Abstract
The hot tritium bombardment technique [Goldanskii, V. I., Kashirin, I. A., Shishkov, A. V., Baratova, L. A. & Grebenshchikov, N. I. (1988) J. Mol. Biol. 201, 567–574] has been applied to measure the exposure of proteins on the ribosomal surface. The technique is based on replacement of hydrogen by high energy tritium atoms in thin surface layer of macromolecules. Quantitation of tritium radioactivity of each protein has revealed that proteins S1, S4, S5, S7, S18, S20, and S21 of the small subunit, and proteins L7/L12, L9, L10, L11, L16, L17, L24, and L27 of the large subunit are well exposed on the surface of the Escherichia coli 70 S ribosome. Proteins S8, S10, S12, S16, S17, L14, L20, L29, L30, L31, L32, L33, and L34 have virtually no groups exposed on the ribosomal surface. The remaining proteins are found to be exposed to lesser degree than the well exposed ones. No additional ribosomal proteins was exposed upon dissociation of ribosomes into subunits, thus indicating the absence of proteins on intersubunit contacting surfaces.
Keywords: ribosomal proteins, protein topography, ribosomal surface, tritium labeling
A detailed understanding of the biochemical processes that the ribosome performs to synthesize protein requires a well-defined structural model of this ribonucleoprotein complex. Significant progress in definition of ribosome morphology has been achieved recently by using energy-filtering cryoelectron microscopy (1, 2). Though this technique has refined the overall conformation of the ribosome, a further elucidation of details of ribosomal structure is still in order. The knowledge of ribosomal surface is of particular significance, as macromolecular ribosomal ligands interact with exposed regions of the particle. Identification of the exposed sites on the surface of the ribosome is important for modelling three-dimensional ribosome structure. Determination of exposure of these sites at different functional states of the ribosome allows to detect conformational rearrangements and movements of ribosomal components during the ribosomal working cycle. Several approaches were applied to reveal the exposed regions on the ribosome: immunoelectron microscopic localization of the protein and rRNA antigenic determinants, chemical and enzymatic modifications of accessible proteins within ribosomal particles, and probing of exposed rRNA regions with oligodeoxyribonucleotides or by footprinting.
An alternative and seemingly the most direct approach for studies of protein topography on the ribosome surface, the so-called hot tritium bombardment technique, was used in this study. This method proposed first for radioactive labeling of organic compounds with tritium (3) was then developed for determination of exposed proteins and their regions on the surface of macromolecular complexes (4–10). The principle of the technique is that tritium atoms, produced by dissociation of tritium gas (3H2) on a heated tungsten wire, bombard biological molecules. This results in the replacement of surface hydrogen by tritium in covalent bonds (including C—H bonds). The energy of tritium atoms can be adjusted so that the labeling is achieved without destruction of a macromolecule. The depth of penetration of reactive tritium atoms into the inner region of a macromolecule has been estimated to be 3 to 5 Å (5, 6). In this way only a thin layer of a molecule surface becomes labeled. It was shown that the labeling is roughly equal for different amino acid residues on condition that they are equally exposed (5, 6). Thus, the radioactivity of a protein within macromolecular complex must be proportional to its accessibility to atomized tritium, i.e. to the exposure of the protein on the surface of a complex. These features of the method allowed to apply it for direct measurements of proteins exposure on the surface of biological macromolecular complexes, such as viruses (7–9) and membranes (10). Application of the hot tritium bombardment technique to ribosomes made it possible to distinguish between the ribosomal proteins that are well exposed on the ribosome surface and that are buried under the surface (11–13). Here, for the first time, we present quantitative data on exposure of proteins on the surface of 70S ribosomes and ribosomal subunits obtained by combining the hot tritium bombardment technique with measurements of tritium radioactivity of each individual ribosomal protein.
MATERIALS AND METHODS
Materials.
Buffer reagents were obtained from Sigma, acrylamide and methylenebisacrilamide were from Fluka, urea was from Bio-Rad, scintillator liquid Supersolve X was from Koch-Light Laboratories (Buck, U.K.), and DNase 1 was from Serva. Autoradiography film Hyperfilm-MP and fluorographic reagent Amplify were purchased from Amersham. Sucrose, acetone, hydrogen peroxide, and acetic acid were from ReaKhim (Moscow, Russia).
Preparation of 70S Ribosomes.
Method of Staehelin et al. (14) was used with modifications. Frozen Escherichia coli MRE-600 cell paste was thawed, suspended in equal volume of buffer A (10 mM MgCl2/10 mM Tris⋅HCl, pH 7.3/50 mM NH4Cl/0.1 mM Na2⋅EDTA/6 mM 2-mercaptoethanol) and disrupted in a French press. DNase was added to the homogenate up to 0.5 μg/ml and the mixture was incubated at 4°C for 20 min. Debris was removed by centrifugation for 2 hr at 23,000 × g. Ribosomes were pelleted from the cell extract for 7 hr at 105,000 × g, resuspended in buffer A, and additionally purified by centrifugation through a sucrose cushion containing 30% sucrose in buffer B (10 mM MgCl2/20 mM Tris⋅HCl pH 7.3/500 mM NH4Cl/0.1 mM Na2⋅EDTA/6 mM 2-mercaptoethanol). The centrifugation was carried out for 19 hr at 143,000 × g, each tube containing 33 ml of sucrose solution and 30 ml of ribosome suspension. The pellets were resuspended in buffer A, and the suspension was clarified by centrifugation for 30 min at 23,000 × g and dialyzed against the same buffer. All procedures were carried out at 5°C unless stated otherwise. The resulting ribosome suspensions at concentrations more than 100 mg/ml were stored at −70°C.
Preparation of total ribosomal protein was done according to (16) by extraction of 70S ribosomes with 67% acetic acid for 1 hr at 4°C in the presence of 0.1 M MgCl2. After removal of rRNA precipitate the solution of total protein was diluted with water and acetic acid to adjust protein concentration to 0.4 mg/ml and concentration of acetic acid to either 5% or 67%. Resulting solutions were stored at −70°C.
Tritium Bombardment Procedure.
Samples of nondissociated 70S ribosomes were prepared by dilution of aliquots of the concentrated ribosome suspension with 1.5 ml of buffer C (10 mM Mg Cl2/20 mM Tris⋅HCl, pH 7.3/100 mM NH4Cl/0.1 mM Na2⋅EDTA/1 mM DTT) to give 2 mg/ml concentration of the particles. The suspension was incubated at 37°C for 15 min before further steps of sample preparation.
The equimolar mixture of ribosomal subunits was prepared similarly: aliquots of the concentrated ribosome suspension were diluted with 1.5 ml of buffer D (1 mM MgCl2/20 mM Tris⋅HCl, pH 7.3/100 mM NH4Cl/0.1 mM Na2⋅EDTA/1 mM DTT). Dissociation of 70S ribosomes occurred in buffer D during incubation at 37°C for 15 min. The final concentration of ribosomal subunits was 2 mg/ml.
Solutions of total ribosomal protein subjected to tritium bombardment contained 0.4 mg/ml of total protein in 2 ml of acetic acid solutions.
Samples for tritium bombardment were prepared according to (12). Suspension of ribosomes, subunits or solution of total ribosomal protein was first frozen in liquid nitrogen drop by drop and the frozen drops were ground with liquid nitrogen in a cooled (−195°C) mortar. This procedure excluded the effect of any preferential orientation of ribosomal particles at the phase boundary. The suspension of sample powder in liquid nitrogen was poured in the reactor flask during its rotation on its axis in horizontal position. The reactor flask was rotating until nitrogen has evaporated and the particles of sample powder adhered to the inner wall of the flask. Immediately after that a vacuum was produced in the reactor with a residual pressure of 10−4 torr (1 torr = 133 Pa). Gaseous tritium was injected to adjust the pressure to 2 × 10−3 torr. Dissociation of tritium occurred on tungsten wire heated to 1,800 K whereas the reactor flask was immersed in liquid nitrogen. The reaction continued until the pressure of tritium gas decreased to 2 × 10−4 torr, that usually took from 7–10 min.
Analysis of Tritium Incorporation into Ribosomal Proteins.
Labeled ribosomes or ribosomal subunits were pelleted by centrifugation in a TLA 100.3 rotor of TL-100 Beckman centrifuge at 100,000 rpm, 4°C, for 40 min to remove labeled water and buffer components.
Labeled ribosomal particles were incubated for 1 hr at 4°C with 67% acetic acid and 0.1 M MgCl2 to extract total protein (16) for further analysis. After removal of rRNA precipitate total ribosomal protein was precipitated with 7.5 volumes of acetone.
When total ribosomal protein was labeled under denaturing conditions, the removal of exchangeable tritium from such preparation was performed by precipitation with 7.5 volumes of acetone immediately after tritium bombardment.
Separation of individual labeled proteins was performed by two-dimensional polyacrylamide/urea gel electrophoresis according to (15), with a gel slab size of 80 × 110 × 1 mm. This procedure allows to separate all ribosomal proteins except S5/L6, S9/S11, S12/L20, and L32/L33. Protein spots stained with Coomassie blue G-250 in 3.6% HClO4 were cut from the gel by metallic tube with sharp edges. The pieces of the gel were solubilized by treatment with 0.15 ml of fresh 30% hydrogen peroxide at 60°C in polypropylene tubes hermetically sealed with screw caps. Formation of transparent homogeneous solution was achieved usually by overnight incubation. Resulting solutions were mixed with 10 ml of Supersolve X scintillation cocktail, the mixtures were kept at room temperature for 10 hr to reduce chemiluminescence to zero before measurements in a scintillation counter. Radioactivity detected in a piece of the same gel without protein ranged from 50–80 cpm. Fluorographic detection of radioactivity in the gel was also applied in some experiments. Hyperfilm-MP for autoradiography and fluorographic reagent Amplify were used.
RESULTS
Tritium Bombardment of Denatured Ribosomal Proteins.
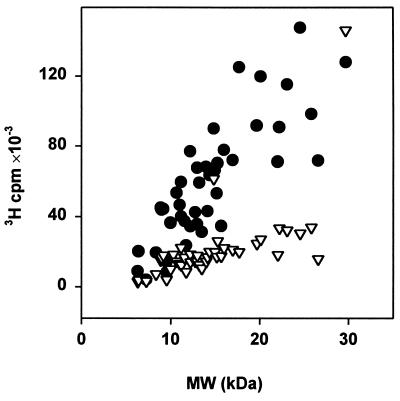
Distribution of tritium label among ribosomal proteins subjected to hot tritium bombardment in denatured state is shown in Fig. 1. Because all of the amino acid residues of denatured protein are expected to be exposed to a more or less equal extent, a protein should incorporate tritium proportionally to its molecular mass when labeled under denaturing conditions. The majority of ribosomal proteins does demonstrate such proportionality. Labeling of proteins in 67% acetic acid is significantly less effective than that in 5% acid. This could be explained by a protective effect of acetic acid molecules that are possibly adsorbed at the surface of denatured protein or by incomplete denaturation in concentrated acetic acid. The protein L2, as well as L11, incorporate tritium equally when labeled in 67% and 5% acetic acid. Identical patterns of tritium distribution were obtained in four independent experiments.
Figure 1.
Labeling of total ribosomal protein by hot tritium bombardment. Proteins were denatured with 67% (▿) or with 5% (•) acetic acid. Tritium radioactivity of each individual protein is plotted against its molecular mass.
The procedure of tritium distribution analysis allows to detect only the radioactivity incorporated in nonexchangeable C—H bonds of protein molecules because all exchangeable tritium was removed during pelleting of ribosomes after bombardment and preparation of samples for electrophoresis. The latter procedure includes spin-column chromatography of extracted ribosomal protein on Sephadex G-25 in 8 M urea (12). Completeness of the removal was checked by an additional spin-column gel-filtration of the same sample: the radioactivity of ribosomal protein remained exactly the same as after the first gel-filtration step (data not shown).
Tritium Bombardment of 70S Ribosomes.
A photograph of a Coomassie-stained two-dimensional gel containing ribosomal proteins is given in Fig. 2 together with fluorogram of the same gel. Proteins were extracted from 70S ribosomes labeled by tritium bombardment under nondissociating conditions, at 10 mM Mg2+ (buffer C). It is seen that just a certain set of proteins becomes radioactive upon tritium bombardment of the 70S ribosome. This shows that not all of the ribosomal proteins are exposed on the ribosome surface. All of the radioactive spots on the fluorogram strictly correspond to the spots of proteins on the stained gel that gives evidence of the absence of degradation or modification products of labeled proteins.
Figure 2.
Separation of individual ribosomal proteins by two-dimensional gel electrophoresis: photograph of Coomassie-stained gel (A) and fluorogram of the same gel (B). Proteins were extracted from 70S ribosomes labeled at 10 mM Mg2+ (buffer C). The gel contains 150 μg of total ribosomal protein.
The integrity of the labeled ribosomes was examined by sucrose gradient centrifugation according to (12). The UV adsorption profile of the gradient corresponded to the radioactivity profile obtained by hot trichloroacetic acid precipitation of the gradient fractions, no aggregates or peaks with sedimentation coefficients other than 70S were detected (not shown). This ensures that the labeling did not lead to destruction of the ribosome.
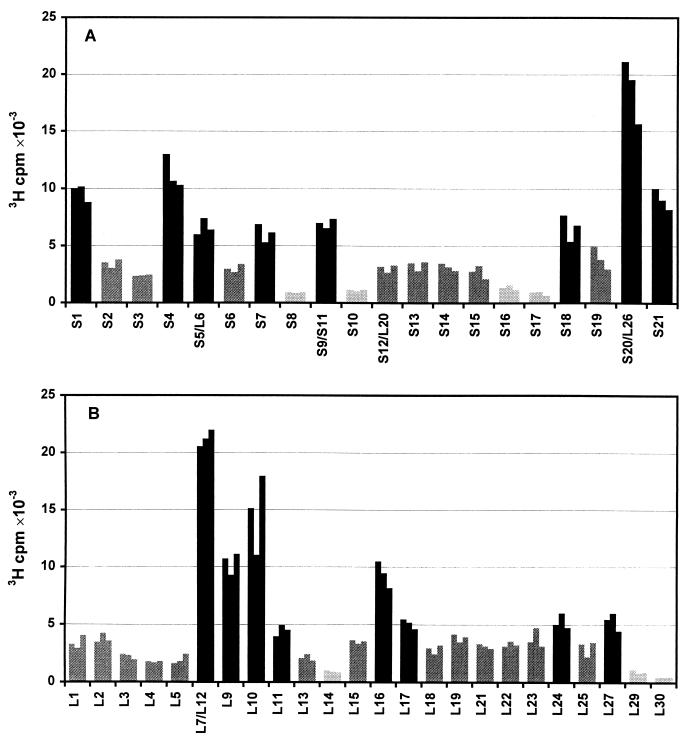
Quantitative data on tritium radioactivity of individual proteins of labeled 70S ribosomes are given in Fig. 3. Solubilization of gel pieces and measurements of radioactivity in homogeneous solutions allow to avoid nonlinearity of fluorographic detection and thus obtain the direct data on protein exposure on the ribosome surface. Proteins S1, S4, S5, S7, S18, S20/L26, and S21 of the small subunit, and proteins L7/L12, L9, L10, L11, L16, L17, L24, and L27 of the large subunit were found to be well exposed on the surface of the E. coli 70 S ribosome. Proteins S8, S10, S12, S16, and S17 of the small subunit, and L14, L20, L29, and L30 of the large subunit did not incorporate tritium to a considerable degree, indicating that these proteins have virtually no groups exposed on the ribosomal surface. The same low level of labeling was detected in our earlier experiments for proteins L31, L32, L33, and L34. The values of tritium radioactivity for the latter four proteins are not presented in this study, because the electrophoresis stopped when these proteins had migrated out of the gel to improve resolution power of the procedure. The remaining proteins are found to be exposed to lesser degree than the well exposed ones.
Figure 3.
Labeling of nondissociated 70S ribosomes at 10 mM Mg2+ (buffer C). Tritium radioactivity of each protein is shown by three bars representing three independent experiments. Black bars, the well-exposed proteins; grey bars, the poorly exposed proteins; dark gray bars, the moderately exposed proteins. (A) Proteins of the 30S subunit; (B) proteins of the 50S subunit.
Although proteins S9 and S11 were not separated by the electrophoretic procedure used, an additional SDS electrophoresis of proteins extracted from the S9/S11 spot has shown that these proteins are labeled to almost the same extent. Thus, proteins S9 and S11 should be considered as poorly exposed proteins, because the radioactivity of the S9/S11 spot comes from both of them. Separation of S9 and S11 was achieved by electrophoresis in a 15% polyacrylamide gel according to Laemmli (17) without heating the sample at 95°C (data not shown).
The earlier experiments on tritium bombardment of isolated 30S subunits has shown that protein S5 is an exposed protein (13), whereas protein L6 is not exposed on the surface of the large subunit (12). Consequently, the radioactivity detected in the S5/L6 spot on the gel of two-dimensional electrophoresis should be attributed to the protein S5. The same set of experiments (12) indicates that the proteins S12 and L20 are not exposed to a considerable degree on the surfaces of ribosomal subunits. This allows to consider the radioactivity of S12/L20 spot as derived from both these proteins and assign them to poorly exposed ones.
It should be mentioned that protein S1 is present in the gel in a significantly lower molar amount than other proteins. Hence, the actual exposure of this protein is much higher than it is measured in our experiments.
Tritium Bombardment of Ribosomal Subunits Mixture.
The subunits were subjected to tritium bombardment in the state of their equimolar mixture obtained by dissociation of 70S ribosomes in 1 mM MgCl2 (buffer D). It is known that vacant 70S ribosomes dissociate completely at this magnesium concentration. Degree of dissociation was checked by analytical centrifugation and found to be complete (data not shown). The integrity of the subunits after tritium bombardment was controlled by sucrose gradient centrifugation as it was done for 70S ribosomes. No destruction of the labeled subunits was detected.
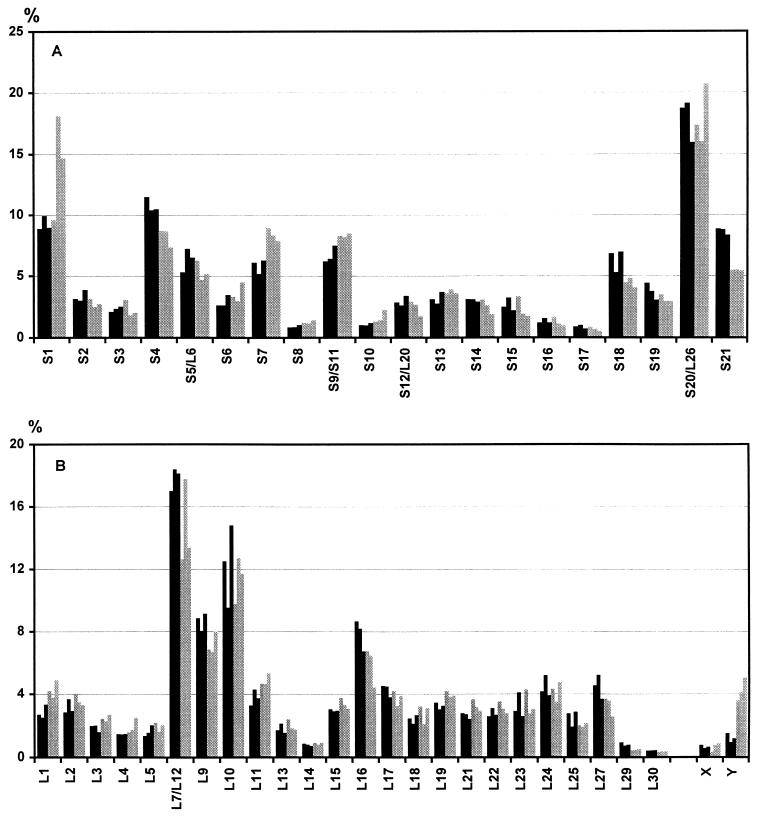
The pattern of tritium distribution among proteins of labeled ribosomal subunits is shown in Fig. 4. It is seen, that the same set of proteins is exposed on the surface of both dissociated and nondissociated ribosomes. Moreover, the degree of exposure of each individual protein within the 70S ribosome is identical to that within ribosomal subunits of the dissociated ribosome. Thus, the dissociation of 70S ribosomes into subunits does not result in the exposure of any additional ribosomal proteins. Consequently, none of the ribosomal protein can be hidden by association of the subunits. This clearly demonstrates the absence of ribosomal proteins on intersubunit contacting surfaces.
Figure 4.
Labeling of the equimolar mixture of ribosomal subunits at 1 mM Mg2+ (buffer D). Radioactivity of each protein is shown by bars in percents of the total radioactivity found in all the protein spots on the gel. Gray bars, proteins labeled within subunits of dissociated ribosomes; black bars, proteins labeled within the nondissociated ribosomes at 10 mM Mg2+. Each bar represents an independent experiment. (A) Proteins of the 30S subunit, and (B) proteins of the 50S subunit.
One additional protein spot, however, becomes highly labeled after dissociation of the 70S ribosome. This is spot Y, which has not been identified as ribosomal protein. The corresponding minor protein may be a component located between the ribosomal subunits. Indeed, the reassociation of ribosomal subunits by increasing MgCl2 concentration up to 10 mM leads to its reshielding (data not shown). Identification of protein Y is now in progress.
DISCUSSION
The previous approaches to mapping the ribosome surface include localization of proteins by immunoelectron microscopy (18–22) as well as chemical and enzymatic modifications of accessible proteins within ribosomal particles (23–28). All of those techniques detect either exposure of a protein antigenic determinant (which can be a very small protein portion) or accessibility of an amino acid residue to appropriate chemical or enzyme. In the strict sense, these approaches of surface mapping are indirect. Moreover, small molecules of modifying reagents can mark inner regions of the ribosome due to diffusion. Finally, the intactness of the ribosome after modification (especially in the case of hydrolytic enzymes) is sometimes questionable as well. Thus, determination of the actual exposure of ribosomal proteins requires more direct approach.
Use of the direct method of hot tritium bombardment has demonstrated that only a certain set of ribosomal proteins is well exposed and there are proteins completely buried inside the ribosome. This result contradicts the immunoelectron microscopic studies where almost all proteins were shown to have antigenic determinants on the surface of ribosomal particles (18–22). Our experiments on hot tritium bombardment show the absence of ribosomal proteins on the interface: no additional proteins become exposed upon dissociation of ribosomes into subunits. This suggests that the contacting surfaces of the ribosomal subunits are built up entirely of ribosomal RNA, whose involvement in intersubunit contacts was demonstrated by chemical and enzymatic modifications of RNA within the 70S ribosome and ribosomal subunits (29–33).
The data on accessibility of ribosomal proteins to tritium atoms can be reconciled with all the results on the topography of the ribosomal proteins on the 30S subunit. The three-dimensional model of the small subunit by Moore and coworkers (34) was somewhat corrected by slight relocation of several proteins according to their exposure: proteins S4, S5, S7, S18, S20, and S21 were raised to the surface of the particle to its external side, opposite to the 50S subunit, while protein S3 was withdrawn from the surface and buried into the particle (13).
The results presented here are based on quantitative determination of radioactivity, that excludes nonlinearity of fluorographic detection. As it is seen by comparison of Figs. 2 and 3, some proteins give almost equally intensive spots on the fluorogram while the actual radioactivity of the proteins differ two or more times (proteins S4 and L7/L12, for example). Hence, one can expect that small changes in protein exposure upon dissociation of the ribosome could not be detected by fluorography. The quantitative analysis of tritium distribution confirms the previous conclusion on the absence of proteins on the ribosomal interface (11, 12). At the same time proteins S13 and L5 referred to as highly exposed (11, 12) should be considered as moderately exposed ones on the basis of the quantitative data presented.
The hot tritium bombardment technique provides the most direct and adequate data on exposure of components of a macromolecular complex on its surface: the method is not destructive, only the surface of a macromolecule becomes labeled and, finally, there is direct proportionality of the degree of labeling to the degree of exposure. Combination of the method with quantitative determination of tritium incorporated gives the complete pattern of protein exposure on the surface of ribosomal particles. This, in turn, opens a way to study interactions of the ribosome with its ligands, such as tRNA, translation factors, and antibiotics, as well as to reveal possible conformational rearrangements of the ribosome.
Acknowledgments
We thank Drs. M. M. Yusupov, A. Kommer, E. V. Makeyev, and V. A. Shirokov for helpful suggestions and comments and for participation in some experiments. Special thanks also to A. N. Turkin and V. Y. Pankratova for technical assistance. This work was supported by Grant 96-04-49759 from the Russian Foundation for Fundamental Research and Russian Academy of Sciences.
References
- 1.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata R K, Agraval R K. Nature (London) 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 2.Stark H, Orlova E V, Rinke-Appel J, Junke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. Cell. 1997;88:19–28. doi: 10.1016/s0092-8674(00)81854-1. [DOI] [PubMed] [Google Scholar]
- 3.Klein R, Scheer M. J Phys Chem. 1958;62:1011–1014. [Google Scholar]
- 4.Shishkov A V, Filatov E S, Simonov E F, Unukovich M S, Goldanskii V I, Nesmeyanov A N. Dokl Akad Nauk SSSR. 1976;228:1237–1239. [PubMed] [Google Scholar]
- 5.Goldanskii V I, Rumyantsev Yu M, Shishkov A V, Baratova L A, Belyanova L P. Mol Biol (Moscow) 1982;16:528–534. [PubMed] [Google Scholar]
- 6.Baratova L A, Goldanskii V I, Rumyantsev Yu M, Unukovich M S, Shishkov A V. Mol Biol (Moscow) 1982;16:117–122. [PubMed] [Google Scholar]
- 7.Goldanskii V I, Kashirin I A, Shishkov A V, Baratova L A, Grebenshchikov N I. J Mol Biol. 1988;201:567–574. doi: 10.1016/0022-2836(88)90638-9. [DOI] [PubMed] [Google Scholar]
- 8.Baratova L A, Grebenshchikov N I, Dobrov E N, Gedrovich A V, Kashirin I A, Shishkov A V, Efimov A V, Jarvekulg L, Radavsky Y L, Saarma M. Virology. 1992;188:175–180. doi: 10.1016/0042-6822(92)90747-d. [DOI] [PubMed] [Google Scholar]
- 9.Baratova L A, Grebenshchikov N I, Shishkov A V, Kashirin I A, Radavsky J L, Jarvekulg L, Saarma M. J Gen Virol. 1992;73:229–235. doi: 10.1099/0022-1317-73-2-229. [DOI] [PubMed] [Google Scholar]
- 10.Tsetlin V I, Alyonycheva T N, Shemyakin V V, Neiman L A, Ivanov V T. Eur J Biochem. 1988;178:123–129. doi: 10.1111/j.1432-1033.1988.tb14437.x. [DOI] [PubMed] [Google Scholar]
- 11.Yusupov M M, Spirin A S. FEBS Lett. 1986;197:229–233. doi: 10.1016/0014-5793(86)80332-5. [DOI] [PubMed] [Google Scholar]
- 12.Yusupov M M, Spirin A S. Methods Enzymol. 1988;164:426–439. doi: 10.1016/s0076-6879(88)64059-6. [DOI] [PubMed] [Google Scholar]
- 13.Spirin A S, Agafonov D E, Kolb V A, Kommer A. Biokhimiya. 1996;61:1366–1368. [PubMed] [Google Scholar]
- 14.Staehelin T, Maglott D, Monro R E. Cold Spring Harbor Symp Quant Biol. 1969;34:39–48. doi: 10.1101/sqb.1969.034.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Kanny J W, Lambert J M, Traut R R. Methods Enzymol. 1979;59:539–550. doi: 10.1016/0076-6879(79)59112-5. [DOI] [PubMed] [Google Scholar]
- 16.Hardy S J S, Kurland C G, Voynow P, Mora G. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Kahan L, Winkelmann D A, Lake J A. J Mol Biol. 1981;145:193–214. doi: 10.1016/0022-2836(81)90340-5. [DOI] [PubMed] [Google Scholar]
- 19.Lake J A, Stryharz W A. J Mol Biol. 1981;153:979–992. doi: 10.1016/0022-2836(81)90462-9. [DOI] [PubMed] [Google Scholar]
- 20.Stöffler G, Stöffler-Meilicke M. Annu Rev Biophys Bioeng. 1984;13:303–330. doi: 10.1146/annurev.bb.13.060184.001511. [DOI] [PubMed] [Google Scholar]
- 21.Stöffler G, Stöffler-Meilicke M. In: Structure, Function, and Genetics of Ribosomes. Hardesty B, Kramer G, editors. New York: Springer; 1986. pp. 28–37. [Google Scholar]
- 22.Oakes M O, Henderson E, Scheinman A, Clark M, Lake J A. In: Structure, Function, and Genetics of Ribosomes. Hardesty B, Kramer G, editors. New York: Springer; 1986. pp. 47–59. [Google Scholar]
- 23.Baunert H G, Sköld S E, Kurland C G. Eur J Biochem. 1978;89:353–359. doi: 10.1111/j.1432-1033.1978.tb12536.x. [DOI] [PubMed] [Google Scholar]
- 24.Lambert J M, Traut R R. J Mol Biol. 1981;149:451–476. doi: 10.1016/0022-2836(81)90481-2. [DOI] [PubMed] [Google Scholar]
- 25.Cover J A, Lambert J M, Norman C M, Traut R R. Biochemistry. 1981;20:2843–2852. doi: 10.1021/bi00513a021. [DOI] [PubMed] [Google Scholar]
- 26.Chiam C L, Wagner R. Biochemistry. 1983;22:1193–1200. doi: 10.1021/bi00274a032. [DOI] [PubMed] [Google Scholar]
- 27.Brimacombe R, Maly P, Zwieb C. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- 28.Lambert J M, Boileau G, Cover J-A, Traut R R. Biochemistry. 1983;22:3913–3920. doi: 10.1021/bi00285a029. [DOI] [PubMed] [Google Scholar]
- 29.Chapman N N, Noller H F. J Mol Biol. 1977;109:131–149. doi: 10.1016/s0022-2836(77)80049-1. [DOI] [PubMed] [Google Scholar]
- 30.Herr W, Noller H F. J Mol Biol. 1979;130:421–432. doi: 10.1016/0022-2836(79)90432-7. [DOI] [PubMed] [Google Scholar]
- 31.Vassilenko S K, Carbon P, Ebel J P, Ehresmann C. J Mol Biol. 1981;152:699–721. doi: 10.1016/0022-2836(81)90123-6. [DOI] [PubMed] [Google Scholar]
- 32.Brow D A, Noller H F. J Mol Biol. 1983;163:27–46. doi: 10.1016/0022-2836(83)90028-1. [DOI] [PubMed] [Google Scholar]
- 33.Meier N, Wagner R. Eur J Biochem. 1985;146:83–87. doi: 10.1111/j.1432-1033.1985.tb08622.x. [DOI] [PubMed] [Google Scholar]
- 34.Capel M S, Engelman D M, Freeborn B R, Kjeldgaard M, Langer J A, Ramakrishnan V, Schindler D G, Schneider D K, Shoenborn B P, Sillers I Y, Yabuki S, Moore P B. Science. 1987;238:1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]