Silencing Hice1 by RNAi treatment invokes significant adverse mitotic effects, including mitotic delay and severe spindle abnormalities, consequently leading to elevated aneuploidy (Fig. 2 and 3). These results support a novel role for Hice1 in mitotic regulation.
Hice1 is important for maintaining the integrity of spindles. In Hice1-depleted cells, spindles are unstable and often appear as aberrant configurations that evolve quickly (within minutes) (Fig. 3 and 4). These abnormalities are in part attributable to the loss of MT-bundling and -stabilizing activities of Hice1 (Fig. 5 to 7). The N-terminal region of Hice1 (Hice1-N1), which is mainly responsible for such activities, contains multiple clusters of lysine- and arginine-rich motifs and hence is highly basic (pI value of 10.04) (Fig. 5F). Importantly, there is no significant homology between Hice1-N1 and the MT-binding domains of known MAPs, suggesting the existence of a novel MT-binding motif. However, it is possible that the similarity may manifest only at the level of tertiary structure, as is the case for Hec1 and EB1 (35). Interestingly, the coiled-coil 1 domain appears to be important for both binding and bundling MTs in vivo but less in vitro. A plausible explanation is that the coiled-coil domain 1 is used for interacting with some other factors to facilitate Hice1 interaction with the MTs or for self-oligomerization to enhance the N-terminal region binding to the MT subunits. The detailed mechanism of this interaction warrants further elucidation.
Given the biochemical activity of Hice1, we speculate that Hice1 may regulate the spindle dynamics by directly binding and modulating MT dynamics, like other known regulators such as TOG, CLASPs, and MCAK (20). The observed MT-bundling and -stabilizing activities of Hice1 are similar to those of TPX2 and HURP (Fig. 6) (15, 32, 33). However, the MT bundles formed by Hice1 appeared thinner but otherwise more compact and longer than those reported for TPX2 and HURP (Fig. 6). Therefore, the mechanisms of bundling/stabilization by these distinct molecules and the biological consequences in cells are likely different. HURP has been shown to be important for the stable MT-kinetochore attachment and MT-chromatin association, in part through the Ran-Importin pathway (15, 33, 37). In contrast, Hice1 is preferentially distributed to the spindle pole regions (Fig. 1; see Fig. S1 in the supplemental material). Therefore, it appears that cells can regionalize distinct bundling activities along the axis of a bipolar spindle to allow a coordinated maintenance of spindle dynamics. Whereas HURP is a downstream effector of the Ran-Importin pathway, the upstream signaling molecules for Hice1 are currently unknown. Moreover, we cannot rule out the possibility that the bundling activity of Hice1 may be in part due to the overexpression of Hice1 in cells or the concentration-dependent oligomerization of His-Hice1 in vitro. Thus, the physiological significance of this activity needs to be further addressed. Interestingly, the potential Hice1 binding partner Hec1 also possesses MT-bundling activity when dimerized with Nuf2 in vitro (3). Whether Hice1 and Hec1 may work together for spindle regulation during mitosis is currently under investigation.
Spindle pole instability is a hallmark phenotype in Hice1-depleted cells. Other than the direct role in regulating MT/spindle dynamics, Hice1 may additionally serve as a mitosis-specific “cohesin” for the spindle poles to safeguard their integrity, in a manner similar to that of Kizuna, a Plk1 target (25), since Hice1 is a component of the centrosome. Alternatively, Hice1 may function to restrain the activity of pole-pulling motors to maintain the equilibrium of antagonizing mechanochemical forces on the spindle poles. Abrogation of these potential regulatory mechanisms could cause fragmentation of the mitotic spindle poles. Spindle pole fragmentation was previously observed in cells defective in Aurora A, TPX2, or Kizuna (6, 10, 25). Whether these molecules are functionally related to Hice1 is not clear at present. Intriguingly, in our preliminary study, we found that Hice1 associates with Aurora A in the same immunocomplex, suggesting that Hice1 may function in the Aurora A regulatory pathway. Additional studies need to be performed to fully unveil the Hice1-associated molecular network in the regulation of MT dynamics, pole cohesion, and/or spindle bipolarity.
The mitotic delay caused by Hice1 depletion is consistent with spindle checkpoint activation. It is possible that residual Hice1 remains in the cell after RNAi treatment or that other molecules may functionally substitute for the absence of Hice1, thus allowing mitotic division to continue in a compromised yet delayed manner.
Homology searching by BLAST suggests that Hice1 has potential homologues in vertebrates but not in lower eukaryotes such as fly, worm, and yeast. We surmise that Hice1 emerges at a later stage in evolution as a higher-eukaryote-specific modulator, thus serving as an additional layer for mitotic spindle regulation. Interestingly, the Hice1 genomic locus (19p13.11) has been shown to undergo amplification in some brain tumors and high-grade serous ovarian cancers (26, 29). Consistently, we observed that Hice1 is overexpressed in several cancer cell lines from the NCI-60 cancer cell panel as well as in additional breast and ovarian cancer cell lines (unpublished data). Given its novel mitotic role, the genomic alteration and overexpression of Hice1 may be relevant to tumorigenesis.
Supplementary Material
Acknowledgments
We thank Chi-Fen Chen, Phang-Lang Chen, Xiaoqin Lin, Ning Ru, and Ulla Bengtsson for technical assistance and suggestions and Lily Khidr for critically reading the manuscript. We are grateful to Tim Yen and Song-Tao Liu for their insightful suggestions.
G. Wu is supported by a postdoctoral fellowship from the Susan G. Komen Breast Cancer Foundation. R. Wei is supported by a DOD predoctoral fellowship. This work is supported by a grant from NIH (CA 107568) to W.-H.L.
The authors declare that they have no competing financial interests.
Footnotes
Published ahead of print on 24 March 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersen, S. S. 2000. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 10261-267. [DOI] [PubMed] [Google Scholar]
- 2.Brinkley, B. R. 2001. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 1118-21. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman, I. M., J. S. Chappie, E. M. Wilson-Kubalek, and A. Desai. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127983-997. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y., Z. D. Sharp, and W. H. Lee. 1997. HEC binds to the seventh regulatory subunit of the 26 S proteasome and modulates the proteolysis of mitotic cyclins. J. Biol. Chem. 27224081-24087. [DOI] [PubMed] [Google Scholar]
- 5.Cho, H. P., Y. Liu, M. Gomez, J. Dunlap, M. Tyers, and Y. Wang. 2005. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol. Cell. Biol. 254541-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Luca, M., P. Lavia, and G. Guarguaglini. 2006. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle 5296-303. [DOI] [PubMed] [Google Scholar]
- 7.Furuta, S., X. Jiang, B. Gu, E. Cheng, P. L. Chen, and W. H. Lee. 2005. Depletion of BRCA1 impairs differentiation but enhances proliferation of mammary epithelial cells. Proc. Natl. Acad. Sci. USA 1029176-9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaglio, T., A. Saredi, J. B. Bingham, M. J. Hasbani, S. R. Gill, T. A. Schroer, and D. A. Compton. 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135399-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaglio, T., A. Saredi, and D. A. Compton. 1995. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 131693-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett, S., K. Auer, D. A. Compton, and T. M. Kapoor. 2002. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 122055-2059. [DOI] [PubMed] [Google Scholar]
- 11.Gruss, O. J., and I. Vernos. 2004. The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 166949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayward, D. G., and A. M. Fry. 2006. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 237155-166. [DOI] [PubMed] [Google Scholar]
- 13.Karsenti, E., and I. Vernos. 2001. The mitotic spindle: a self-made machine. Science 294543-547. [DOI] [PubMed] [Google Scholar]
- 14.Kline-Smith, S. L., and C. E. Walczak. 2004. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell 15317-327. [DOI] [PubMed] [Google Scholar]
- 15.Koffa, M. D., C. M. Casanova, R. Santarella, T. Kocher, M. Wilm, and I. W. Mattaj. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16743-754. [DOI] [PubMed] [Google Scholar]
- 16.Kops, G. J., B. A. Weaver, and D. W. Cleveland. 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5773-785. [DOI] [PubMed] [Google Scholar]
- 17.Lee, G., R. L. Neve, and K. S. Kosik. 1989. The microtubule binding domain of tau protein. Neuron 21615-1624. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. Y., Y. Obata, M. Yoshida, E. Stockert, B. Williamson, A. A. Jungbluth, Y. T. Chen, L. J. Old, and M. J. Scanlan. 2003. Immunomic analysis of human sarcoma. Proc. Natl. Acad. Sci. USA 1002651-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396643-649. [DOI] [PubMed] [Google Scholar]
- 20.Maiato, H., J. DeLuca, E. D. Salmon, and W. C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 1175461-5477. [DOI] [PubMed] [Google Scholar]
- 21.Merdes, A., K. Ramyar, J. D. Vechio, and D. W. Cleveland. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87447-458. [DOI] [PubMed] [Google Scholar]
- 22.Mishima, M., S. Kaitna, and M. Glotzer. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 241-54. [DOI] [PubMed] [Google Scholar]
- 23.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 221-32. [DOI] [PubMed] [Google Scholar]
- 24.O'Regan, L., J. Blot, and A. M. Fry. 2007. Mitotic regulation by NIMA-related kinases. Cell Div. 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshimori, N., M. Ohsugi, and T. Yamamoto. 2006. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 81095-1101. [DOI] [PubMed] [Google Scholar]
- 26.Park, J. T., M. Li, K. Nakayama, T. L. Mao, B. Davidson, Z. Zhang, R. J. Kurman, C. G. Eberhart, M. Shih Ie, and T. L. Wang. 2006. Notch3 gene amplification in ovarian cancer. Cancer Res. 666312-6318. [DOI] [PubMed] [Google Scholar]
- 27.Pihan, G. A., and S. J. Doxsey. 1999. The mitotic machinery as a source of genetic instability in cancer. Semin. Cancer Biol. 9289-302. [DOI] [PubMed] [Google Scholar]
- 28.Raemaekers, T., K. Ribbeck, J. Beaudouin, W. Annaert, M. Van Camp, I. Stockmans, N. Smets, R. Bouillon, J. Ellenberg, and G. Carmeliet. 2003. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 1621017-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roversi, G., R. Pfundt, R. F. Moroni, I. Magnani, S. van Reijmersdal, B. Pollo, H. Straatman, L. Larizza, and E. F. Schoenmakers. 2006. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene 251571-1583. [DOI] [PubMed] [Google Scholar]
- 30.Saffin, J. M., M. Venoux, C. Prigent, J. Espeut, F. Poulat, D. Giorgi, A. Abrieu, and S. Rouquier. 2005. ASAP, a human microtubule-associated protein required for bipolar spindle assembly and cytokinesis. Proc. Natl. Acad. Sci. USA 10211302-11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawin, K. E., K. LeGuellec, M. Philippe, and T. J. Mitchison. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359540-543. [DOI] [PubMed] [Google Scholar]
- 32.Schatz, C. A., R. Santarella, A. Hoenger, E. Karsenti, I. W. Mattaj, O. J. Gruss, and R. E. Carazo-Salas. 2003. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 222060-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sillje, H. H., S. Nagel, R. Korner, and E. A. Nigg. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16731-742. [DOI] [PubMed] [Google Scholar]
- 34.van Vugt, M. A., and R. H. Medema. 2005. Getting in and out of mitosis with Polo-like kinase-1. Oncogene 242844-2859. [DOI] [PubMed] [Google Scholar]
- 35.Wei, R. R., J. Al-Bassam, and S. C. Harrison. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 1454-59. [DOI] [PubMed] [Google Scholar]
- 36.Wittmann, T., M. Wilm, E. Karsenti, and I. Vernos. 2000. TPX2, a novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 1491405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong, J., and G. Fang. 2006. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, G., W. H. Lee, and P. L. Chen. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. J. Biol. Chem. 27530618-30622. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, W. M., A. Seki, and G. Fang. 2006. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 173881-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.