Abstract
Bves is an integral membrane protein with no determined function and no homology to proteins outside of the Popdc family. It is widely expressed throughout development in myriad organisms. Here, we demonstrate an interaction between Bves and guanine nucleotide exchange factor T (GEFT), a GEF for Rho-family GTPases. This interaction represents the first identification of any protein that has a direct physical interaction with any member of the Popdc family. Bves and GEFT are shown to colocalize in adult skeletal muscle. We also demonstrate that exogenous expression of Bves reduces Rac1 and Cdc42 activity levels while not affecting levels of active RhoA. Consistent with a repression of Rac1 and Cdc42 activity, we show changes in speed of cell locomotion and cell roundness also result from exogenous expression of Bves. Modulation of Rho-family GTPase signaling by Bves would be highly consistent with previously described phenotypes occurring upon disruption of Bves function in a wide variety of model systems. Therefore, we propose Bves as a novel regulator of the Rac1 and Cdc42 signaling cascades.
Keywords: Popdc, cell motility, GEF
Bves (blood vessel epicardial substance) is the most studied member of the Popdc family, which is a group of evolutionarily conserved transmembrane proteins. Bves was discovered by our laboratory in 1999, and is widely expressed throughout development and adulthood in many different species. All three developing germ layers (1), cardiac muscle (2–6), skeletal muscle (2, 6, 7), neural tissues (1, 2), epicardium (1, 8–12), epithelial components of the eye (13), and smooth muscle (1, 6) have been demonstrated to express bves. Although expression of the Bves protein is now resolved, few definitive indications of molecular function exist.
Several possibilities for potential Bves function have been described. Epithelial integrity of cultured corneal cells is severely decreased by knockdown of Bves protein by using morpholino oligonucleotides, possibly via an interaction with ZO-1 at the tight junction (14). Perturbation of Bves function has also been shown to disrupt proper migration of epithelial components of the early Xenopus embryo (15) and affect wound healing of epithelia in scratch assays (13). Mice null for the bves gene are delayed in regeneration of skeletal muscle upon injury (7). A recent report demonstrates that Bves knockdown by using antisense RNA during Drosophila oogenesis results in failure of pole cells to migrate properly to the anterodorsal side of the embryo (16). Despite these studies, no direct molecular mechanism for any of these phenotypes exists at this time.
In an effort to ascribe molecular function, we conducted a yeast two-hybrid screen and identified guanine nucleotide exchange factor T (GEFT) (17) as a protein that interacts with the cytoplasmic portion of Bves. GEF proteins modulate activity of small GTPases, specifically the Rho family of GTPases, Rac1 and Cdc42, in the case of GEFT (17–20). GEFs stimulate the exchange of GDP for GTP, thereby activating Rho small GTPases. The small GTPases, Rac1 and Cdc42, have myriad effects on cell behavior, including control of proliferation, differentiation, cell motility, and gene expression (21, 22). Given the known functions of GEFs and Bves in regulation of cell differentiation and motility, this relationship was studied in detail.
The present study is the first to demonstrate direct physical interaction of Bves with any protein and linkage to any known molecular pathway. We further describe this interaction by determining that transfection of a truncated version of Bves decreases Rac1 and Cdc42 activity, and that transfection of this Bves truncation or full-length Bves also decreases motility of NIH 3T3 cells in real-time assays. Modulation of GEFT function by Bves provides molecular explanation of phenotypes previously observed with disruption of Bves and is critical for future investigation of the function of this protein.
Results
The Cytoplasmic C Terminus of Bves Interacts with GEFT.
A yeast two-hybrid screen was used to isolate Bves-interacting proteins from an embryonic mouse heart library. The cytoplasmic carboxyl terminal portion of Bves (amino acids 115–358) was used for this screen. This region of Bves contains the uncharacterized Popdc domain (23–25). Using a yeast two-hybrid screen with cDNAs expressed in the embryonic mouse heart, we isolated 104 interacting proteins when the carboxyl terminus (amino acids 115–358) of mBves was used as bait. Two independent clones were isolated that contained the coding sequence for amino acids 46–344 of the mouse GEFT protein. Both of these clones passed the false-positive screening process. As previous experiments have shown defects in cell motility/interaction (13–15), we chose to pursue this interaction further.
Deletion Analysis of Interacting Domains.
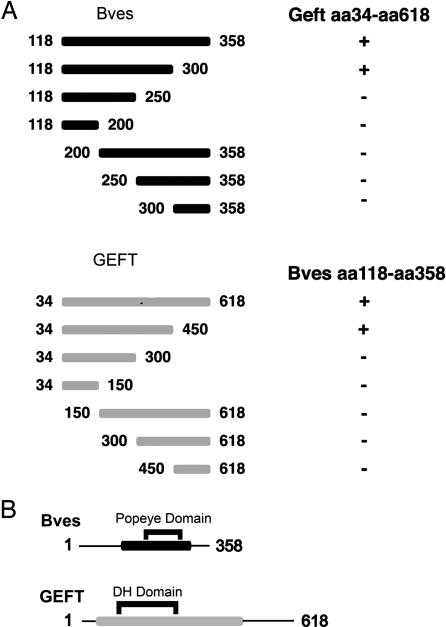
To determine which regions of the Bves and GEFT proteins were responsible for the interaction revealed by the yeast two-hybrid screen, a deletion analysis further using the yeast two-hybrid method was used. A series of truncations of the cytoplasmic portion of Bves revealed that the portion of the protein between amino acids 250 and 300 is critical for interaction with GEFT (Fig. 1A).
Fig. 1.
Interaction domain analysis. (A) Bves and GEFT truncations were generated by using PCR for further definition of interacting domains. By using a yeast two-hybrid strategy, Bves truncations (black bars) were screened against full-length GEFT (amino acids 34–618) for interaction. GEFT truncations (gray bars) were screened against the intracellular C terminus (amino acids 118–358) of Bves. Results of matings listed on right: + signifies growth on selective media, whereas − indicates no growth observed upon mating. (B) Minimal interaction domain of Bves (black bar) contains the Popdc domain, whereas minimal interaction domain of GEFT (gray bar) contains the Dbl homology domain.
The truncation analysis to determine the region of the GEFT protein responsible for interaction with Bves revealed that the portion of the protein between amino acid 300 and amino acid 400 is necessary for interaction with Bves (Fig. 1A). However, further analysis of the results of these studies revealed that these regions (amino acids 250–300 of Bves and amino acids 300–450 of GEFT) are not sufficient for the Bves–GEFT interaction to occur, as neither amino acids 250–300 of Bves or amino acids 300–450 of GEFT interact with the other full-length interacting partner. Thus, the data presented here demonstrate that the amino acid 250–300 region of Bves and the amino acid 300–450 region of GEFT are necessary but not sufficient for the interaction between these two proteins.
Bves and GEFT Colocalize in Muscle Cells.
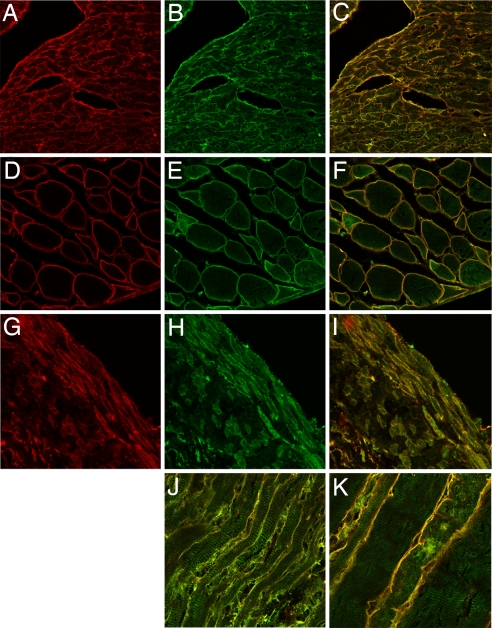
As both Bves and GEFT are highly expressed in muscle (2, 6, 20, 26), we next examined their localization in mouse hindlimb muscle, the heart, and intestinal smooth muscle. As shown in Fig. 2 F and K, Bves and GEFT colocalize at the cell membrane in skeletal muscle. Localization is not complete, as GEFT staining is also observed in myofibrils (26) (Fig. 2K). A similar situation is observed in cardiac muscle, where intense overlap of staining is observed at the cell surface, with additional GEFT reactivity with the contractile apparatus (Fig. 2 C and J). Finally, intestinal smooth muscle also showed the same intensity of staining at the cell surface (Fig. 2I). These data indicate that Bves and GEFT codistribute within cells.
Fig. 2.
Colocalization of Bves and GEFT in cross and transverse sections of mouse cardiac, skeletal, and smooth muscle. Bves, shown in red (A, D, and G), is primarily distributed at the cell periphery in cardiac (A–C, J), skeletal (D–F, K), and smooth muscle (G–I). GEFT (B, E, and H) also has distribution at the cell membrane in these muscle types, but it displays a broader intracellular localization at the myofibrils. Merged images are shown in C, F, I, J, and K.
Biochemical Verification of mBves–mGEFT Interaction.
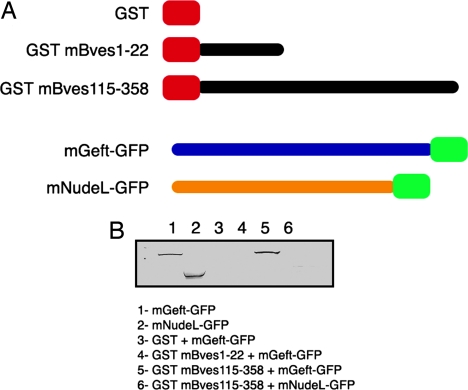
Using a GST-pulldown strategy, our laboratory biochemically confirmed the Bves–GEFT interaction revealed by the yeast two-hybrid screen. Prokaryotic GST fusion protein expression constructs of Bves were generated, whereas GEFT-GFP fusion protein expression plasmids were generated for use in mammalian cells. COS-7 cells were transfected with the GEFT-GFP expression plasmid, and protein was harvested and incubated with GST-mBves Sepharose. Fig. 3B demonstrates that mBves specifically pulls down GEFT protein, whereas no interaction is detected by using GFP-NudeL protein as a negative control. NudeL is a microtubule-binding protein unrelated to Bves function (27, 28). It should also be noted that the Dbl homology (DH) domain (29), which is the domain of GEFT responsible for nucleotide exchange activity with GTPases, falls within the region that we have found to be necessary for GEFT–Bves interaction. This domain is present in all Dbl family members; thus, we tested two other members of this family, Vav1 and Cool1, for interaction with Bves by using the GST-pulldown method. We were unable to precipitate Vav1 above background [supporting information (SI) Fig. S1A]. However, we were able to pull down Cool1 above background, but not to the degree that GEFT was reactive (Fig. S1B). Thus, our results indicate that Bves interacts preferentially with mGEFT in this assay and corroborate the yeast two-hybrid analyses.
Fig. 3.
GST-Bves pulldown of GEFT. GST-Bves fusion proteins representing the extracellular N terminus and cytoplasmic C terminus of Bves were tested for interaction with GEFT-GFP and NudeL-GFP. Representative mobilities of mGEFT-GFP and mNudeL-GFP are provided (lanes 1 and 2). No reactivity is observed in lanes containing isolates from GST/mGEFT-GFP (lane 3), GST-mBves1–22/mGEFT-GFP (lane 4), or GST-mBves 115–358/mNudeL-GFP pulldowns (lane 6), indicating that these proteins do not interact. A band representing mGEFT-GFP is clearly seen in the lane containing isolate from the GST-mBves115–358/mGEFT-GFP pulldown (lane 5).
Exogenous Expression of mBves Affects Activation of Rac and Cdc42.
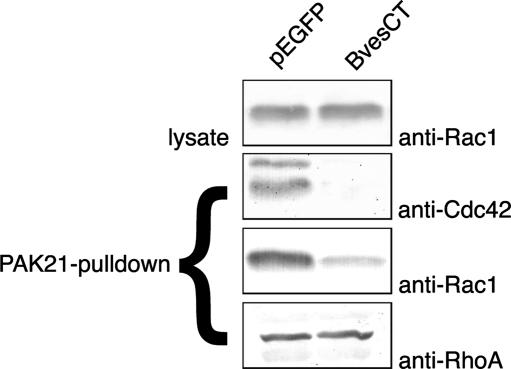
Having demonstrated that mBves interacts with mGEFT, we next sought to determine whether mBves expression changes activity levels of the Rac and Cdc42 GTPases. As the PAK-21 protein binds to only activated (GTP-bound) forms of active GTPases (30, 31), we used a PAK-21 pulldown approach to assay for GTPase activity upon transfection of mBves constructs. NIH 3T3 cells were transfected with pEGFP-mBvesCT-myc or pEGFP-C3 vector as a control. Lysates were harvested and subjected to PAK-21 pulldown. Amounts of GTP-bound Rac1, Cdc42, and RhoA were determined by SDS-PAGE followed by immunoblotting by using published methodologies. Whole-cell lysates from each sample were also immunoblotted to verify that similar amounts of protein were used for each pulldown experiment, and each assay was performed in triplicate. As seen in Fig. 4, transfection of mBves-CT markedly reduced the amount of active Rac1 and Cdc42, whereas the amount of active RhoA remained unchanged. As GEFT has previously been shown to bind and preferentially activate Rac1 and Cdc42, as opposed to RhoA (17), this result is consistent with Bves modulation of Rho-family GTPase activity through an interaction with GEFT.
Fig. 4.
Transfection of the carboxyl terminus of Bves reduces Rac1 and Cdc42 activity in NIH 3T3 cells, but does not affect the amount of active RhoA. Cells were transfected with either pEGFP (control) or pEGFP-BvesCT (amino acids 118–358 of mouse Bves). Lysates were harvested, and PAK-21 pulldowns were performed. Samples from pulldowns were loaded and blotted with anti-Rac1, anti-Cdc42, and anti-RhoA antibodies to determine relative amounts of isolated active proteins. The amount of isolated Rac1 and Cdc42 is significantly reduced upon truncated Bves expression, whereas the amount of active RhoA remains unchanged. Cell lysates were loaded and blotted with anti-Rac1 to verify equivalent amounts of total Rac1 were present in samples used for assay.
mBves Decreases Movement Speed of NIH 3T3 Cells and Increases Cell Roundness.
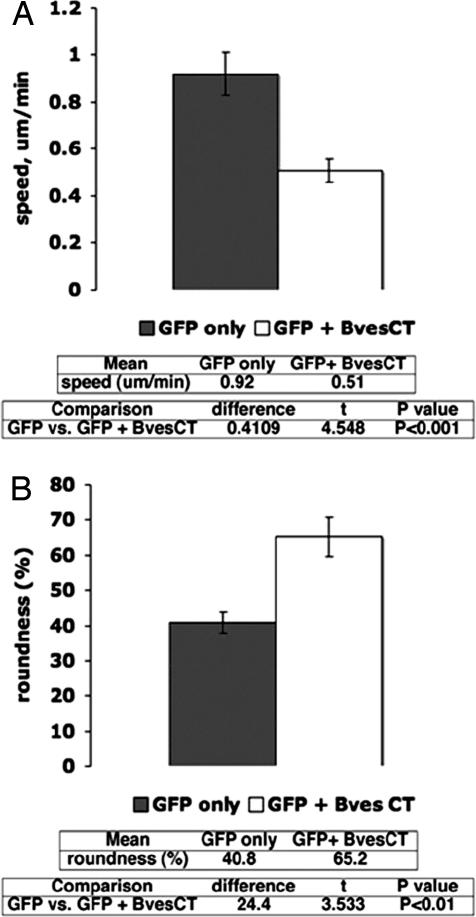
Having determined that expression of Bves-CT reduces the amount of active Rac1 and Cdc42 in NIH 3T3 cells, we next sought to determine whether transfection of Bves-CT has an effect on cell motility. Previous studies have determined that reduction of Rac1 and Cdc42 activity results in a decrease in cell movement (32–34). Time-lapse imaging was carried out with cells cotransfected with a GFP marker plasmid together with a plasmid expressing Bves-CT (SI Text and time-lapse Movie S1 and Movie S2). Parallel transfections with GFP marker plasmid alone were carried out as controls. As summarized in Fig. 5A, cells expressing Bves-CT showed markedly (≈45%) reduced speed of cell locomotion (total path length/time) compared with control cells transfected with a GFP-only expression plasmid. In contrast, there was no significant change in the directionality of cell movement (net path length/total path length) with the transfection of Bves-CT (data not shown).
Fig. 5.
Transfection of Bves-CT reduces motility and increases roundness of NIH 3T3 cells. (A) Upon transfection of Bves-CT, a significant reduction in motility speed (white bar) is observed in comparison to cells transfected with a GFP only expressing plasmid (gray bar) (Raw data are given as Movie S1 and Movie S2). (B) Roundness of cells measured in real-time as described in Materials and Methods. Software analysis of cellular area and perimeter allows determination of cell roundness. Upon transfection of Bves-CT, an increase in roundness (white bar) of ≈25% is observed in comparison to cells transfected with GFP only (gray bar). Error bars represent SEM, significance determined by using standard Student's t test.
As Rac1 and Cdc42 are known to regulate lamellipodial and filopodial cell protrusions (22), we further examined cell protrusive activity by using time-lapse imaging to measure positive and negative cytoplasmic flow and quantitate the overall roundness of the cell. Positive cytoplasmic flow is the net area of cell protrusions, whether from lammelopodial and/or filopodial extensions, whereas negative cytoplasmic flow represents the net area of cytoplasmic retractions. Our analyses showed no net change in positive or negative cytoplasmic flow, indicating no net change in the overall level of cell protrusive activity. Nevertheless, Bves-transfected cells showed an increase in roundness (Fig. 5B). Roundness, which is calculated from the measured area and perimeter length of a cell, quantifies how efficiently the measured perimeter encompasses the cellular area, with maximum roundness corresponding to a perfect circle. We found that cells expressing exogenous Bves-CT were ≈25% more round than cells transfected with a GFP marker plasmid alone (Fig. 5B).
Subsequent experiments conducted at Vanderbilt University by our laboratory further support our findings. Disruption of Bves/GEFT function inhibits or delays the differentiation of skeletal myogenic cells in vitro (Fig. S2). Taken together, these findings demonstrate that exogenous expression of full-length Bves or the cytoplasmic carboxyl terminus of Bves negatively regulates cell movement.
Discussion
Bves is a protein expressed in a variety of tissue types throughout development. Several phenotypes have been reported in vivo and in vitro when Bves protein levels are decreased, but no molecular mechanism for these observations has been determined to this point. The present data establish a direct physical interaction with any protein and link Bves to an established molecular pathway.
Upon knockdown of Bves expression in gastrulating Xenopus laevis, defects in epithelial morphogenesis and cell movements have been observed (15). Global inactivation of the murine Bves gene leads to defects in skeletal muscle repair by satellite cells (7), whereas knockdown of Bves in cultured epithelia results in defects in wound healing (13). Finally, RNA interference analysis in Drosophila inhibits germ cell migration (16). The described interaction with a component of the Rac1/Cdc42 signaling pathway may provide the first molecular mechanism to explain these cellular/embryonic phenotypes observed upon alteration of Bves expression levels previously described in the literature.
Bves Interacts with GEFT, a Modulator of Rho GTPase Signaling.
Bves interacts with GEFT, which has previously been shown to affect cell proliferation, foci formation (17), neurite outgrowth (18, 19), differentiation, and skeletal muscle regeneration (20), presumably through modulation of the Rho GTPase activity. The motility of cells is controlled by Rho GTPases through regulation of filopodial and lamellipodial extension, as well as polymerization of actin (22). Here, we show that when a Bves truncation is transfected into NIH 3T3 cells, movement and roundness of these cells are dramatically affected. We also show here that exogenous overexpression of truncated Bves reduces the amount of active Rac and Cdc42 when expressed in NIH 3T3 cells. These results strongly support our hypothesis that Bves modulates the Rac/Cdc42 activity through an interaction with GEFT.
Modulation of Rac/Cdc42 Activity by Bves Is Consistent with Observed Bves Knockdown/Knockout Phenotypes.
Control of GTPase activity via an interaction with GEFs could provide an explanation for previously observed phenotypes that currently lack mechanistic explanation. Numerous studies have demonstrated the critical role for Rac/Cdc42 during gastrulation and convergent extension of Xenopus (35–39). Our laboratory previously described a defect in epithelial migration upon knockdown of Bves expression by using morpholino oligonucleotides on developing Xenopus embryos. Specifically, Bves depletion results in randomization of cell movements during convergent/extension. This process is largely dictated by changes in cell shape, as cells radially intercalate, converge toward the midline, and extend toward the blastopore. Cells unable to alter their morphology remain rounded and fail to undergo gastrulation properly. Perturbation of Rac/Cdc42 activity by Bves knockdown would seem a plausible explanation for this phenotype.
Similarly, previous work has shown that knockdown of Bves expression in cultured corneal epithelial cells disrupted epithelial integrity and delayed healing of epithelial sheets upon wounding via scratch assay. Rac1 and Cdc42 are required for wound healing and epithelial sheet integrity (40–46). Again, the phenotypes observed upon disregulation of normal Bves levels are consistent with a role for Bves in control of Rac/Cdc42 signaling.
Additionally, the Brand laboratory noted that in Bves-null animals, skeletal muscle regeneration is delayed upon injury. Rac/Cdc42 has been shown to affect skeletal muscle regeneration (20, 47), and regeneration depends on process extenstion and myoblast motility (48, 49). Inhibition of myogenesis after expression of mutated Bves (Fig. S2) is consistent with these published findings.
Potential Mechanisms of Bves Modulation of Rac/Cdc42 Activity.
The discovery of an interaction between Bves and GEFT leads us to potential models for Bves function. Future investigations by our laboratory will attempt to determine which, if any, of these current models represent the actual mechanism through which Bves generates the previously observed phenotypes. In the first model, Bves would control the nucleotide binding ability of GEFT. As shown previously, Bves preferentially localizes to the plasma membrane (6, 10, 14). GEFT contains a pleckstrin homology domain, which has been demonstrated to localize Dbl family GEFs to the membrane (50, 51). As demonstrated here, the intracellular carboxyl terminus of Bves interacts with the DH domain of GEFT, which is the portion of GEFT responsible for interaction with the nucleotide-binding pocket of GTPases. This GEF–GTPase interaction leads to a conformational change in the nucleotide-binding pocket of the GTPase, which stimulates GDP release (52). A Bves–GEFT interaction may serve as a negative regulator of GEFT activity, thereby leading to decreased activation of GTPase signaling. Thus, the expression of truncated Bves may lead to aberrant blockage of this active site, causing the experimental results presented here. Another potential model for Bves regulation of GTPase signaling through GEFT interaction is one in which Bves controls the proper localization of GEFT to active sites of GTPase activity. As Rac and Cdc42 activity have been previously reported to be highest at the leading edges of motile cells (22), proper localization of GEF proteins to the plasma membrane in these areas is critical for proper control of cellular motility. Bves may serve to localize GEFT to this leading edge, allowing them to catalyze nucleotide exchange of GTPases. This model is also consistent with the results described here. Exogenous expression of truncated Bves may disrupt this controlled localization, resulting in a decrease in overall GTPase activity.
In summary, we have determined that Bves interacts with GEFT, a member of the Dbl family of GEFs, and that exogenous expression of Bves leads to a decrease in active levels of Rac1 and Cdc42. This represents the first direct molecular interaction elucidated for the Bves protein and provides a current link to a characterized cellular pathway. These results are consistent with previously observed phenotypes and provide a molecular context for future investigation of Bves function.
Materials and Methods
Yeast Two-Hybrid Screen and Deletion Analysis.
The cytoplasmic portion (Bves-CT, amino acids 115–358) of mouse bves was used to screen a 17.5-day mouse heart library. Deletion analysis of Bves-CT and Geft was standard. Details are provided in the SI Text.
GST-Pulldown of Dbl Family Members.
Glutathione beads were prepared as described in the SI Text. mGEFT was amplified from pCMVTag-2b-mGEFT and cloned in frame with GFP of pEGFP-C1 (Clontech) to generate pEGFP-mGEFT. pEGFP-mNudeL1 was generated previously by our laboratory (53). pcDNA3-N-myc-Cool1 was a gracious gift from Richard Cerione (Cornell University, Ithaca, NY), and pC.HA-Vav1 was obtained from Addgene.
COS-7 cells transfected with pEGFP-mGEFT, pC.HA-Vav1, pcDNA3-N-myc-Cool1, or pEGFP-mNudeL1 were grown to confluence in 10-cm dishes. Protein was extracted as described above. Cells were incubated on ice for 30 min with gentle agitation, scraped off the plate, and centrifuged for 30 min at 18,000 × g at 4°C. Cell lysate was removed from the pellet and retained.
Lysate was precleared by incubation with a 20-μl bed volume of glutathione-Sepharose 4B for 2 h at 4°C, after which beads were spun down, and lysate was removed. Glutathione-Sepharose 4B bound with GST constructs was then added to the lysate and incubated overnight at 4°C. Sepharose conjugates were captured by using centrifugation and washed five times with 100 μl PBS, and bound protein was eluted with 20 μl of 1× SDS sample buffer, boiled for 3 min, and loaded onto an 10% SDS-PAGE gel. Western blotting was performed by using standard methods. Blots were developed by using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche) and scanned into digital format (Hewlett-Packard).
Rac1/Cdc42 Activation Assay.
COS-7 cells were transfected as described with pEGFP-C3 vector and pEGFP-mBvesCT-myc (amino acids 115–358) expression vectors. Cells were then harvested in magnesium-containing lysis buffer 2 days after transfection, and lysates were sonicated for 5 sec and centrifuged for 30 min at 18,000 × g at 4°C following the manufacturer's specifications for the Rac/Cdc42 Assay Reagent Kit (Upstate Cell Signaling) (54). A total of 10 μg Rac/Cdc42 assay reagent was added to 600 μl protein lysate and gently rocked at 4°C for 30 min. PAK-21–agarose conjugates were collected by centrifugation for 5 sec at 14,000 × g at room temperature and washed three times with 500 μl magnesium-containing lysis buffer, and bound protein was eluted in 25 μl SDS-PAGE sample buffer. Western blotting of these samples and of 10 μl of the original lysate as a loading control was performed by using standard protocols.
Motility Assays.
The intracellular C terminus of Bves (amino acids 115–358) was cloned into a mammalian expression construct (pCMV-myc). NIH 3T3 cells were cotransfected by using Lipofectamine 2000 at 95% confluency with pCMV-myc-BvesCT and pEGFP-C1 (as a tracer for transfected cells), GFP-Bves, or with pEGFP-C1 alone. Cells were split to ≈10% confluency 2 days after transfection. For monitoring the velocity of cell motility (total path length/time), cells in 10-cm2 dishes were placed on the 37°C heated stage of a Leica DMIRE2 inverted microscope. Time-lapse images were captured by using a Hamamatsu ORCA ER Digital Camera. Images were captured every 60 sec over a 45-min interval by using a ×10 objective. Quantitative motion analysis was carried out by using Dynamic Image Analysis Software (Solltech). The outline of each cell was traced frame by frame, and using these tracings the Dynamic Image Analysis Software calculated the speed of cell movement by tracking the change in position of the cell centroid for each frame. All data from these experiments were evaluated by ANOVA using Statview (SAS Institute).
Experiments evaluating the effects of exogenous expression of the carboxyl terminus of Bves were conducted at the Cell Imaging Shared Resource at Vanderbilt University. Cells were transfected as described and split to ≈10% confluence in 24-well culture plates (Nalge). Plates were placed on the 37°C heated stage of an inverted Nikon TE300 widefield microscope with automated stage for acquisition of multiple fields or view. Images were captured every 2 min for 30 min by using a ×20 objective. Quantitative motion analysis was carried out as described above by using Metamorph software (Molecular Devices). Data were evaluated by using Microsoft Excel.
Cell Roundness Assay.
In addition to the motility data rendered from the analysis of exogenous expression of GFP-Bves described above, the cell tracings were also used to investigate the relative roundness of cells transfected with either GFP-Bves or GFP alone. The roundness of these cells was calculated by using the Eq. 100 × 4π (area/perimeter2) (55). This equation provides a measurement of how efficiently a given amount of perimeter encloses an area: a circle has the largest area for any given perimeter with a roundness of 100%. Accordingly, the greater the number of cell protrusions, the lower the roundness. All data obtained from quantitative assessments were evaluated by ANOVA by using Statview as above.
Supplementary Material
Acknowledgments.
We thank members of the laboratory for critical reading and discussion of the manuscript, Dr. Mingyao Liu of Texas A&M University (College Station, TX) for the GEFT plasmid used as template for cDNA cloning, and Dr. Richard Cerione of Cornell University (Ithaca, NY) for the pcDNA3-N-myc-Cool1 construct. This work was supported by National Institutes of Health Grants HL37675 and HL079050 (to D.M.B.), HL079050 (to T.K.S.), and HL07411 and HL79050 (to H.A.H.) and by American Heart Association Grant 0415252B (to T.K.S.). Portions of the cell imaging presented here were performed at the Vanderbilt Cell Imaging Shared Resource.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802345105/DCSupplemental.
References
- 1.Osler ME, Bader DM. Bves expression during avian embryogenesis. Dev Dyn. 2004;229:658–667. doi: 10.1002/dvdy.10490. [DOI] [PubMed] [Google Scholar]
- 2.Andree B, et al. Isolation and characterization of the novel popeye gene family expressed in skeletal muscle and heart. Dev Biol. 2000;223:371–382. doi: 10.1006/dbio.2000.9751. [DOI] [PubMed] [Google Scholar]
- 3.DiAngelo JR, Vasavada TK, Cain W, Duncan MK. Production of monoclonal antibodies against chicken Pop1 (BVES) Hybrid Hybridomics. 2001;20:377–381. doi: 10.1089/15368590152740789. [DOI] [PubMed] [Google Scholar]
- 4.Hitz MP, Pandur P, Brand T, Kuhl M. Cardiac specific expression of Xenopus Popeye-1. Mech Dev. 2002;115:123–126. doi: 10.1016/s0925-4773(02)00085-0. [DOI] [PubMed] [Google Scholar]
- 5.Reese DE, Bader DM. Cloning and expression of hbves, a novel and highly conserved mRNA expressed in the developing and adult heart and skeletal muscle in the human. Mamm Genome. 1999;10:913–915. doi: 10.1007/s003359901113. [DOI] [PubMed] [Google Scholar]
- 6.Smith TK, Bader DM. Characterization of Bves expression during mouse development using newly generated immunoreagents. Dev Dyn. 2006;235:1701–1708. doi: 10.1002/dvdy.20739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andree B, Fleige A, Arnold HH, Brand T. Mouse Pop1 is required for muscle regeneration in adult skeletal muscle. Mol Cell Biol. 2002;22:1504–1512. doi: 10.1128/mcb.22.5.1504-1512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ Res. 2002;91:761–768. doi: 10.1161/01.res.0000038961.53759.3c. [DOI] [PubMed] [Google Scholar]
- 9.Reese DE, Zavaljevski M, Streiff NL, Bader D. Bves: A novel gene expressed during coronary blood vessel development. Dev Biol. 1999;209:159–171. doi: 10.1006/dbio.1999.9246. [DOI] [PubMed] [Google Scholar]
- 10.Wada AM, Reese DE, Bader DM. Bves: Prototype of a new class of cell adhesion molecules expressed during coronary artery development. Development. 2001;128:2085–2093. doi: 10.1242/dev.128.11.2085. [DOI] [PubMed] [Google Scholar]
- 11.Wada AM, Smith TK, Osler ME, Reese DE, Bader DM. Epicardial/mesothelial cell line retains vasculogenic potential of embryonic epicardium. Circ Res. 2003;92:525–531. doi: 10.1161/01.RES.0000060484.11032.0B. [DOI] [PubMed] [Google Scholar]
- 12.Vasavada TK, DiAngelo JR, Duncan MK. Developmental expression of Pop1/Bves. J Histochem Cytochem. 2004;52:371–377. doi: 10.1177/002215540405200308. [DOI] [PubMed] [Google Scholar]
- 13.Ripley AN, Chang MS, Bader DM. Bves is expressed in the epithelial components of the retina, lens, and cornea. Invest Ophthalmol Vis Sci. 2004;45:2475–2483. doi: 10.1167/iovs.04-0013. [DOI] [PubMed] [Google Scholar]
- 14.Osler ME, Chang MS, Bader DM. Bves modulates epithelial integrity through an interaction at the tight junction. J Cell Sci. 2005;118:4667–4678. doi: 10.1242/jcs.02588. [DOI] [PubMed] [Google Scholar]
- 15.Ripley AN, Osler ME, Wright CVE, Bader DM. Xbves is a regulator of epithelial movement during early Xenopus laevis development. Proc Natl Acad Sci USA. 2006;103:614–619. doi: 10.1073/pnas.0506095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S, Zhao D, Bownes M. Blood vessel/epicardial substance (bves) expression, essential for embryonic development, is down regulated by Grk/EFGR signalling. Int J Dev Biol. 2007;51:37–44. doi: 10.1387/ijdb.052108sl. [DOI] [PubMed] [Google Scholar]
- 17.Guo X, et al. A Rac/Cdc42-specific exchange factor, GEFT, induces cell proliferation, transformation, and migration. J Biol Chem. 2003;278:13207–13215. doi: 10.1074/jbc.M208896200. [DOI] [PubMed] [Google Scholar]
- 18.Bryan B, et al. GEFT, a Rho family guanine nucleotide exchange factor, regulates neurite outgrowth and dendritic spine formation. J Biol Chem. 2004;279:45824–45832. doi: 10.1074/jbc.M406216200. [DOI] [PubMed] [Google Scholar]
- 19.Bryan BA, Cai Y, Liu M. The Rho-family guanine nucleotide exchange factor GEFT enhances retinoic acid- and cAMP-induced neurite outgrowth. J Neurosci Res. 2006;83:1151–1159. doi: 10.1002/jnr.20814. [DOI] [PubMed] [Google Scholar]
- 20.Bryan BA, et al. Modulation of muscle regeneration, myogenesis, and adipogenesis by the Rho family guanine nucleotide exchange factor GEFT. Mol Cell Biol. 2005;25:11089–11101. doi: 10.1128/MCB.25.24.11089-11101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 22.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 23.Brand T. The Popeye domain-containing gene family. Cell Biochem Biophys. 2005;43:95–103. doi: 10.1385/CBB:43:1:095. [DOI] [PubMed] [Google Scholar]
- 24.Breher SS, et al. Popeye domain containing gene 2 (Popdc2) is a myocyte-specific differentiation marker during chick heart development. Dev Dyn. 2004;229:695–702. doi: 10.1002/dvdy.20015. [DOI] [PubMed] [Google Scholar]
- 25.Osler ME, Smith TK, Bader DM. Bves, a member of the Popeye domain-containing gene family. Dev Dyn. 2006;235:586–593. doi: 10.1002/dvdy.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Souchet M, et al. Human p63RhoGEF, a novel RhoA-specific guanine nucleotide exchange factor, is localized in cardiac sarcomere. J Cell Sci. 2002;115:629–640. doi: 10.1242/jcs.115.3.629. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Y, et al. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman GR, Cerione RA. Signaling to the Rho GTPases: Networking with the DH domain. FEBS Lett. 2002;513:85–91. doi: 10.1016/s0014-5793(01)03310-5. [DOI] [PubMed] [Google Scholar]
- 30.Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 31.Chiang SH, et al. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature. 2001;410:944–948. doi: 10.1038/35073608. [DOI] [PubMed] [Google Scholar]
- 32.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 33.Itoh RE, et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 35.Habas R, Dawid IB, He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwan KM, Kirschner MW. A microtubule-binding Rho-GEF controls cell morphology during convergent extension of Xenopus laevis. Development. 2005;132:4599–4610. doi: 10.1242/dev.02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyakoshi A, Ueno N, Kinoshita N. Rho guanine nucleotide exchange factor xNET1 implicated in gastrulation movements during Xenopus development. Differentiation. 2004;72:48–55. doi: 10.1111/j.1432-0436.2004.07201004.x. [DOI] [PubMed] [Google Scholar]
- 38.Ren R, Nagel M, Tahinci E, Winklbauer R, Symes S. Migrating anterior mesoderm cells and intercalating trunk mesoderm cells have distinct responses to Rho and Rac during Xenopus gastrulation. Dev Dyn. 2006;235:1090–1099. doi: 10.1002/dvdy.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahinci E, Symes K. Distinct functions of Rho and Rac are required for convergent extension during Xenopus gastrulation. Dev Biol. 2003;259:318–335. doi: 10.1016/s0012-1606(03)00206-9. [DOI] [PubMed] [Google Scholar]
- 40.Fenteany G, Janmey PA, Stossel TP. Signaling pathways and cell mechanics involved in wound closure by epithelial cell sheets. Curr Biol. 2000;10:831–838. doi: 10.1016/s0960-9822(00)00579-0. [DOI] [PubMed] [Google Scholar]
- 41.Kimura K, Kawamoto K, Teranishi S, Nishida T. Role of Rac1 in fibronectin-induced adhesion and motility of human corneal epithelial cells. Invest Ophthalmol Vis Sci. 2006;47:4323–4329. doi: 10.1167/iovs.05-1508. [DOI] [PubMed] [Google Scholar]
- 42.Kofron M, Heasman J, Lang SA, Wylie CC. Plakoglobin is required for maintenance of the cortical actin skeleton in early Xenopus embryos and for cdc42-mediated wound healing. J Cell Biol. 2002;158:695–708. doi: 10.1083/jcb.200202123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malliri A, van Es S, Huveneers S, Collard JG. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J Biol Chem. 2004;279:30092–30098. doi: 10.1074/jbc.M401192200. [DOI] [PubMed] [Google Scholar]
- 44.Nobes CD. Rho GTPases and cell migration-fibroblast wound healing. Methods Enzymol. 2000;325:441–449. doi: 10.1016/s0076-6879(00)25464-5. [DOI] [PubMed] [Google Scholar]
- 45.Stramer B, et al. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J Cell Biol. 2005;168:567–573. doi: 10.1083/jcb.200405120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woolner S, Jacinto A, Martin P. The small GTPase Rac plays multiple roles in epithelial sheet fusion–dynamic studies of Drosophila dorsal closure. Dev Biol. 2005;282:163–173. doi: 10.1016/j.ydbio.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- 48.Carlson BM. The regeneration of skeletal muscle. A review. Am J Anat. 1973;137:119–149. doi: 10.1002/aja.1001370202. [DOI] [PubMed] [Google Scholar]
- 49.Carlson BM, Faulkner JA. The regeneration of skeletal muscle fibers following injury: A review. Med Sci Sports Exerc. 1983;15:187–198. [PubMed] [Google Scholar]
- 50.Russo C, et al. Modulation of oncogenic DBL activity by phosphoinositol phosphate binding to pleckstrin homology domain. J Biol Chem. 2001;276:19524–19531. doi: 10.1074/jbc.M009742200. [DOI] [PubMed] [Google Scholar]
- 51.Vanni C, et al. Regulation of proto-Dbl by intracellular membrane targeting and protein stability. J Biol Chem. 2002;277:19745–19753. doi: 10.1074/jbc.M111025200. [DOI] [PubMed] [Google Scholar]
- 52.Rossman KL, et al. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 2002;21:1315–1326. doi: 10.1093/emboj/21.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soukoulis V, et al. Cytoplasmic LEK1 is a regulator of microtubule function through its interaction with the LIS1 pathway. Proc Natl Acad Sci USA. 2005;102:8549–8554. doi: 10.1073/pnas.0502303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor SJ, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- 55.Stites J, et al. Phosphorylation of the Dictyostelium myosin II heavy chain is necessary for maintaining cellular polarity and suppressing turning during chemotaxis. Cell Motil Cytoskeleton. 1998;39:31–51. doi: 10.1002/(SICI)1097-0169(1998)39:1<31::AID-CM4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.