Abstract
Tissue microarray technology and immunohistochemical techniques have become a routine and indispensable tool for current anatomical pathology diagnosis. However, manual quantification by eye is relatively slow and subjective, and the use of digital image analysis software to extract information of immunostained specimens is an area of ongoing research, especially when the immunohistochemical signals have different localization in the cells (nuclear, membrane, cytoplasm). To minimize critical aspects of manual quantitative data acquisition, we generated semi-automated image-processing steps for the quantification of individual stained cells with immunohistochemical staining of different subcellular location. The precision of these macros was evaluated in 196 digital colour images of different Hodgkin lymphoma biopsies stained for different nuclear (Ki67, p53), cytoplasmic (TIA-1, CD68) and membrane markers (CD4, CD8, CD56, HLA-Dr). Semi-automated counts were compared to those obtained manually by three separate observers. Paired t-tests demonstrated significant differences between intra- and inter-observer measurements, with more substantial variability when the cellular density of the digital images was > 100 positive cells/image. Overall, variability was more pronounced for intra-observer than for inter-observer comparisons, especially for cytoplasmic and membrane staining patterns (P < 0.0001 and P = 0.050). The comparison between the semi-automated and manual microscopic measurement methods indicates significantly lower variability in the results yielded by the former method. Our semi-automated computerized method eliminates the major causes of observer variability and may be considered a valid alternative to manual microscopic quantification for diagnostic, prognostic and therapeutic purposes.
Keywords: antibodies, image analysis, immunohistochemistry, pathology diagnosis, tumour markers
Introduction
Due to the low cost and the small amounts of tissue required, the integrated use of tissue microarray technology (TMA) and immunohistochemical (IHC) techniques for the detection of specific cellular antigens has replaced biochemical and molecular methods of analysis. Their results correlate closely with those of the old methods (Aasmundstad et al. 1992; Podhajsky et al. 1997; Bhatnagar et al. 1999; Simone et al. 2000). TMA has become a routine and indispensable tool for current anatomical pathology diagnosis that has enabled immunohistochemical studies to be made of multiple markers under homogeneous conditions that are inexpensive to carry out. The results obtained with this TMA have been validated for diagnostic, prognostic and therapeutic purposes in different epithelial carcinomas (Rosen et al. 2004; Cao et al. 2007; Li et al. 2007; Sandlund et al. 2007; Tubbs et al. 2007) but also in haematological malignancies, including follicular lymphoma (Alvaro et al. 2006a,b; Lee et al. 2006), diffuse large B-cell lymphoma (Zettl et al. 2003; Zu et al. 2005; de Jong et al. 2007), and Hodgkin's lymphoma (Hedvat et al. 2001; Alvaro et al. 2005; Alvaro-Naranjo et al. 2005; Bosch Princep et al. 2005).
In the evaluation and quantification of positively immunostained cells, the most prevalent method is the manual counting performed by the pathologist using a conventional microscope. However, the time necessary to quantify larger samples, the interpretation of immunostains, and the reproducibility among observers is not always optimal (Taylor, 2000; Seidal et al. 2001; Rhodes et al. 2002; Leong & Leong, 2004). This is basically due to a number of variables that influence antigen staining in paraffin-embedded tissues. The principal extrinsic factors of variability are related to the specimen and include the clone, the dilution of the antibody, the detection system and chromogen, the antigen retrieval method, and the external/internal reference standards (Taylor, 2006). All these extrinsic factors can be modified and controlled as part of the technique, unlike the intrinsic factors related to the tissue sample, which are extremely difficult to standardize between laboratories, or even within the same laboratory (Leong, 2004). These intrinsic factors include the type of fixative, fixation time, tissue processing, and the level of antigen expression and preservation. Quantification methods range from simple counting of stained cells to more complex grading of their intensity on an arbitrary scale. Under these conditions, the specific and precise quantification of stained cells is subject to evaluation by the human eye, which has difficulties in distinguishing subtle differences in staining intensity, especially at the extremes of the continuous colour scale.
As a means of avoiding subjectivity, a number of automated image analysis programs suitable for use in histopathology have recently become commercially available. Used in conjunction with a microscope and a digital camera, these programs can detect, quantify and classify immunostained cells in digital images based on colour, size, and shape of these cells (Gil & Wu, 2003). These systems of processing images are based on a method of segmentation that divides images according to areas of interest and evaluates every pixel of the image to determine whether it belongs to these areas. Although these systems have improved the levels of sensitivity, precision, reproducibility and standardization of these kinds of measurements, they generally require user interaction for adequate object selection, modification of object boundaries, and selection of thresholds. On the other hand, various semi-automated processes have been developed to identify individual stained cells (the stain being taken up mainly by the nuclei) in different tissue samples and to quantify them using cellular shape descriptors. Routines (macros) written for commercially available image analysis software have been used to perform semi-automated image analysis of these nuclei (Veltri et al. 2000; Mofidi et al. 2003; Nabi et al. 2004; Singh et al. 2005). However, markers used for anatomical pathologic diagnosis are not confined to the nuclei alone but can also be located in the membrane or disseminated throughout the cytoplasm in granules. These various immunostaining patterns are a critical aspect of the acquisition of reproducible quantitative data, but no reliable fully automated method has yet been described that is applicable to these subcellular locations of proteins (Camp et al. 2002; Leong & Leong, 2004).
Image-Pro® Plus (Media Cybernetics, Silver Springs, MD, USA) is a commercially available digital image software used for quantification in several immunohistochemical studies (Media Cybernetics, 2002). However, to our knowledge, no study has verified the convenience of this software to detect and to quantify different subcellular localization of immunohistochemical signals at the same time. This software runs within Image Tool® (University of Texas, Health Sciences centre, San Antonio, TX, USA) and provides tools that make it possible to acquire, display, edit, analyse, process, compress, save and print grey scale and colour images. One of the most important features of this software is the built-in scripting capabilities that allow the user to record repetitive tasks and playback saved scripts to automate image analysis (macro).
Using the version 5.0 of the Image-Pro Plus software, we have developed several specific macros based on the automation of colour-specific thresholds (for hue, saturation and intensity) and morphological features in such a way that the algorithms segment and quantify immunohistochemically marked infiltrated cells inside the reactive background of tissue sections of Hodgkin's lymphoma (HL) patients. The set of macros allows the quantification of the three patterns (membrane, cytoplasm, nuclear) of staining independently of the type of antibodies used. The final purpose of this study is to validate a semi-automated method, designed and developed in our laboratory for digital microscopic images, and to compare it with the measurements of independent observers using a manual quantification method in immunohistochemistry.
Materials and methods
TMA construction and immunohistochemistry
The present study has been carried out on samples of classical Hodgkin's lymphoma randomly selected and identified from the Spanish Hodgkin's Lymphoma Study group between 1994 and 1998. TMAs were constructed from different cHL cases selected on the basis of the availability of suitable formalin-fixed and paraffin-embedded tissue. All samples were fixed and processed using the conventional histological techniques of the different centres, with inclusion in paraffin. The selected tissue sections represent the richest areas of Hodgkin and Reed Sternberg (H/RS) cells marked in the paraffin blocks. Cylinders of 1 mm diameter from different areas were included in each case. Tissue sections were prepared as previously described (Garcia et al. 2003). Briefly, TMA blocks were sectioned at a thickness of 3 µm, dried for 16 h at 56 °C before being dewaxed in xylene and rehydrated through a graded ethanol series, washed in water and finally in phosphate-buffered saline (PBS). Antigen was retrieved by heat treatment in a pressure cooker or by pronase digestion (Dako, Carpinteria, CA, USA), as necessary. Automatic immunostaining was done with Horizon TechMate, as previously described (Alvaro-Naranjo et al. 2005). Briefly, TMAs were incubated with the appropriate dilution of the primary antibodies: CD4 (clone 1F6, Dako), CD8 (clone 1A5, Novocastra, Newcastle upon Tyne, UK), CD56 (clone 1B6, Novocastra), HLA-Dr (clone TAL1B5, Dako), CD68 (clone KP1, Dako), TIA-1 (clone 2G9, Master Diagnostica, Granada, Spain), Ki67 (clone MIB-1, Dako) and p53 (clone DO-7, Novocastra). After washing, TMAs were incubated with the secondary antibody, conjugated to peroxidase (Dako EnVision+™, Dako Corporation). Reagents in the DAKO EnVision™ system include peroxidase block, labelled polymer, and buffered substrate/DAB + chromogen. Upon oxidation, DAB chromogen forms a brown end-product at the site of the target antigen. Finally, tissue arrays were counterstained with haematoxylin, dehydrated, and mounted.
Image capture
Using a Zeiss Axioskop 2 plus standard light microscope (Carl Zeiss, Goettingen, Germany), representative areas of TMAs spots were selected with a 10–20× objective with reference to the presence of H/RS cells with an appropriate inflammatory background. These selected areas were captured through the 40× objective (Achroplan 40X/0.65 ∞/0.17) with a Coolsnap digital camera (Coolsnap, RS Photometrics, Tucson, AZ) coupled to the microscope. Digitized images have a resolution of 1392 × 1040 pixels with RGB 24 True Colour format and were saved in uncompressed TIFF format. Different levels of illumination can cause significant differences in the measurements of images. Thus, the same range of illumination values was used to ensure the greatest reproducibility. The present study considers a total of 196 images: 100 images of the membrane staining pattern for each CD4 (n = 30), CD8 (n = 32), CD56 (n = 19), and HLA-Dr (n = 19), 66 images of the cytoplasmic staining pattern for CD68 (n = 28), and TIA-1 (n = 38), and 30 images of the nuclear staining pattern for Ki-67 (n = 15), and p53 (n = 15).
Processing for manual microscopic quantification
For the manual microscopic quantification, the different images were opened with the Image-Pro Plus 5.0 program and positive cells were quantified using the ‘manual tagging’ command available in the software. The cells of the infiltrate that stained positive for CD4, CD8, CD56, HLA-Dr, CD68, TIA-1, Ki-67 and p53 were counted directly on the screen by placing markers on the image with the mouse. The numbers of positive cells was displayed in the Manual Tag View Menu, which was updated automatically. According to a consensus of positivity defined previously in our laboratory, three different observers (a biologist and two pathologists) have quantified each image manually on two occasions, separated by an interval of 1 month. Finally, the results were exported to Excel 2002 files.
Processing for semi-automated quantification
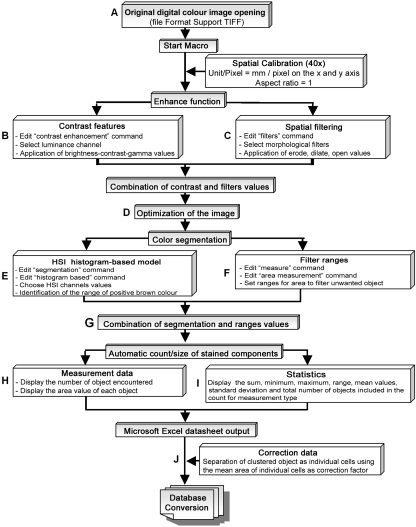
The number of positive cells and the total stained area were quantified semi-quantitatively with the Image-Pro Plus 5.0 software. This was initially programmed for different routines (multistep macros) that enable the evaluation of the different immunohistochemical staining patterns (membrane, nuclear, and cytoplasmic DAB, all of which are brown in colour). To design the different macros, the parameters were determined by the ‘split-sample technique’, which takes 10 images of each staining pattern as a training set and all other images as a test set. The ‘ground truth’ data of the colour and the morphological parameters, which were incorporated in the different macros, were acquired from images of the training set. The accuracy of the method was evaluated by applying the macro on images of the test set. The automated procedure starts after the manual acquisition of the image and finish after the automatic transfer of the data to a Microsoft Excel 2002 worksheet (semi-automated). The subsequent stages of the image analysis algorithms are illustrated in Fig. 1. All the parameters included during the process were adapted individually to analyse the different staining patterns of the photographed images.
Fig. 1.
Schematic representation of computer algorithms considered for the detection and quantification of positively immunostained cells in paraffin-embedded tissues. (A) Manual loading of image onto the screen. (B) Application of ‘contrast enhancement’ algorithms. (C) Application of ‘spatial filtering’ algorithms if needed. (D) Obtaining of the optimized image. (E) Application of the ‘segmentation’ algorithms. (F) Application of the measurement parameters. (G) Determination of the positive objects. (H,I) Transfer of global data on Excel worksheet. (J) Determination of final number of positive stained individual cells with special algorithms implemented in the worksheet. The final algorithm models may be used to evaluate nuclear-, cytoplasmic-, and membrane-stained structures.
After manual loading onto the screen of the Image-Pro Plus program (Fig. 1A), images were calibrated in size to standardize the measurement scale. The real area of each analysed field was 19.690 µm2. In a second step (Fig. 1B), the ‘contrast enhancement’ command was applied with predetermined values of brightness (illumination on the images), contrast (degree of difference between the lightest and darkest areas on the image), and gamma (distribution of the brightness across the intensity spectrum of the image) in the white luminance channel (achromatic channel). These different values, previously determined with the training set images, were applied for each pixel and were generally sufficient to eliminate the contribution of background staining, to exclude cellular debris, and to highlight the colour of the positive staining. Nevertheless, for cytoplasmic stained markers, a special adaptation of the images was necessary to unify the different positive granules in the cytoplasm in an object alone. The ‘spatial filtering’ command (Fig. 1C) used in our macros applied predetermined values for the morphological filters erosion, dilation, and opening. These filters are used to reduce bright areas and enlarge the contours of dark ones (i.e. erosion) or its complement, to reduce the dilation (i.e. reduce dark zones and enlarge the brightness ones). The opening is used to extract the bright regions; the final result is the reunification of all these dispersed stained points. The resulting ranges of the ‘Contrast Enhancement’ and ‘Spatial Filtering’ thresholds implemented in the respective macros allow optimization of the appearance of the image (Fig. 1D) without altering the true immunoreactivity of the cases (data not shown).
In a third step, the colour, size and shape of the stained infiltrate cells were identified. Under the circumstances of HL stained tissue section, the ‘ground truth’ data should refer to data that have been validated using biological knowledge or tests. The ‘ground truth’ pixels range values of DAB deposits of the positive stained cells are provided by the observers from images of the training set using the HSI Histogram-based model (Hue-Saturation-Intensity) of the ‘Segmentation’ command (Fig. 1E). The collection of these ‘ground truth’ data enables us to calibrate the different thresholds of the segmentation data (for the different subcellular localizations), and helps in the creation of the algorithms and the analysis of all the other images of the test set. The thresholds pertaining to these commands allow simultaneous colour and morphological segmentation of positive elements. Subsequently, the area ranges of the separate cells were selected to exclude smaller and larger artefacts (Fig. 1F). The thresholds pertaining to these commands were also included in the respective macros and, using the ‘Count/size’ command, were applied individually on photographed images. This process automatically calculated the number and area of the positive stained elements such as individual cells and clusters (Fig. 1G). All the macros were used individually to analyse the different staining patterns of the photographed images. The global data obtained from these macros (Fig. 1H and I) were transferred automatically to a Microsoft Excel 2002 worksheet. The calculation time of each macro amounted to 5–6 s per photographed image. The global number of positive elements includes isolated cells but also aggregation of positive cells (clusters). To identify the number of elements that constitute these clusters, various cut-offs were previously established. For this purpose, the mean areas of individual positive cells were calculated for each marker on the basis of a large number of digital images. Positive elements greater than this mean were considered to be clustered infiltrates and were separated as individual cells by dividing their areas by the mean area obtained for individual cells (Fig. 1J). The results were then added to those obtained for the individual positive cells.
Statistical analysis
The intra-observer reproducibility (concordance of the measurements of the same image by a single observer), inter-observer reproducibility (concordance between two or more observers in the measurements of the same image), and the agreement between the manual and the semi-automatic methods were evaluated using the SPSS Statistical program (v. 11.1, SPSS Inc., Chicago, IL, USA).
To assess the level of agreement between the results from paired combinations of observers and the semi-automatic method, statistics for linear regression, Spearman's correlation coefficient, paired t-tests, intra-class correlation coefficient (ICC), and the Bland-Altman analysis were calculated. The ICC was derived from a random-effects two-way analysis of variance with an index of agreement ranging from 0 (no agreement) to 1 (perfect agreement). The Bland-Altman analysis assumes that neither system is a ‘gold standard’ and represents only a comparison of the two methods. In this analysis, the corresponding graphs present the difference between the paired measurements plotted against the average of the two values. If two determinations agree, then the difference between every pair determination should vary randomly around zero.
The conditional probabilities of observing a difference between paired measurements obtained by the manual and semi-automated methods were estimated using the Kaplan-Meier procedure. A cut-off value representing a difference of 10 cells/image was considered acceptable for our immunohistochemical quantification. This value was obtained from a consideration of the means of the differences between each pair of observers and each paired method when digital images contained fewer than 100 cells/image (range of difference from 1 to 12 cells).
Results
Intra-observer analysis of manual quantification
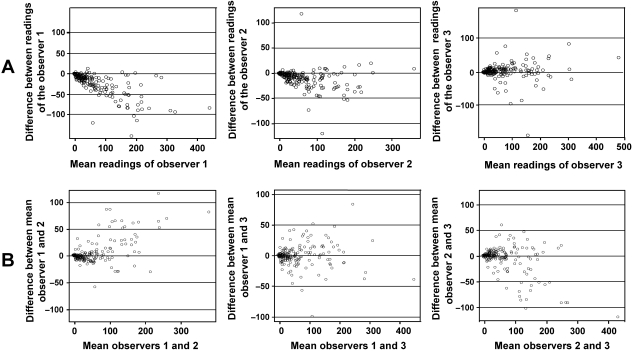
Spearman's correlation test revealed high and positive correlations between each pair of determinations for the three observers (r = 0.966 for the two readings of observer 1, r = 0.960 for the two readings of observer 2 and r = 0.930 for the two readings of observer 3, P < 0.0001). Nevertheless, the graphical representation of the Bland-Altman analysis (Fig. 2A) shows that the differences are not homogeneous and that there is variability between the two readings of observers 1 and 2. It is also important to mention that the dispersion of the data was greater when the digital image had more than the arbitrary value of 100 positive cells/image.
Fig. 2.
Bland-Altman representations of intra-observer measurements (A: first reading vs. second reading for each observer) and inter-observer measurements (B: mean of two readings of one observer vs. mean of two readings of other observer).
Paired t-tests were carried out to establish by how much the second determination was likely to differ from the first and therefore whether the two determinations were interchangeable (Table 1). The results indicate that, overall, the mean difference between the two determinations was significantly different (P < 0.0001) for observers 1 and 2, who both tended to score more positive cells in the second determination than in the first. The mean difference for observer 3 was not significantly different (P = 0.085), although this observer scored fewer positive cells during the second determination. The mean differences for the three staining patterns were statistically different for observers 1 and 2. Except for the cytoplasmic pattern, the mean differences of the membrane and nuclear patterns of observer 3 were not statistically different.
Table 1.
Analysis of intra-observer manual measurements
| Pair readings/patterns | Mean difference | SD | 95% CI of difference | P-value* |
|---|---|---|---|---|
| Observer 1: reading 1 vs. reading 2 | ||||
| Total | –22.20 | 27.461 | –26.07 to –18.33 | < 0.0001 |
| Membrane | –30.18 | 31.352 | –36.40 to –23.96 | < 0.0001 |
| Cytoplasm | –16.15 | 18.390 | –20.67 to–11.63 | < 0.0001 |
| Nuclear | –8.90 | 21.710 | –17.01 to –0.79 | 0.033 |
| Observer 2: reading 1 vs. reading 2 | ||||
| Total | –9.25 | 18.094 | –11.80 to –6.70 | < 0.0001 |
| Membrane | –10.54 | 23.561 | –15.21 to –5.87 | < 0.0001 |
| Cytoplasm | –11.05 | 9.691 | –13.43 to –8.66 | < 0.0001 |
| Nuclear | –1.00 | 3.543 | –2.32 to 0.32 | 0.133 |
| Observer 3: reading 1 vs. reading 2 | ||||
| Total | 3.46 | 28.000 | –0.49 to 7.40 | 0.085 |
| Membrane | 0.97 | 32.966 | –5.57 to 7.51 | 0.769 |
| Cytoplasm | 8.79 | 25.411 | 2.54 to 15.03 | 0.007 |
| Nuclear | 0.03 | 4.064 | –1.48 to 1.55 | 0.964 |
Paired t-test. CI, confidence interval.
Although the intra-class correlation coefficients present an excellent degree of agreement, these results were not statistically applicable because the conditions of equality of variances were not satisfied.
In summary, these results indicated intra-observer differences in measurements for the three staining patterns, from small differences between the two determinations of nuclear staining to major differences between the two determinations of cytoplasmic and membrane staining.
Inter-observer analysis of manual quantification
Regression of the mean of paired determinations of one observer on the mean of paired determinations of other observer also reveals high and positive correlations (r = 0.964 for observer 1 vs. observer 2, r = 0.0972 for observer 1 vs. observer 3 and r = 0.0942 for observer 2 vs. observer 3, P < 0.0001). The Bland-Altman analysis, illustrated in Fig. 2B, shows the dispersion of the data around the mean difference between the different observers. Again, the variability between measurements was greater when the digital image contained more than 100 positive cells/image.
The mean inter-observer difference was generally lower than found in the intra-observer evaluation (Table 2). Observer 3 tended to score more positive cells than did observers 1 and 2. The mean differences between observers 1 and 2 and between observers 2 and 3 were statistically significant (P = 0.003 and P = 0.002, respectively). No significant difference was observed between observers 1 and 3 (P = 0.473).
Table 2.
Analysis of inter-observer manual measurements
| Pair observers/patterns | Mean difference | SD | 95% CI of difference | P-value* |
|---|---|---|---|---|
| Observer 1 vs. Observer 2 | ||||
| Total | 4.85 | 22.605 | 1.67 to 8.04 | 0.003 |
| Membrane | 12.94 | 28.581 | 7.27 to 18.61 | < 0.0001 |
| Cytoplasm | –4.90 | 7.707 | –6.80 to –3.01 | 0.000 |
| Nuclear | –0.65 | 6.382 | –3.03 to 1.73 | 0.581 |
| Observer 1 vs. Observer 3 | ||||
| Total | –0.91 | 17.731 | –3.41 to 1.59 | 0.473 |
| Membrane | 1.68 | 20.729 | –2.44 to 5.79 | 0.421 |
| Cytoplasm | –3.30 | 15.463 | –7.10 to 0.50 | 0.087 |
| Nuclear | –4.27 | 7.967 | –7.24 to –1.29 | 0.006 |
| Observer 2 vs. Observer 3 | ||||
| Total | –5.76 | 25.854 | –9.40 to –2.12 | 0.002 |
| Membrane | –11.27 | 33.377 | –17.89 to –4.64 | 0.001 |
| Cytoplasm | 1.60 | 13.902 | –1.82 to 5.02 | 0.354 |
| Nuclear | –3.62 | 5.104 | –5.52 to –1.71 | 0.001 |
Paired t-test. CI, confidence interval.
The intra-class correlation coefficients showed excellent agreement but the condition of equality of variances was not satisfied. In summary, the inter-observer comparisons demonstrated constancy in the differences noted for the nuclear pattern, but the differences for the cytoplasmic and membrane patterns were markedly lower than those found in the intra-observer comparison.
Comparison with semi-automated quantification with Image-Pro Plus
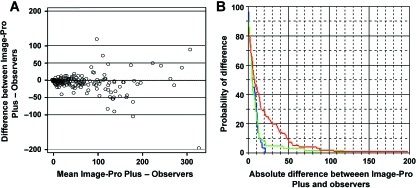
With our semi-automatic process, the same macro applied to the same image always gave the same results, even when it was run by different users. Plotting the global mean of the manual microscopic scores against the results obtained with the image analysis program illustrates a very high and positive correlation coefficient (r = 0.980, P < 0.001). The Bland-Altman analysis, as illustrated in Fig. 3A and Fig. 4A(in black), indicates that the differences between the semi-automated and the manual methods appear closer to zero than the manual microscopic differences alone (Fig. 4A, in red). Nevertheless, the discordance between the manual and semi-automated methods persisted for images containing more than 100 positive cells/image.
Fig. 3.
Comparison of manual microscopic (mean of all observers) and semi-automated methods by Bland-Altman graph (A) and Kaplan-Meier curves (B). Nuclear-staining pattern (in blue), cytoplasm-staining pattern (in green), membrane-staining pattern (in red).
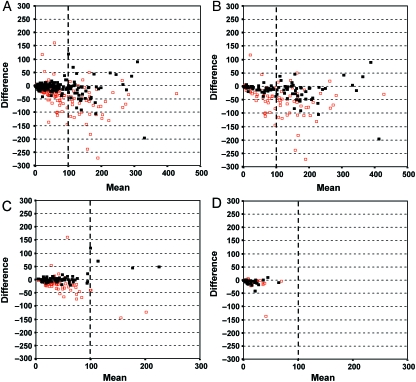
Fig. 4.
Superimposed Bland-Altman graphs comparing manual microscopic and semi-automated methods (in black), and first and second manual microscopic readings (in red) (A). The differences between the semi-automated and the manual methods appear closer to zero than the differences between manual microscopic readings for membrane (B), cytoplasmic (C) and nuclear (D) patterns of staining.
Considering the different patterns of staining, the conditional probability of observing differences between the semi-automated and the manual methods was evaluated using the Kaplan-Meier method (Fig. 3B). The curves indicated a difference greater than 10 cells/image in 30% of cases for the nuclear and cytoplasmic patterns, and in 47% of cases for the membrane pattern. Nevertheless, superimposing the graphs comparing the manual microscopic and semi-automated methods with manual microscopic method alone shows that the differences between the semi-automated and the manual methods appear closer to zero than the differences between manual microscopic evaluations alone. This is especially true for membrane (Fig. 4B) and cytoplasmic patterns (Fig. 4C). Although the information about the nuclear pattern is visibly limited (Fig. 4D), the previous results of our group confirm the concordance between the automated and the manual methods for this kind of staining pattern (Lopez et al. 2008).
Discussion
The manual microscopic quantification of immunostained cells is the most frequently used method, and is the main source of discrepancies in immunohistochemical interpretations that lead to contradictory reports. This study aimed to compare and optimize the manual microscopic and semi-automated methods for quantifying the frequency of positive DAB-immunostained cells in HL TMA in function of the subcellular distribution of the staining.
The most important disadvantage associated with the manual evaluation and interpretation of immunostains on tissue samples is the variability of the cell counts obtained, which arises from the large number of variables that influence the cell count, such as antigen staining, heterogeneity of stain intensities, variation in cell size, shape, and distribution (clusters). The findings underline the necessity of developing a more reproducible method of quantification that eliminates the major causes of this variability. Different investigations have addressed different aspects of the problem of reproducibility in immunohistochemical methods, focusing upon improved sample preparation (fixation), more effective methods of antigen retrieval, and the development of external standards and controls (Taylor, 2006). Furthermore, the difficulties of distinguishing subtle differences in the staining intensity by eye do not permit a precise quantification of immunohistochemical stained cells. This subjectivity makes it difficult to establish criteria for cell positivity and has given rise to different scoring definitions (Seidal et al. 2001; Rubin et al. 2004; Taylor, 2006; Yaziji & Barry, 2006; de Jong et al. 2007). More concretely, the subjectivity of the determination of positivity arising from the interpretation of DAB intensity appears clearly implicated as a cause of variable intra- and inter-observer precision. Our results confirm the existence of intra- and inter-observer variations observed for manual microscopic digital image analysis. This issue has already been addressed in other published studies but has not produced a consensual conclusion, probably due to the different methods of staining and analysis employed. Whereas some authors have demonstrated that inter-observer differences are greater than intra-observer differences (McCarty et al. 1985; Johnsson et al. 1994; Mosedale et al. 1996; Johansson et al. 2001), others have published results indicating the opposite relationship, and have observed no significant differences between the two (Jagoe et al. 1991; Johnsson et al. 1994). The fact that the variability of our results is less pronounced in inter-observer than intra-observer comparisons suggests that it would be preferable if this kind of quantification were done once by several pathologists rather than repeatedly by the same pathologist.
The special attention to the precise subcellular localization of the immunohistochemical signals is essential for properly interpreting immunostains and distinguishing genuine positive staining (Cheuk & Chan, 2004). In this study, the information concerning the nuclear markers is more limited that for the two other patterns of staining. This is because traditional biologic nuclear markers that influence the outcome of HL are mainly limited to Ki67 and p53 and because the number of positive nuclei does not exceed 100 nuclei/image. Nevertheless, we have previously published a work that demonstrates that the automated method represents an objective and accurate quantification system for nuclear markers. The concordance of these results with the manual method was approximately 90%, independently of the tissue specimen and the degree of difficulty observed with the digital images (Lopez et al. 2008). In the present study, our results indicate that the quantification of immunostained cells by different pathologists produces smaller differences in estimates for markers located in the nucleus and cytoplasm. This could be explained by the ease of visualization of DAB-staining in these two locations. Membrane staining may be more difficult to identify, especially when the digital images contain groups of positive cells (clusters) or positive granules in the cytoplasm of the cells. When the results were considered positive or negative for the diagnosis, the intra- and inter-observer variability had little effect on the results. In contrast, when the threshold levels of positivity and cut-off values affected the diagnosis, the prognosis or the treatment, the greater variability observed in images with more than 100 positive cells/image had a substantial impact on the result.
To eliminate subjectivity in the interpretation of positive staining, a number of automated methods of separation and quantification of immunohistochemical staining have been investigated (Montironi et al. 1996; Lehr et al. 1997; Smejkal & Shainoff, 1997; Kohlberger et al. 1999; Ma & Lozanoff, 1999; Vilaplana & Lavialle, 1999; Ruifrok & Johnston, 2001; King et al. 2002; McGinley et al. 2002). These studies have principally focused on the method for differentiating positive DAB-nuclear staining from other haematoxylin staining (i.e. nuclear and background staining) (Goto et al. 1992; Huang et al. 1996; Ruifrok, 1997; King et al. 2002; Brey et al. 2003). To our knowledge, the development and standardization of the algorithms that allow the detection and differentiation of this positive staining independently of the location of the DAB deposit in the cell have not been sufficiently investigated. The software Image-Pro Plus incorporates the standard image processing functions and multiple measurements following calibration. The software allows digital images to be segmented using the RGB (red, green, blue) or the HSI (hue, saturation, intensity) Histogram-based models. Our previous automated process that was developed to quantify different nuclear markers in digital images with various degree of complexity (Lopez et al. 2008) has been implemented with parameters of segmentation determined with the RGB Histogram-based model. In these precise conditions, the RGB model enables us to avoid overlapping in the ranges of positive and negative DAB colour values of the different positive nuclear markers. However, when the objective is to evaluate the DAB colour in membrane or in cytoplasm, the RGB model is the least suitable option to separate the DAB-stained pixels from the background. Effectively, for these precise localizations, the selection of brown deposits by setting a threshold on the different three channels has been seen to be complicated by the interference of the counterstaining (haematoxylin), mainly in the blue channel (Kuyatt et al. 1993; Ruifrok, 1997; King et al. 2002). Under these conditions, and as the HSI model has been previously described to allow the separation of DAB-stained areas from background staining with a low level pixel misclassification (Brey et al. 2003), we have developed our macros with this colour space. Concretely, the standard modifications of the threshold have been realized firstly on the blue channel values and the images have then been segmented with the HSI colour space. The various sub-cellular locations of DAB-staining required special filtering to eliminate impulse noise (using mean and median enhancement filters) and different morphological filters to dilate and smooth the edges of the objects to unify the positive pixels. The complete scripting language and the capabilities of extension of the program, without the need of additional language expertise, permitted us to include the automation of the different steps described in this paper in an algorithm referred as a macro. Under these conditions, the problems associated with subjectivity of scoring with the naked eye are clearly avoided, and the same macro applied to the same image always gives the same results.
The interest and the complexity of this new methodology are demonstrated by the comparative study with the manual evaluation of the same digital images. Currently, there is controversy as to which is the better statistical method for determining concordance between distinct quantification methods. The plotting of differences of the semi-automated vs. the manual system per observer (Bland-Altman method) permits the interpretation of the results as a function of the differences between the values obtaining using the two quantification methods and depends on the magnitude of the variables that are being analysed (Bland & Altman, 1995). Our results indicate that there was a better concordance between the semi-automated and the manual methods than between intra- and inter-observer evaluations alone. This indicates that our semi-automated method is a reliable alternative method for the quantitative analysis of immunostains, especially for membrane and cytoplasmic patterns. Nevertheless, the differences observed remain important when digital images contain more than 100 positive cells. If the level of difference needs to be related to the total number of positive cells in each image, in most cases featuring high cellular density, the variability in the estimates is not generally of clinical importance. However, the parameters of segmentation need to be improved due to the fact that, for some specific markers, the exact number may determine the treatment that has to be applied.
The accuracy and precision of these macros for the detection and quantification of the immunostained cells have permitted the identification and evaluation of different components of the reactive immune microenvironment present in different haematological malignancies including follicular lymphoma (Alvaro et al. 2006a,b) and Hodgkin's lymphoma (Alvaro et al. 2005; Bosch et al. 2005). These results have been correlated with the clinical-biological features of these patients and have been demonstrated to have a significant impact on their outcome in the same order as results obtained with a manual evaluation (Alvaro-Naranjo et al. 2005).
In conclusion, the development of specific macros with the commercial available software Image-Pro Plus is a valid method for the manual microscopic quantification. Although improvements related to the speed and overall assessment of image complexities are warranted, the use of our semi-automated method offers clear advantages over the manual method in terms of improved precision, repeatability and objective evaluation of HL immunostains. Further improvements in the rapidity and avoidance of subjective assessments of the different sub-cellular locations of the staining appear to be required.
Acknowledgments
This work was supported by grants FIS 04/1440, 04/1467, and 05/1527 from the Ministerio de Ciencia y Tecnología, Spain. We are grateful to Maria del Mar Barberà, Bàrbara Tomàs, Vanesa Gestí, Ana Suñé, and Marc Iniesta for their technical assistance.
References
- Aasmundstad TA, Haugen OA, Johannesen E, Hoe AL, Kvinnsland S. Oestrogen receptor analysis: correlation between enzyme immunoassay and immunohistochemical methods. J Clin Pathol. 1992;45:125–129. doi: 10.1136/jcp.45.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro T, Lejeune M, Salvado MT, et al. Outcome in Hodgkin's lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin Cancer Res. 2005;11:1467–1473. doi: 10.1158/1078-0432.CCR-04-1869. [DOI] [PubMed] [Google Scholar]
- Alvaro T, Lejeune M, Camacho FI, et al. The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma. Haematologica. 2006a;91:1605–1612. [PubMed] [Google Scholar]
- Alvaro T, Lejeune M, Salvado MT, et al. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J Clin Oncol. 2006b;24:5350–5357. doi: 10.1200/JCO.2006.06.4766. [DOI] [PubMed] [Google Scholar]
- Alvaro-Naranjo T, Lejeune M, Salvado-Usach MT, et al. Tumor-infiltrating cells as a prognostic factor in Hodgkin's lymphoma: a quantitative tissue microarray study in a large retrospective cohort of 267 patients. Leuk Lymphoma. 2005;46:1581–1591. doi: 10.1080/10428190500220654. [DOI] [PubMed] [Google Scholar]
- Bhatnagar J, Tewari HB, Bhatnagar M, Austin GE. Comparison of carcinoembryonic antigen in tissue and serum with grade and stage of colon cancer. Anticancer Res. 1999;19:2181–2187. [PubMed] [Google Scholar]
- Bland JM, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085–1087. doi: 10.1016/s0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- Bosch Princep R, Lejeune M, Salvado Usach MT, Jaen Martinez J, Pons Ferre LE, Alvaro Naranjo T. Decreased number of granzyme B+ activated CD8+ cytotoxic T lymphocytes in the inflammatory background of HIV-associated Hodgkin's lymphoma. Ann Hematol. 2005;84:661–666. doi: 10.1007/s00277-005-1051-3. [DOI] [PubMed] [Google Scholar]
- Brey EM, Lalani Z, Johnston C, et al. Automated selection of DAB-labeled tissue for immunohistochemical quantification. J Histochem Cytochem. 2003;51:575–584. doi: 10.1177/002215540305100503. [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liu D, Tavanapong W, Wong J, Oh J, de Groen PC. Computer-aided detection of diagnostic and therapeutic operations in colonoscopy videos. IEEE Trans Biomed Eng. 2007;54:1268–1279. doi: 10.1109/TBME.2007.890734. [DOI] [PubMed] [Google Scholar]
- Cheuk W, Chan JK. Subcellular localization of immunohistochemical signals: knowledge of the ultrastructural or biologic features of the antigens helps predict the signal localization and proper interpretation of immunostains. Int J Surg Pathol. 2004;12:185–206. doi: 10.1177/106689690401200301. [DOI] [PubMed] [Google Scholar]
- Garcia JF, Camacho FI, Morente M, et al. Hodgkin and Reed-Sternberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue microarrays. Blood. 2003;101:681–689. doi: 10.1182/blood-2002-04-1128. [DOI] [PubMed] [Google Scholar]
- Gil J, Wu HS. Applications of image analysis to anatomic pathology: realities and promises. Cancer Invest. 2003;21:950–959. doi: 10.1081/cnv-120025097. [DOI] [PubMed] [Google Scholar]
- Goto M, Nagatomo Y, Hasui K, Yamanaka H, Murashima S, Sato E. Chromaticity analysis of immunostained tumor specimens. Pathol Res Pract. 1992;188:433–437. doi: 10.1016/S0344-0338(11)80033-6. [DOI] [PubMed] [Google Scholar]
- Hedvat CV, Jaffe ES, Qin J, et al. Macrophage-derived chemokine expression in classical Hodgkin's lymphoma: application of tissue microarrays. Mod Pathol. 2001;14:1270–1276. doi: 10.1038/modpathol.3880473. [DOI] [PubMed] [Google Scholar]
- Huang X, Chen S, Tietz EI. Immunocytochemical detection of regional protein changes in rat brain sections using computer-assisted image analysis. J Histochem Cytochem. 1996;44:981–987. doi: 10.1177/44.9.8773563. [DOI] [PubMed] [Google Scholar]
- Jagoe R, Steel JH, Vucicevic V, et al. Observer variation in quantification of immunocytochemistry by image analysis. Histochem J. 1991;23:541–547. doi: 10.1007/BF01041181. [DOI] [PubMed] [Google Scholar]
- Johansson AC, Visse E, Widegren B, Sjogren HO, Siesjo P. Computerized image analysis as a tool to quantify infiltrating leukocytes: a comparison between high- and low-magnification images. J Histochem Cytochem. 2001;49:1073–1079. doi: 10.1177/002215540104900902. [DOI] [PubMed] [Google Scholar]
- Johnsson A, Olsson C, Anderson H, Cavallin-Stahl E. Evaluation of a method for quantitative immunohistochemical analysis of cisplatin-DNA adducts in tissues from nude mice. Cytometry. 1994;17:142–150. doi: 10.1002/cyto.990170206. [DOI] [PubMed] [Google Scholar]
- de Jong D, Rosenwald A, Chhanabhai M, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications – a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2007;25:805–812. doi: 10.1200/JCO.2006.09.4490. [DOI] [PubMed] [Google Scholar]
- King TW, Brey EM, Youssef AA, Johnston C, Patrick CW., Jr Quantification of vascular density using a semiautomated technique for immunostained specimens. Anal Quant Cytol Histol. 2002;24:39–48. [PubMed] [Google Scholar]
- Kohlberger PD, Breitenecker F, Kaider A, et al. Modified true-color computer-assisted image analysis versus subjective scoring of estrogen receptor expression in breast cancer: a comparison. Anticancer Res. 1999;19:2189–2193. [PubMed] [Google Scholar]
- Kuyatt BL, Reidy CA, Hui KY, Jordan WH. Quantitation of smooth muscle proliferation in cultured aorta. A color image analysis method for the Macintosh. Anal Quant Cytol Histol. 1993;15:83–87. [PubMed] [Google Scholar]
- Lee AM, Clear AJ, Calaminici M, et al. Number of CD4+ cells and location of forkhead box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J Clin Oncol. 2006;24:5052–5059. doi: 10.1200/JCO.2006.06.4642. [DOI] [PubMed] [Google Scholar]
- Lehr HA, Mankoff DA, Corwin D, Santeusanio G, Gown AM. Application of Photoshop-based image analysis to quantification of hormone receptor expression in breast cancer. J Histochem Cytochem. 1997;45:1559–1565. doi: 10.1177/002215549704501112. [DOI] [PubMed] [Google Scholar]
- Leong AS. Quantitation in immunohistology: fact or fiction? A discussion of variables that influence results. Appl Immunohistochem Mol Morphol. 2004;12:1–7. doi: 10.1097/00129039-200403000-00001. [DOI] [PubMed] [Google Scholar]
- Leong FJ, Leong AS. Digital imaging in pathology: theoretical and practical considerations, and applications. Pathology. 2004;36:234–241. doi: 10.1080/00313020410001692576. [DOI] [PubMed] [Google Scholar]
- Li SH, Li CF, Sung MT, et al. Skp2 is an independent prognosticator of gallbladder carcinoma among p27(Kip1)-interacting cell cycle regulators: an immunohistochemical study of 62 cases by tissue microarray. Mod Pathol. 2007;20:497–507. doi: 10.1038/modpathol.3800762. [DOI] [PubMed] [Google Scholar]
- Lopez C, Lejeune M, Salvado MT, et al. Automated quantification of nuclear immunohistochemical markers with different complexity. Histochem Cell Biol. [Epub ahead of print] [DOI] [PubMed]
- Ma W, Lozanoff S. A full color system for quantitative assessment of histochemical and immunohistochemical staining patterns. Biotech Histochem. 1999;74:1–9. doi: 10.3109/10520299909066470. [DOI] [PubMed] [Google Scholar]
- McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS., Sr Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109:716–721. [PubMed] [Google Scholar]
- McGinley JN, Knott KK, Thompson HJ. Semi-automated method of quantifying vasculature of 1-methyl-1-nitrosourea-induced rat mammary carcinomas using immunohistochemical detection. J Histochem Cytochem. 2002;50:213–222. doi: 10.1177/002215540205000209. [DOI] [PubMed] [Google Scholar]
- Media Cybernetics. Image-Pro Plus – applications notes. Silver Spring: Media Cybernetics; 2002. URL: http://www.mediacy.com/action.htm.
- Mofidi R, Walsh R, Ridgway PF, et al. Objective measurement of breast cancer oestrogen receptor status through digital image analysis. Eur J Surg Oncol. 2003;29:20–24. doi: 10.1053/ejso.2002.1373. [DOI] [PubMed] [Google Scholar]
- Montironi R, Diamanti L, Thompson D, Bartels HG, Bartels PH. Analysis of the capillary architecture in the precursors of prostate cancer: recent findings and new concepts. Eur Urol. 1996;30:191–200. doi: 10.1159/000474169. [DOI] [PubMed] [Google Scholar]
- Mosedale DE, Metcalfe JC, Grainger DJ. Optimization of immunofluorescence methods by quantitative image analysis. J Histochem Cytochem. 1996;44:1043–1050. doi: 10.1177/44.9.8773570. [DOI] [PubMed] [Google Scholar]
- Nabi G, Seth A, Dinda AK, Gupta NP. Computer based receptogram approach: an objective way of assessing immunohistochemistry of androgen receptor staining and its correlation with hormonal response in metastatic carcinoma of prostate. J Clin Pathol. 2004;57:146–150. doi: 10.1136/jcp.2003.010520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podhajsky RJ, Bidanset DJ, Caterson B, Blight AR. A quantitative immunohistochemical study of the cellular response to crush injury in optic nerve. Exp Neurol. 1997;143:153–161. doi: 10.1006/exnr.1996.6354. [DOI] [PubMed] [Google Scholar]
- Rhodes A, Jasani B, Anderson E, Dodson AR, Balaton AJ. Evaluation of HER-2/neu immunohistochemical assay sensitivity and scoring on formalin-fixed and paraffin-processed cell lines and breast tumors: a comparative study involving results from laboratories in 21 countries. Am J Clin Pathol. 2002;118:408–417. doi: 10.1309/97WN-W6UX-XJWT-02H2. [DOI] [PubMed] [Google Scholar]
- Rosen DG, Huang X, Deavers MT, Malpica A, Silva EG, Liu J. Validation of tissue microarray technology in ovarian carcinoma. Mod Pathol. 2004;17:790–797. doi: 10.1038/modpathol.3800120. [DOI] [PubMed] [Google Scholar]
- Rubin MA, Zerkowski MP, Camp RL, et al. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164:831–840. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruifrok AC. Quantification of immunohistochemical staining by color translation and automated thresholding. Anal Quant Cytol Histol. 1997;19:107–113. [PubMed] [Google Scholar]
- Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- Sandlund J, Hedberg Y, Bergh A, Grankvist K, Ljungberg B, Rasmuson T. Evaluation of CD31 (PECAM-1) expression using tissue microarray in patients with renal cell carcinoma. Tumour Biol. 2007;28:158–164. doi: 10.1159/000102980. [DOI] [PubMed] [Google Scholar]
- Seidal T, Balaton AJ, Battifora H. Interpretation and quantification of immunostains. Am J Surg Pathol. 2001;25:1204–1207. doi: 10.1097/00000478-200109000-00013. [DOI] [PubMed] [Google Scholar]
- Simone NL, Remaley AT, Charboneau L, et al. Sensitive immunoassay of tissue cell proteins procured by laser capture microdissection. Am J Pathol. 2000;156:445–452. doi: 10.1016/S0002-9440(10)64749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Kim D, Mohler JL. Java Web Start based software for automated quantitative nuclear analysis of prostate cancer and benign prostate hyperplasia. Biomed Eng Online. 2005;4:31. doi: 10.1186/1475-925X-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smejkal GB, Shainoff JR. Enhanced digital imaging of diaminobenzidene-stained immunoblots. Biotechniques. 1997;22:462. doi: 10.2144/97223bm20. [DOI] [PubMed] [Google Scholar]
- Taylor CR. The total test approach to standardization of immunohistochemistry. Arch Pathol Lab Med. 2000;124:945–951. doi: 10.5858/2000-124-0945-TTTATS. [DOI] [PubMed] [Google Scholar]
- Taylor CR. Quantifiable internal reference standards for immunohistochemistry: the measurement of quantity by weight. Appl Immunohistochem Mol Morphol. 2006;14:253–259. doi: 10.1097/00129039-200609000-00001. [DOI] [PubMed] [Google Scholar]
- Tubbs RR, Swain E, Pettay JD, Hicks DG. An approach to the validation of novel molecular markers of breast cancer via TMA-based FISH scanning. J Mol Histol. 2007;38:141–150. doi: 10.1007/s10735-006-9076-z. [DOI] [PubMed] [Google Scholar]
- Veltri RW, Partin AW, Miller MC. Quantitative nuclear grade (QNG): a new image analysis-based biomarker of clinically relevant nuclear structure alterations. J Cell Biochem Suppl. 2000;(Suppl 35):151–157. doi: 10.1002/1097-4644(2000)79:35+<151::aid-jcb1139>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Vilaplana J, Lavialle M. A method to quantify glial fibrillary acidic protein immunoreactivity on the suprachiasmatic nucleus. J Neurosci Methods. 1999;88:181–187. doi: 10.1016/s0165-0270(99)00016-3. [DOI] [PubMed] [Google Scholar]
- Yaziji H, Barry T. Diagnostic immunohistochemistry: what can go wrong? Adv Anat Pathol. 2006;13:238–246. doi: 10.1097/01.pap.0000213041.39070.2f. [DOI] [PubMed] [Google Scholar]
- Zettl A, Meister S, Katzenberger T, et al. Immunohistochemical analysis of B-cell lymphoma using tissue microarrays identifies particular phenotypic profiles of B-cell lymphomas. Histopathology. 2003;43:209–219. doi: 10.1046/j.1365-2559.2003.01702.x. [DOI] [PubMed] [Google Scholar]
- Zu Y, Steinberg SM, Campo E, et al. Validation of tissue microarray immunohistochemistry staining and interpretation in diffuse large B-cell lymphoma. Leuk Lymphoma. 2005;46:693–701. doi: 10.1080/10428190500051844. [DOI] [PubMed] [Google Scholar]