Abstract
This paper outlines prospects for applying the emerging techniques of synthetic biology to the field of anatomy, with the aim of programming cells to organize themselves into specific, novel arrangements, structures and tissues. There are two main reasons why developing this hybrid discipline – synthetic morphology – would be useful. The first is that having a way to engineer self-constructing assemblies of cells would provide a powerful means of tissue engineering for clinical use in surgery and regenerative medicine. The second is that construction of simple novel systems according to theories of morphogenesis gained from study of real embryos will provide a means of testing those theories rigorously, something that is very difficult to do by manipulation of complex embryos. This paper sets out the engineering requirements for synthetic morphology, which include the development of a library of sensor modules, regulatory modules and effector modules that can be connected functionally within cells. A substantial number of sensor and regulatory modules already exist and this paper argues that some potential effector modules have already been identified. The necessary library may therefore be within reach. The paper ends by suggesting a set of challenges, ranging from simple to complex, the achievement of which would provide valuable proofs of concept.
Keywords: biotechnology, genetic engineering, synthetic biology, synthetic morphology, tissue engineering
Introduction
Anatomy is a mainly analytical science, being concerned with determining natural morphology and how it develops. Unlike subjects such as chemistry and solid-state physics, anatomy has so far lacked a strong synthetic aspect. In this article, I will argue that recent developments in basic developmental biology and in synthetic biology can now be combined to provide a foundation for a ‘synthetic morphology’, in which cells can be programmed to organize themselves into specific, designed arrangements, structures and tissues. This idea of internally programmed self-organization stands in contrast to the externally imposed manipulations of surgery, which is the closest thing we currently have to a synthetic application of anatomy. Synthetic morphology, if it can be brought into being, has the potential to expand dramatically the range of possibilities of tissue engineering, both extra- and intracorporeal. It also has the potential to play a very important role in the verification of the results of basic developmental biology: theories of morphogenesis are routinely deduced from observation of normal and mutant embryos, but only when the theories are applied to the creation of novel, designed forms will they have been properly tested. As the physicist Richard Feynmann once said, ‘What I cannot create, I do not understand’ (Gleick, 1992).
This review concentrates on the first steps towards useful synthetic morphology, rather than on its potential long-term applications. It is intended to provoke discussion about how we might progress to the first, crude, proof-in-principle demonstrations of synthetic morphological systems. These first demonstrations will probably just be populations of engineered cells that have the ability to organize themselves in very simple ways. They might, for example, arrange themselves at a set distance from the source of a signalling molecule, or they might clump together in response to a signal, or they might clump together but disperse when their population reaches a particular size. They would do this not because this behaviour is a natural property of the parent cell type, but because they have been engineered with synthetic gene circuits in which this behaviour has been designed.
It should be noted that this approach is quite distinct from the ‘reprogramming’ of stem cells, for example for the purposes of regenerative medicine. The guiding principle of stem cell manipulation is that the genome of these cells already contains the complete ‘genetic programme’ for making all of the cell types in an embryonic and adult body. The (rather poorly chosen) word ‘reprogramming’ is used in this context to mean setting the state of the stem cells to some desired state of their existing developmental–genetic programme (e.g. the state of gene expression that corresponds to being a neural progenitor cell). The principle of the work described in this paper, by contrast, is to create entirely novel genetic programmes that do not already exist in any cell. To be clear, these novel programmes may use basic existing cell biological components common to all cells (e.g. guided self-assembly of actin filaments), but not ‘developmental’ modules that are present in only some times and places in embryos. If a normal mammalian tissue is desired, it is sensible to use stem cell approaches: synthetic morphology is intended to create structures that do not exist in any normal developmental programme. It should also be noted that there is no reason why the development of morphologies in designed systems should be based closely on how similar structures develop in evolved systems when another way would be more efficient. Part of the point of synthetic morphology is that it provides a means of escaping evolved life's historically fixed constraints.
At least within the proof-of-principle systems discussed in this article, the formation of novel structures will take place in culture. Initially, these structures will be very simple, and will consist, for example, of aggregates of cells (two- or three-dimensional, depending on the culture system used), or aligned chains of cells, or arrays of alternating cell types, or arrangements of cells in one state surrounded by those in another state, or sheets of cells that fold or enclose space. Cells will make decisions about changes of state, morphology and cell–cell interaction in response to their environment and in response to signals emanating from other cells: this autonomous decision-making is the basis of their ability to organize themselves without direct intervention by an experimeter. The cells will therefore need systems to achieve actual morphogenesis (cell adhesion, cell shape, motility, etc), mechanisms to detect the environment and signals, the means to produce signals, and a mechanism for intergating environmental and state information and using it to control morphogenesis (Fig. 1). The rest of this article describes possible ways in which these mechanisms can be realized with existing biotechnology.
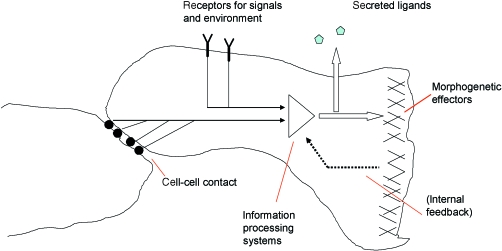
Fig. 1.
An overview of the types of mechanism needed for synthetic morphology. In the case illustrated, signals from cell–cell contacts and receptors for a secreted ligand have been combined by information-processing systems to produce two responses, one morphogenetic (cell motility) and the other the production of a new signalling ligand.
It will be important that the first proof-of-principle demonstrations of synthetic morphology hold to a general design strategy that will be extensible to larger and more complex (and ultimately useful) projects. The design strategy advocated here is based closely on ideas that have proved successful in mechanical and electronic engineering. The most important of these ideas are standardization of components and modularity of construction. The dramatic expansion of mechanics in the nineteenth century depended on the development of interchangeable, standardized components (nuts, bolts, pipes, wheels, gears, bearings, etc.) that were available ‘off the shelf’ and could be used in different ways to produce different structures and machines. This freed inventors, quite literally, from the need to ‘reinvent the wheel’ for each application. Similarly, the rise of the electronics industry relied on the production of a variety of standard components (resistors, capacitors, plugs, valves, transistors, logic gates, etc.) that could be connected together in different ways to produce different machines such as radio receivers, gramophones or computers, with only a tiny number of components, if any, having to be custom-designed for each application. In both fields, modular devices can be connected to produce modular sub-assemblies (gearboxes, amplifiers, etc.) that can in turn be connected in different ways for different purposes.
The probable complexity of engineering that will be involved in synthetic morphology, coupled with the large number of morphologies that may be wanted, argues strongly for a similar approach to be taken from the outset: economy with money and time demands that the necessary (genetic) engineering be done with standardized components that can be interconnected in different ways. The field of existing engineering that is formally closest to synthetic morphology, in terms of what it sets out to do, is arguably robotics because robots process information to effect changes of form or arrangement in three-dimensional space. In robotics, modules can be divided into three broad classes (Wiener, 1948). The class that tends to be thought about earliest in the design phase, because they actually perform the mechanical task required of the robot, is that of ‘effector modules’. These modules consist of the motors, levers, etc., that actually produce shape change. They are controlled by the class that tends to be designed next in the process, the ‘information- processing modules’ that control the effector modules according to a combination of internal programmes and external information. These modules are provided with information by what is usually the last class to be considered during design, the ‘sensory modules’, (pressure detectors, light detectors, etc.) that provide data for the information processing modules (Fig. 2).
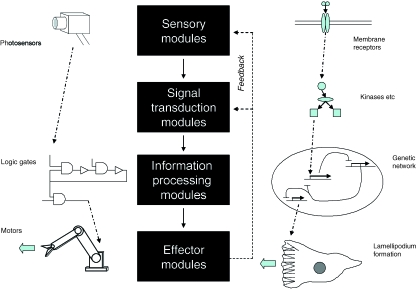
Fig. 2.
A comparison of the types of modules already used in robotics (left column) and proposed for synthetic morphology (right column). In the cases illustrated, a light causes a robot arm to move, and a signalling molecule causes a cell to move. In both cases, the information processing modules drawn, constructed from NAND gates and gene promoters, respectively, include a set–reset memory latch.
It seems reasonable that synthetic morphology will also require modules of these three types. Effector modules will control such things as cell shape, motility, interaction and replication. Information processing modules will control effector modules according to sensory data and stored memory. Sensory modules will detect such things as growth factors, cytokine gradients and other cells. In both systems, therefore, there are three types of module, which are (named in the order used when the robots were described above) (1) effector, (2) information processing and (3) sensory (Fig. 2). In biological systems, these types of modules can be more closely entwined than in typical engineering, with one element being used for more than one function (microfilaments, for example, play a role in both sensory and effector systems). In this article, the splitting of biological systems into these formal units is therefore a simplification for the sake of clear discussion. That said, entwined functions will normally be avoided in synthetic morphology, to limit the danger of cross-talk (see below).
Eukaryotic biology is not normally described in terms of engineering modules. It is therefore not immediately obvious, from the ordinary language of biology, whether the knowledge and biotechnology to develop these modules exists. In the rest of this article, I shall argue that it does and that a substantial number of useful modules exist already.
Morphogenetic effector modules
Following the general strategy outlined above, a set of ideal morphogenetic effector modules would have two key properties: (1) they should, between them, cover most of the elementary morphogenetic mechanisms known to be available to cells and tissues, and (2) it should be possible to switch them on and off by manipulating a single master control gene, for ease of coupling them to information processing modules. The protein encoded by this master control gene should interact directly with the constitutive morphogenetic machinery of the cell, and not require any ‘developmental programmes’ in the host genome.
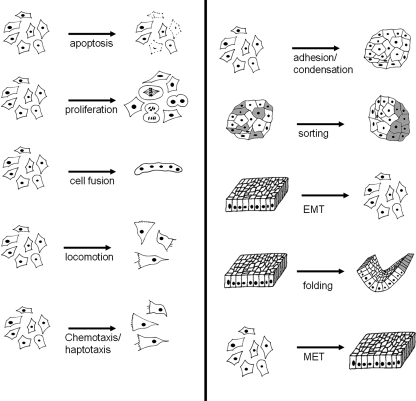
Conventional study of normal animal development has revealed a range of about ten basic morphogenetic mechanisms that are used over and over again at different times and places in the embryo. These mechanisms are: elective cell death, cell proliferation, cell fusion, cell locomotion (chemotaxis, haptotaxis) adhesion, condensation, sorting, epithelial-to-mesenchymal transition (EMT), mesenchymal-to-epithelial transition (MET) and epithelial folding (Davies, 2005) (Fig. 3). In developing embryos, these mechanisms are invoked in precise combinations and sequences to create anatomical form. The development of a renal nephron, for example, involves proliferation, condensation, MET, apoptosis, proliferation and folding in that sequence (Davies & Bard, 1998).
Fig. 3.
Ten basic cellular mechanisms of animal morphogenesis. The development of most animal tissues, organs and bodies occurs by a combination of these events, each acting to a strictly controlled extent and in a strictly controlled sequence. Using these events as morphogenetic effector modules should therefore give synthetic morphology a great range of possible designed anatomies.
Morphogenetic mechanisms involve a large number of cellular components that interact at a variety of scales. Proteins form complexes by self-assembly, the probability of which is modulated by other proteins that generally operate in feedback loops, set up by yet other proteins, to organize structure and behaviour (Davies, 2005). Basic research into cell biology, usually done for reasons unrelated to synthetic morphology, has demonstrated that the molecular-scale processes that are responsible for each of the mechanisms in Fig. 3 can be induced to occur in cultured animal cells by the forced expression of a single specific gene, which will be referred to in this paper as the mechanism's ‘driver’ gene. This is typically a non-constitutive signalling molecule, adhesion molecule or transcription factor that can, when expressed or activated, organize the directed self-assembly of housekeeping proteins that are constitutively present in the cell (actin, for example). Some driver genes and the cells in which they have so far been shown to work are shown in Table 1. Not all drivers have been shown to work in the same cell lines yet, which is a current weakness of the information in the table. Helpfully, however, some have been shown to work in Chinese hamster ovary (CHO) cells, which happen to have been much used for the testing of synthetic information processing modules (see below). Here, it will be assumed that each mechanism will work in all of these cells, although it is acknowledged that finding this out experimentally is an important priority because any mechanism that requires a peculiar and cell-type-specific set of cytoplasmic factors to be present will not be very useful and will need to be replaced.
Table 1.
Potential ‘morphogenetic modules’ that can be activated by an identified master regulator (‘driver’, in the table)
| Morphogenetic event | Driver | Shown in | Reference(s) |
|---|---|---|---|
| Apoptosis | Nedd2 | NIH-3T3 (fibroblasts), N18 (neuronal lineage) | (Kumar et al. 1994) |
| Cell proliferation | P27kip (to switch off) (presence of survival factors is assumed) | Rat1 (fibroblasts) | (Vlach et al. 1996) |
| Cell fusion | C. elegans EFF-1, or cytomegalovirus gH/gL glycoproteins | Sf9 cells (insect), CHO cells (ovary) | (Podbilewicz et al. 2006) (Kinzler & Compton, 2005) |
| Cell locomotion | CAS/Crk | COS cells (fibroblast) | (Klemke et al. 1998) |
| Chemotaxis | Transfection with CCR4, in a gradient of CKLF1 | HEK293 cells (kidney, probably neuronal lineage) | (Wang et al. 2006) |
| Haptotaxis | External fibronectin gradient (CHO) | CHO cells | (Rhoads & Guan, 2007) |
| External collagen gradient (3T3) | NIH-3T3 cells | (Sells et al. 1999) | |
| Cell–cell adhesion/ condensation | E-cadherin | L cells (fibroblasts) | (Nagafuchi et al. 1987) |
| Cell sorting | E- or P-cadherin in different cell types, or the same cadherin in different amounts. | L cells (fibroblasts) | (Nose et al. 1988; Friedlander et al. 1989; Collares-Buzato et al. 1998) |
| MDCK cells (epithelium) probably | |||
| Epithelial→Mesenchymal transition | LMP1 | MDCK cells (epithelium) | (Horikawa et al. 2007) |
| Epithelial folding (by apical constriction) | Shroom | MDCK cells (epithelial), Xenopus blastomeres | (Haigo et al. 2003; Hildebrand, 2005) |
The ‘cell types’ column lists cells in which the process has been shown to work. Most examples require only internal factors, but the two concerned with guidance – chemotaxis and haptotaxis – require external guiding gradients of the diffusible or matrix molecules mentioned in the table. These might be produced by other cells in the system (which could, for example, secrete CKLF-1).
It should be stressed that these basic morphogenetic events are probably activated in real embryos by a variety of different genes and, furthermore, that some of the genes listed in Table 1 are very unlikely to be much used for this purpose in real embryos. For synthetic morphology, this does not matter: all we need is one reliable way to activate each process, and attempting to emulate the vast numbers of pathways in real, evolved, animals would work against the engineering principle of using standardized modules wherever possible.
The modules listed in Table 1 therefore provide at least the first crude basis for a library of effectors that can be switched on by the activation of a single gene by the information processing modules, and are thus a means for decisions made by information processing modules to be realized in changes of anatomical form. It should be noted that, in terms of the engineering analogies in the Introduction, the modules in Table 1 are not intended to correspond to basic components (transistors, resistors, etc), but rather to higher-level assemblies of these.
Information processing modules
The role of information-processing modules will be to integrate information about a cell's current environment, including cell-to-cell signalling, with information about previous states (‘memory’), and to trigger appropriate effector modules. In the simplest of cases, where there is a direct link between one signal and one morphogenetic response, there may not be a need for information processing modules. Where signals have to be combined, however, and especially where they have to be integrated with memory about previous states or events, information processing modules will be required.
There are two broad ways in which information can be processed (computed), analog and digital. Analog computation has the advantage that it is faster than digital (comparing machines built from the same mechanical, electronic, optical or biological technology) and that complex mathematical calculations, particularly calculus, can be performed by technically simple units. It has the disadvantage, however, that it has poor immunity to noise and that modules tend to be custom-designed rather than general-purpose [although some electronic analogue computers of the 1950s–1970s were almost general-purpose machines (Lang, 2000)]. Memory can also be difficult to implement. Digital computation, using Boolean algebra, tends to use standard units and shows great immunity to noise, which is why almost all computation is nowadays digital and analog computation exists only in niche applications such as fast signal processing (Small, 2001). For biological engineering, the advantages of noise immunity and easy modularity are joined by the ease with which digital mechanisms can be tested; the standard methods for detecting gene expression, such as PCR or Western blotting, are markedly non-linear and it is much easier to verify an on/off state choice than to make an accurate measurement of an analog concentration. For these reasons, existing work in synthetic biology has been concentrated on building digital information-processing modules, and these form a natural resource for synthetic morphology.
Boolean algebra, the combinations of inputs to set an output by rules such as ‘output is on if input A AND input B is on’, is the foundation for most digital computing (whether mechanical, electronic or biotechnological). Several information-processing genetic networks, based on Boolean algebra, have already been developed for both prokaryotes and eukaryotes. To save space, this section will illustrate each type of module with only eukaryotic examples whenever possible. It should be noted that many of the proof-of-principle modules described below have been designed, for ease of testing, to respond to small molecules that can be applied to cells by an experimenter. Many of these happen to be antibiotics, because several gene control systems that are sensitive to specific antibiotics have been well characterized and are easy to work with, but the antimicrobial action of these molecules is irrelevant to the eukaryotic host cells and they are just used as signals. They were chosen because they do not naturally occur in the cells, so there is no risk of unwanted cross-talk with normal cellular physiology.
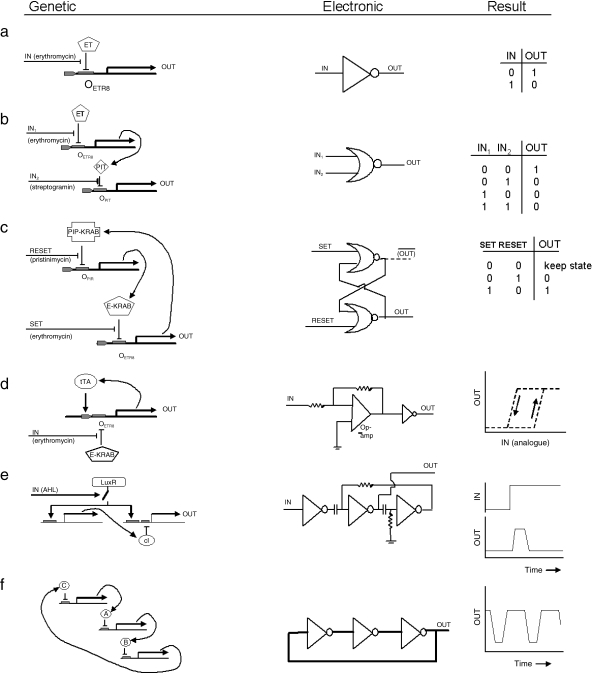
The genetic equivalents of electronic Boolean logic gates can be constructed by combining promoter elements upstream of an output gene. A simple NOT gate (inverter: output is on when input is off and vice versa) can be constructed by placing an output gene under the control of a minimal promoter that also contains the ETR8 operator site, to which the erythromycin-blockable transcriptional activator ET can bind (Fig. 4a). In the absence of erythromycin, ET binds its ETR8 operator site and the output gene is ‘on’. In the presence of erythromycin, ET is incapable of binding ETR8 and the output gene is ‘off’. A NOR gate [output = NOT (A OR B)] can be constructed by connecting two of these biological gates in series, so that the erythromycin-controlled promoter gate drives expression of the streptogramin-blockable transcriptional activator PIT, and the ultimate output gene is placed under the control of a minimal promoter containing the PIR operator site to which PIT can bind (Fig. 4b). In the absence of either erythromycin or streptogramin, PIT is made and the output gene is ‘on’. In the presence of erythromycin, PIT is never made so cannot activate the ouput gene, while in the presence of streptogramin PIT cannot act even if it is made. The output is therefore ‘on’ when neither erythromycin NOR streptogramin is present. The possibility of constructing NOR gates is important, because it can be formally proved that any system of computational logic can be constructed solely by the use of NOR gates [de Morgan's theorem shows that any logic element can be constructed from NOR and NOT, and as connecting the two inputs of a NOR gate together makes it behave as a NOT gate, any logic circuit can be constructed from NOR gates (Devlin, 2004)]. It is important to note that the logic gates described above are not mere conjecture, but have already been constructed and shown to work in CHO cells (Kramer et al. 2004a).
Fig. 4.
A comparison of the genetic and electronic implementations of (a) a NOT gate, (b) a NOR gate, (c) a set–reset latch, (d) a Schmidt trigger, (e) a pulse generator and (f) a ring oscillator. The function of each logic circuit is depicted in the truth tables and graphs in the right-hand column. The line above ‘OUT’ in Fig. 3c indicates a complementary output, that will be 0 when OUT is 1, and vice versa.
As well as requiring Boolean logic, information processing systems require a facility for storing state information in a memory. Although memory can be stored by direct modification of DNA in prokaryotes (Blenkiron et al. in press), the simplest way to construct memory elements in mammalian cells is probably to follow the history of electronics and to construct it by combining logic gates. A simple memory unit, the set–reset latch, can be constructed from two NOR gates (Fig. 4c). A biological version can be constructed by placing the erythromycin-blockable transcriptional repressor, E-KRAB, under the control of a constitutively active promoter that can be repressed by the pristinamycin-blockable repressor PIP-KRAB, and vice versa. In this network, each repressor tries to repress the other gene and thus to allow its own production to continue. There are therefore two stable states, E-KRAB ‘on’ and PIP-KRAB ‘off’, and E-KRBA ‘off’ and PIP-KRAB ‘on’. In this particular system, they can be toggled by adding either erythromycin or pristinimycin to the system, but they could also be toggled, in principle, by adding further elements to their promoters so that a repressor produced as the output from another module forces the latch to go into a particular state (e.g. by repressing transcription of E-KRAB). Again, it should be stressed that this latch has actually been constructed and shown to work in CHO cells (Kramer et al. 2004b).
The information-processing modules described above work ‘digitally’ but, if interfaced with sensory modules, they will need to respond to continuously variable ‘analog’ signals and make an ‘on’/’off’ decision based on threshold(s) of these signals. In electronics, this is solved by devices such as the Schmidt trigger, the digital output of which switches from ‘off’ to ‘on’ at a critical threshold of analog input, and remains ‘on’ until the analog input falls below a slightly lower threshold (this hysteresis ensures stability and avoids vacillation in the presence of noisy inputs). Artificial gene networks have been developed that mimic this behaviour. In one, a gene encoding the transcriptional activator tTA is placed under the control of a promoter that is itself activated by tTA but inhibited by the constitutively expressed erythromycin-blockable transcriptional repressor, E-KRAB: repression is dominant over activation (Fig. 4d). In the absence of erythromycin, E-KRAB is active and represses production of tTA so that this protein is present only at very low levels. Very low levels of erythromycin do not affect this situation significantly, but once enough erythromycin is present to allow transcription of tTA to occur, levels of tTA rise sharply and positive feedback turns the module fully ‘on’. Small reductions in the concentration of erythromycin are not enough to switch the module off again, because the increased concentration of tTA has increased the activity of the promoter beyond its original level so it needs harder repression. There is therefore considerable hysteresis in the system and it switches off at a much lower concentration of erythromycin than was needed to switch it on. Again, this system has been constructed and verified in CHO cells (Kramer & Fussenegger, 2005).
In prokaryotes, more complex modules have been constructed that turn ‘on’ at a particular concentration of external signal and off again at a higher concentration, so that they are ‘on’ only within a concentration range. This behaviour could be used, for example, to cause cells to respond at a particular point on a morphogen gradient. Modules such as these can be adapted to operate in the temporal instead of the spatial domain. Some of the components used in the prokaryotic band-detecting module mentioned above have been connected to form a module that responds to a rising signal of the signalling moleule acyl homoserine lactone (AHL) by producing an ‘on’ pulse and then returning to ‘off’ even if the stimulus remains (Basu et al. 2004). In the module (Fig. 4e), the output gene is activated by AHL-bound LuxR but inhibited by cI (which will always ‘win’), the transcription of which is itself activated by AHL-bound LuxR. In the absence of AHL the output of the module is ‘off’. AHL causes transcription of the output gene and of cI to be turned ‘on’, but once enough time has elapsed for cI to be transcribed and translated, this molecule switches the output gene off again, even though AHL continues to be present.
Synthetic gene networks can be used to initiate, as well as to respond to, signals in the temporal domain. A well-known prokaryotic module that achieves this works very much like a ‘ring oscillator’ (an oscillator that consists of identical elements connected in a ring) in electronics (Elowitz & Leibler, 2000). Three promoters, each controlling the expression of a transcriptional repressor, are connected in a loop so that A represses B, B represses C and C represses A (Fig. 4f). Activity at promoter A represses B (with a time delay as the repressor protein downstream of promoter A is synthesized) and the repression of B allows activation of C (with a time delay as the repressor protein downstream of promoter B decays). The activation of C then represses promoter A (with a time delay as the repressor protein downstream of promoter A is synthesized). After all of these time delays, the activity has therefore moved ‘backwards’ through the network, so that C is active and A repressed. This situation is just as unstable, and presently B will be active and C repressed, and sometime later the state will have returned to A being active; as long as the rates are synthesis and decay of the proteins are chosen carefully, the system continues to oscillate.
The main advantage of the transcription-based modules described above is that they can be well-insulated from interference by endogenous cellular systems. Their main disadvantage is that they are relatively slow (tens of minutes per stage). Cytoplasmic signal transduction systems, based on chemical reactions such as phosphorylation, can process information much more quickly (seconds) but they are harder to insulate. Where fast processing is required, it may be necessary to use signalling proteins from phylogenetically distant sources, to minimize the risk of unwanted cross-talk.
Many of the systems described above have been constructed as proofs of principle, and therefore respond to simple experimentally applied molecules such as antibiotics. They could, however, respond just as easily to changes in production of the transcription factors themselves, and these could of course be produced as the outputs of other modules. These simple logic modules can therefore be connected and cascaded into more complex machines, although at the cost of time delays and a need for a large number of transcription factors. This is the price of modular construction: the alternative price of using more efficient units designed only for the task in hand (which is more evolution's way) is the lack of engineering flexibility in allowing standard modules to be connected. The final outputs of information-processing modules can be activation of driver genes that control the morphogenetic effector modules described above. They can also be used to control the production of signalling molecules (e.g. cytokines) that will interface with the sensory modules described below.
Sensory modules
Sensory modules are needed for two purposes: to detect signals generated by cells of the system, and to monitor physical or chemical aspects of the environment. Both aspects will be considered here, cell–cell signalling first.
For synthetic morphological systems that operate independently and not in the context of a host body, signals and their sensors may be chosen from normal mammalian growth factors and receptors that are not normally expressed by the engineered cells. CHO cells, for example, do not normally express TrkA neurotrophin receptors but if they are transfected with genes encoding one of these then they become sensitive to nerve growth factor (NGF) or its chemical mimics (Wilkie et al. 2001). Signals and receptors could either be secreted molecules, intended to operate over a long range as NGF would, or membrane-bound molecules such as Notch/Delta and Eph/Ephrin, which operate only between cells that are in contact with one another.
With this approach, however, it is essential to ensure that none of the cell states of the synthetic system risks activation of the normal genes encoding these growth factors. This risk can be avoided by using signalling systems that do not exist in the host genome (and this approach will have to be used for any synthetic system that has to operate in the context of a host animal). Suitable signalling systems can sometimes be obtained from organisms of another phylum or kingdom. An early example of cross-kingdom transfection was the transfection of a functional mammalian G-protein-coupled receptor and a G protein subunit, responsive to agonists of the β-adrenergic pathway, into yeast cells (King et al. 1990). The result was that the yeast cells would respond to these agonists as if they were responding to their own pheromones. Yeast has also been engineered with receptors from plants, to make the host cells responsive to plant cytokinins (Chen & Weiss, 2005). There is every reason to suppose that the reciprocal transfections could be done, so that mammalian cells could be rendered sensitive to exotic signalling molecules such as cytokinins. These could, of course, be synthesized by other cells by transfection of appropriate synthetic machinery, thus setting up paracrine cell–cell signalling (or autocrine signalling if this is desired).
Expression of signalling molecules (of whatever origin) can be coupled to information-processing modules by placing a gene encoding a secreted growth factor, or a gene encoding an enzyme responsible for synthesizing a small signalling molecule, under the control of a promoter that is regulated by an output of the information-processing modules. Unlike the wires used in electronic engineering, which transmit signals from point to point, diffusible ligands broadcast signals to all cells that are equipped with appropriate receptors. In a complex system that has cells in different states that need to communicate for different purposes, it may be necessary to use a variety of ligand–receptor systems to eliminate the risk of cross-talk and interference (for some purposes, all ligands may be diffusible, whereas for others some may be membrane-bound so that they affect only adjacent cells). Cells may express the receptors for all of the ligands all of the time, or may modulate their expression of receptors (under the control of information-processing modules) according to cell state. It is also possible for some signals to work by inhibiting others – for example the secretion of a soluble form of a receptor to compete with the membrane-bound form, or of a function-blocking antibody by one cell type can be used to interfere with signalling between other cells.
Detectors of the physico-chemical environment have already been transferred between prokaryotic organisms and connected to host responses. A dramatic example is provided by the engineering of Escherishia coli with light receptors to create lawns of bacteria that operate rather like photographic film (Levskaya et al. 2005). Work in prokaryotes is also leading the way in the rational design of receptors for chemical ligands for which no known biological receptors exist (Looger et al. 2003).
Methods for placing reporter genes under the control of specific mammalian receptor-driven signal transduction pathways are commonplace. Promoters have been constructed that place mammalian genes under the control of the NFAT pathway or the glucocorticoid pathway (Mattheakis et al. 1999). Some of pathway-regulated promoters are even available in ‘kit’ form; examples include control by the NF-κB pathway (e.g. the NF-κB/293/GFP™ kit from System Biosciences) and by the canonical Wnt pathway (e.g. ‘TopFlash’ and ‘FopFlash’, from Upstate Biotechnology). The construction of membrane-bound forms of normally secreted protein ligands could be used to adapt a signalling system for detecting cell–cell contact. Kits are also available for controlling mammalian gene expression by small, non-protein molecules that do not normally occur in mammals, although they are synthesized by some other organisms and the synthetic machinery could be placed in mammalian cells. Examples of such small molecule controllers of gene expression are cumate (Q-biogene's ‘Q-mate’ kit), ecdysone (New England Biolabs’‘Rheoswitch’ kit) and cyclic AMP (George et al. 1997). These molecules could allow long-range signalling that will not interfere with mammalian physiological systems.
Challenges and proofs of principle
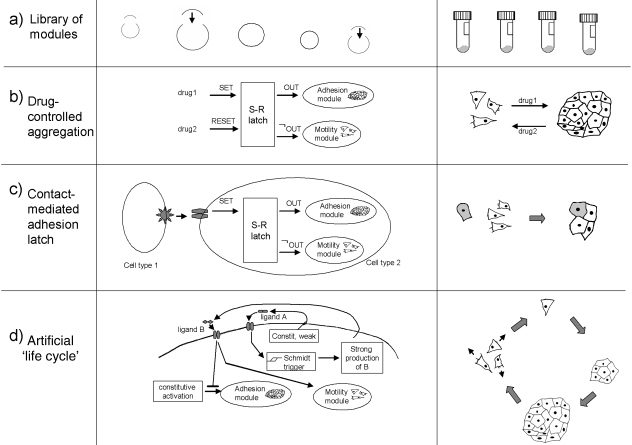
Synthetic morphology is likely to be a complex endeavour, adding as it does layers of complication on what are already difficult areas of genetic engineering. A set of proofs of principle, ranging from the comparatively easy to the very stretching, might therefore be useful to demonstrate the potential of the field before really useful, but very difficult, feats of tissue engineering are attempted. In this sense, ‘proof of principle’ means a demonstration that the assumptions made in this paper, or modified versions of them made in the light of experience, are valid and that synthetic morphological systems really can work in living cells. The last section of this paper therefore lists and discusses some possible milestones (Fig. 5).
Fig. 5.
Challenges and proofs of principle. (a) construction and testing of a library of morphogenetic effector modules, (b) a drug-controlled, latchable switch between single-cell and ‘tissue’ behaviour, (c) a contact-mediated switch between motility and adhesion and (d) one possible route to an artificial multicellular→unicellular life-cycle.
The proofs of principle suggested here are all intended to work in the simple environment of a Petri dish or culture flask in routine culture media, and to use single, simple engineered cell lines such as the fibroblast lines mentioned in Table 1. In such an environment, the morphology and behaviour (locomotion, adhesion, etc.) of genetically engineered cells would be easy to observe and record, and easy to compare with un-engineered control cells from the same cell line. Extracellular molecules, such as inducers of gene expression, would also be simple to apply. This environment has the advantage of immunity from the effects of other living cells and is therefore good for testing the basic function of synthetic systems. What would be ideal, in the long term, would to be create a ‘minimal mammalian cell’ (Murtas, 2007), the genome of which has lost its developmental programme entirely so that it is a blank slate free of the risk of unexpected, endogenous developmental responses to introduced programmes. This, however, would be far into the future [at the moment, the much simpler project of producing a minimal bacterium is stretching biotechnology to its limits (Forster & Church, 2006)].
The first experiments should be designed to verify the action of single morphogenetic effector modules, controlled at first by direct application of an extracellular inducer such as the antibiotics mentioned in the section on information-processing molecules. Morphological assays, which might include time-lapse studies on motility, clustering assays (comparing the scatter of cells induced to activate an adhesion mechanism with controls), and assays of cell multiplication or death or of sorting of marked cells, would be used to verify the function of appropriate morphogenetic effector modules in a variety of cell types. Once this has been done, more complex – and more autonomous – control systems can be built and introduced, the same assays being used to monitor their behaviour. At any stage, the techniques of immunostaining, reverse transcriptase PCR or Western blotting can be used to verify correct function of different elements of an engineered mechanism: a microarray could be used to monitor the entire system at a given timepoint.
The first experiments described above are designed to help with the most pressing practical problem in synthetic morphology, namely the construction and testing of a library of morphogenetic effector modules (in a standard plasmid/BAC form), each controlled by one driver gene, that have been shown to work reliably in a range of cultured cells. The systems listed in Table 1 would be a good starting point, but some may need to be replaced if it becomes clear that they are not reliable in many cell types. The achievement of this will be a major milestone in the very beginning of synthetic morphology. The building of sensory and information-processing libraries is also important, but this is taking place anyway for other applications (Drubin et al. 2007). Direct connection of sensory to motility effector modules could also be used to control the shapes of groups of cells by the use of external boundaries consisting of molecules capable of activating the sensory module and thus shutting down motility.
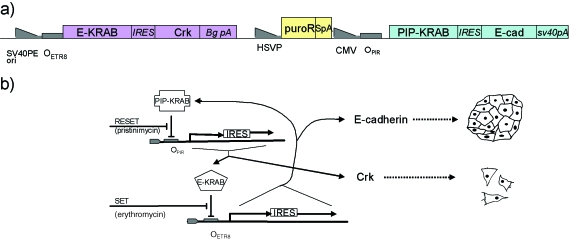
The next step might be the placing of these morphogenetic modules under the control of an information-processing module, for example the set–reset latch shown in Fig. 4(c). This could produce, for example, simple proof-of-principle mechanisms such as a cell line that can be flipped between migratory and sedentary behaviours by short pulses of a setting and resetting molecule in the medium (Fig. 5b). Placing an adhesion effector molecule under this control would make the step from engineering single-cell behaviour to engineering the collective bechaviour of cells (as adhesion would promote the formation of a multicellular mass, a requirement for the eventual formation of tissues). If adhesion and motility effectors were coupled to opposite outputs of the set–reset latch [the OUT and the NOT(OUT) in Fig. 4c], the difference between the motile, single-cell arrangement and the adhering, social arrangement would be exaggerated. A possible design for a construct to implement this is depicted in Fig. 6, although a final design can only be arrived at by careful experimentation as outlined in the figure legend.
Fig. 6.
Example of a possible genetic design for the drug-controlled aggregation illustrated in Fig. 5(b). (a) One possible layout of necessary transcribed elements (colours) and control elements (grey). In the version depicted, the promoters for the three transcribed elements are different (SV40PEori is the strong promoter/enhancer/origin of replication from SV40, CMV is the promoter/enhancer from cytomegalovirus and HSVP the weak promoter from herpes simplex virus, used to drive a resistance gene for selection of transfected cells). Using different promoters minimizes the risk of recombination, but carries the risk that the activity of the promoters may not be sufficiently balanced so that mutant forms may have to be screened for equal activity. Also to mimimize the risk of recombination, the use of multiple promoters of the same type has been avoided by the use of internal ribosomal entry sites (IRES) to produce two proteins from one transcript. The elements are insulated from one another by strong polyadneylation sites (BgpA from beta globin, SpA a synthetic form produced by Promega, and SV40pA from SV40): these also promote stability of mRNA. (b) The function of the construct shown in (a), as a network diagram.
Cell–cell communication could then be coupled to these systems. The first, simplest proofs of principle might be achieved by making the systems described above respond not to pulses of drugs in the medium but rather to a sensor for a membrane-bound ligand on the second cell type. Contact with the second cell type, in a mixed culture, could then mediate the switching of the first cell type from one behaviour to the other. This could be refined still further, by using just one cell type and engineering ligand expression to work in parallel with one of the effectors. In this way, for example, cells could be motile until they meet a cell that is in its adhesive state, in which case they will switch and join the growing aggregate of adhesive cells. If both migration and proliferation were to be switched firmly off when adhesion were activated, the system would then generate a branched fractal structure by a process analagous to diffusion-mediated aggregation (Vicsek, 1983; Mandelbrot, 1997; Oancea, 2007).
Differences in adhesion molecule expression can organize aggregated cells into inner and outer layers (Townes & Holtfreter, 1955). Coupling a contact-mediated signalling system that shows competition and lateral inhibiton [such as the Notch–Delta system (Muskavitch, 1994; Lewis, 1998)] to set a latch that controls cell adhesion molecule expression will cause initially identical cells in an aggregate to segregate into an inner cell group and an outer layer, creating a multilayered ‘tissue’. The system could either be allowed to operate only within a window of time (presence of drug, or endogenous timer) or could operate continuously as cells multiply. The size of aggregates would, however, be limited by diffusion paths of food, oxygen and waste products to and from the innermost cells.
Futher complex steps along this road could be taken to produce what is, in effect, an artificial multicelllular ‘organism’ with a life cycle. Consider a cell engineered with two morphogenetic effectors, cell–cell adhesion and motility, connected so that when one is off the other is on (as described above). This cell is also capable of producing two diffusible ligands, A and B. Ligand A is produced at a low level, constitutively. The Ligand B is produced only when a Schmidt trigger module (Fig. 4d) coupled to a sensor module for ligand A detects that the concentration of ligand A has reached a critical threshold: once production of B is activated, it is produced efficiently and in large quantities. A sensor for ligand B is coupled to regulatory elements so that, in the absence of ligand B, cells are adhesive and non-motile and in its presence they are motile and non-adhesive. Cell multiplication occurs constitutively. When a cell exists in isolation, it will be adhesive and non-motile, and will make small amounts of ligand A, which will diffuse away easily into the bulk medium. As the cell multiplies, it will found a bulky, three-dimensional colony of adhesive cells, in the centre of which the concentration of ligand A will start to rise (because the diffusion path away from the central cells, producing ligand A, is now partially obstructed by the aggregated cells). When the colony has reached a critical size, the concentration of ligand A in its core will exceed the threshold, strong production of ligand B will begin and all of the cells receiving this ligand (the whole colony) will switch from being adhesive to being non-adhesive and highly motile. The colony will therefore break up. Once they have scattered, however, the now-separated cells will experience low concentrations of A, production of B will cease and each cell will be able to found a new colony of its own. The ‘life cycle’ will therefore begin again.
Challenges such as these have no direct application, but if primitive synthetic morphology modules can be connected to create the first artificial life cycle, as described above, the potential power of the approach will have been demonstrated vividly enough that there will a firm foundation for taking the field on from making these little toys to doing something genuinely useful. Clinically useful products will probably centre on structures that are needed but that are outside the normal developmental repertoire (and therefore outside stem cell-based approaches, as explained in the Introduction). The first to be developed will probably be extracorporeal components of life-support machines of various types (because extracorporeal applications invoke fewer safety and ethical concerns). The addition of relatively crude extracoporeal cell culture systems to dialysis machines is already improving the function of renal replacement technology (Humes et al. 2002) and artificial livers depend on extrenal bioreactors filled with hepatocytes (Sauer et al. 2002). The use of synthetic morphological techniques to make self-organizing cell systems that are optimized for life in these bioreactors could be a major step forward. It is important to note that even great improvements in stem cell techniques are unlikely to make these metabolic support machines obsolete, as regenerative medicine requires time (for stem cells to integrate and grow) but a patient with acute liver failure needs support at once.
If ethical and safety concerns were to be adequately addressed, synthetic morphology may one day even be used intracorporeally. Clearly, for such application it will be critical to ensure that engineered cells are all in a uniform and known starting state, and for structures beyond the range of easy nutrient diffusion, it may be necessary to engineer in systems (such as VEGF synthesis) to encourage the provision of a host blood supply. It may also be necessary to implment systems that reliably detect the boundary of synthetic and host tissues, so that the synthetic cells do not metastasize. Examples might be cells engineered to make connections between the nervous system and terminals on artificial limbs or artificial sensors, and cells engineered to make best possible use of engineered matrices developed for tissue engineering, such as that used in the recent development of a bioartificial heart that works, though as yet produces about 2% of the pumping force of a natural one (Ott et al. 2008). Outside the clinical area, there may also be a range of applications of synthetic morphology using ‘lower’ eukaryotic organisms such as fungi to optimize their morphology for environmental applications such as bioremediation (by optimising surface area, flow rates, etc.).
It would be a mistake, however, to judge synthetic morphology solely by the medical or industrial utility of its products. Friedrich Wöhler made his impact on biology by discovering chemical methods for synthesizing acetic acid and urea (in 1824 and 1828, respectively) not because these substances could not be obtained from biological sources (they could, from wine and urine), but because an ability to synthesize biological molecules verified contemporary understanding of biological chemistry, at the time developing in competition with vitalism. In the spirit of the Feyman quotation in the Introduction to this paper, creating a working morphogentic system de novo, without relying on pre-existing differences in cell properties, would demonstrate that the relevant aspects of morphogenesis were properly understood. This would be a major advance for the basic science of anatomy. By contrast, it may be that synthetic approaches turn out to be impossible to realize with our current understanding because, although we have an inventory of the parts of living cells, we do not yet sufficiently understand the subtlety of their connection (Rosen, 2001; Serrano, 2007). In this case, we will at least have learned about deficiencies in our current understanding; we can then guide future research accordingly.
Acknowledgments
I would like to thank Dr Mathieu Unbekandt for his reading of a draft of this manuscript, and for his suggestions.
References
- Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron M, Arvind DK, Davies JA. Design of an irreversible DNA memory element. Natural Computing. 2007;6:403–411. [Google Scholar]
- Chen MT, Weiss R. Artificial cell–cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol. 2005;23:1551–1555. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- Collares-Buzato CB, Jepson MA, McEwan GT, Hirst BH, Simmons NL. Co-culture of two MDCK strains with distinct junctional protein expression: a model for intercellular junction rearrangement and cell sorting. Cell Tissue Res. 1998;291:267–276. doi: 10.1007/s004410050996. [DOI] [PubMed] [Google Scholar]
- Davies JA, Bard JB. The development of the kidney. Curr Top Dev Biol. 1998;39:245–301. doi: 10.1016/S0070-2153(08)60458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. Mechanisms of Morphogenesis. New York: Academic Press; 2005. [Google Scholar]
- Devlin K. Sets, Functions and Logic. New York: Chapman and Hall; 2004. [Google Scholar]
- Drubin DA, Way JC, Silver PA. Designing biological systems. Genes Dev. 2007;21:242–254. doi: 10.1101/gad.1507207. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Forster AC, Church GM. Towards synthesis of a minimal cell. Mol Syst Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander DR, Mege RM, Cunningham BA, Edelman GM. Cell sorting-out is modulated by both the specificity and amount of different cell adhesion molecules (CAMs) expressed on cell surfaces. Proc Natl Acad Sci USA. 1989;86:7043–7047. doi: 10.1073/pnas.86.18.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SE, Bungay PJ, Naylor LH. Functional coupling of endogenous serotonin (5-HT1B) and calcitonin (C1a) receptors in CHO cells to a cyclic AMP-responsive luciferase reporter gene. J Neurochem. 1997;69:1278–1285. doi: 10.1046/j.1471-4159.1997.69031278.x. [DOI] [PubMed] [Google Scholar]
- Gleick J. Genius: Richard Feynman and Modern Physics. Boston: Little, Brown & Co.; 1992. [Google Scholar]
- Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr Biol. 2003;13:2125–2137. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J Cell Sci. 2005;118:5191–5203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- Horikawa T, Yang J, Kondo S, et al. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007;67:1970–1978. doi: 10.1158/0008-5472.CAN-06-3933. [DOI] [PubMed] [Google Scholar]
- Humes HD, Fissell WH, Weitzel WF, et al. Metabolic replacement of kidney function in uremic animals with a bioartificial kidney containing human cells. Am J Kidney Dis. 2002;39:1078–1087. doi: 10.1053/ajkd.2002.32792. [DOI] [PubMed] [Google Scholar]
- King K, Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Control of yeast mating signal transduction by a mammalian beta 2-adrenergic receptor and Gs alpha subunit. Science. 1990;250:121–123. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- Kinzler ER, Compton T. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J Virol. 2005;79:7827–7837. doi: 10.1128/JVI.79.12.7827-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. CAS/Crk coupling serves as a ‘molecular switch’ for induction of cell migration. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng. 2004a;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004b;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc Natl Acad Sci USA. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kinoshita M, Noda M, Copeland NG, Jenkins NA. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- Lang GF. Analog was not a computer trademark. Sound and Vibration. 2000;August:16–24. [Google Scholar]
- Levskaya A, Chevalier AA, Tabor JJ, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- Mandelbrot B. The Fractal Geometry of Nature. New York: Freeman; 1997. [Google Scholar]
- Mattheakis LC, Olivan SE, Dias JM, Northrop JP. Expression of cre recombinase as a reporter of signal transduction in mammalian cells. Chem Biol. 1999;6:835–844. doi: 10.1016/s1074-5521(99)80130-6. [DOI] [PubMed] [Google Scholar]
- Murtas G. Question 7: construction of a semi-synthetic minimal cell: a model for early living cells. Orig Life Evol Biosph. 2007;37:419–422. doi: 10.1007/s11084-007-9090-5. [DOI] [PubMed] [Google Scholar]
- Muskavitch MA. Delta-notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A, Shirayoshi Y, Okazaki K, Yasuda K, Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature. 1987;329:341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Oancea S. A quantitative analysis of red blood cell aggregation from bovine blood. Romanian J Biophys. 2007;17:205–209. [Google Scholar]
- Ott HC, Matthiesen TS, Goh SK, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B, Leikina E, Sapir A, et al. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev Cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Rhoads DS, Guan JL. Analysis of directional cell migration on defined FN gradients: role of intracellular signaling molecules. Exp Cell Res. 2007;313:3859–3867. doi: 10.1016/j.yexcr.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen R. Essays on Life Itself. New York: Columbia University Press; 2001. [Google Scholar]
- Sauer IM, Neuhaus P, Gerlach JC. Concept for modular extracorporeal liver support for the treatment of acute hepatic failure. Metab Brain Dis. 2002;17:477–484. doi: 10.1023/a:1021938708670. [DOI] [PubMed] [Google Scholar]
- Sells MA, Boyd JT, Chernoff J. p21-activated kinase 1 (Pak1) regulates cell motility in mammalian fibroblasts. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L. Synthetic biology: promises and challenges. Mol Syst Biol. 2007;3:158. doi: 10.1038/msb4100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JS. The Analogue Alternative: the Electric Analogue Computer in Britain and the USA, 1930-1975. Lndon: Routledge; 2001. [Google Scholar]
- Townes P, Holtfreter J. Directed movements and selective adhesion of embryonic amphibian cells. J Exp Zool. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- Vicsek T. Fractal modles for diffusion controlled aggregation. J Phys A. 1983;16:L647–L652. [Google Scholar]
- Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. Embo J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Yang X, et al. Chemokine-like factor 1 is a functional ligand for CC chemokine receptor 4 (CCR4) Life Sci. 2006;78:614–621. doi: 10.1016/j.lfs.2005.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener R. Cybernetics. New York: Wiley; 1948. [Google Scholar]
- Wilkie N, Wingrove PB, Bilsland JG, et al. The non-peptidyl fungal metabolite L-783,281 activates TRK neurotrophin receptors. J Neurochem. 2001;78:1135–1145. doi: 10.1046/j.1471-4159.2001.00504.x. [DOI] [PubMed] [Google Scholar]