Abstract
Despite significant advances in identifying signaling molecules that induce cardiogenesis in mammals, the transcription factors that control the onset of cardiac myocyte gene expression have remained elusive. Candidates include the zinc finger transcription factors GATA binding proteins 4 and 6 (GATA4, GATA6). The individual loss of either protein in mice results in lethality prior to the onset of heart development due to defects in the extra-embryonic endoderm; however, when this extra-embryonic deficiency is circumvented using tetraploid embryo complementation, cardiac myocyte differentiation initiates normally. Here we show that these factors have redundant roles in controlling the onset of cardiac myocyte differentiation. As a consequence, Gata4−/−Gata6−/− embryos completely lack hearts, although second heart field progenitor cells are still generated. Our data support a model whereby GATA4 or GATA6 are essential for expression of the network of transcription factors that regulate the onset of cardiac myocyte gene expression during mammalian development.
Keywords: tetraploid, heart development, transcription factors, GATA4, GATA6
INTRODUCTION
Studies in Drosophila melanogaster, Danio rerio and Xenopus laevis have implicated several families of transcription factors in initiating cardiac myocyte gene expression in response to inductive signals, including the Nkx, Tbx, Mef, and GATA families as well as SRF (Olson and Schneider, 2003). However, no single mutation has been identified that blocks cardiac myocyte differentiation in mammals, suggesting that functional redundancy between factors exists. The GATA binding proteins are zinc finger transcription factors that have been implicated in regulating the onset of cardiac myocyte differentiation (Pikkarainen et al., 2004). GATA4, 5 and 6 are all expressed in cardiac progenitor cells, and all three proteins, although individually dispensable for cardiac myocyte formation, have critical roles in regulating heart development in zebrafish and frogs (Holtzinger and Evans, 2005; Peterkin et al., 2003; Reiter et al., 1999). More recently, morpholino-mediated depletion of combinations of GATA factors has resulted in preventing aspects of cardiac specification in Zebrafish and Xenopus suggesting that the GATA factors have redundant but essential roles in controlling early stages of heart development in these species (Holtzinger and Evans, 2007; Peterkin et al., 2007). Elucidating the contribution the GATA factors make to the development of the mammalian heart has been challenging. Gene knockout studies in mice demonstrated that GATA5 was dispensable for embryonic development (Molkentin et al., 2000), while loss of either GATA4 or GATA6 resulted in a developmental arrest during gastrulation due to a requirement for these factors in the extra-embryonic endoderm (Koutsourakis et al., 1999; Kuo et al., 1997; Molkentin et al., 1997; Morrisey et al., 1998). When this early embryonic lethality was circumvented by providing GATA4 or GATA6 null embryos with wild type extra-embryonic endoderm, formation of the early heart and cardiac myocyte gene expression appeared relatively normal, although subtle deficiencies in maturation of the myocytes were identified (Narita et al., 1997b; Watt et al., 2004; Zhao et al., 2005). GATA4 and GATA6 are highly conserved, are capable of binding identical nucleotide sequences in genomic DNA, and regulate expression of similar target genes (Morrisey et al., 1997; Nemer and Nemer, 2003; Pikkarainen et al., 2004). We therefore sought to test the proposal that these factors had redundant functions in controlling mammalian cardiac myocyte differentiation and predicted that if both factors were simultaneously disrupted, the onset of cardiac development would be blocked.
MATERIALS AND METHODS
Generation of Gata4−/− Gata6−/− ES cell lines, embryoid bodies and embryos
The production of Gata4−/− and Gata6−/− ES cells have been described previously (Watt et al., 2004; Zhao et al., 2005). To generate Gata4−/−Gata6−/− ES cells, we first produced a targeting vector, pGATA4loxPDT, that contains a Neo-tk cassette flanked by two loxP sites (sites a and b) from plasmid pHR-1 that was inserted into the SmaI site 85bp upstream of Gata4 exon 3 (Watt et al., 2004). In addition, a single loxP site (site c) from plasmid pHR-1 was inserted into the BamHI site between exons 5 & 6. Negative selection was provided by the diptheria toxin gene. Cre mediated recombination between the two outermost loxP sites deletes both zinc finger domains and the transactivation domain of GATA4 resulting in complete loss of function and the absence of detectable protein (Watt et al., 2004). We generated a 3′probe, used for Southern blot analysis, by PCR from genomic DNA using the primers g4-192 5′ ATGAAAACAGCTTCCCACCC 3′ and g4-193 5′ AGACTGGCCCTAAGCTATTG 3′. This probe is located between exon 6 and the downstream SacI and EcoRI sites and identifies 11kb SacI (not shown) and 3kb EcoRI wild type Gata4 DNA fragments (Fig. 1a, c). R1 ES cells were electroporated with the pGATA4loxPDT vector and grown in 350μg/ml G418 for 7 days to select for transformants. Homologous recombination was predicted to produce the Gata4 loxPNeo allele that generated a unique 5kb SacI fragment due to the addition of a SacI site within Neo-tk and a unique 2kb EcoRI fragment due to the addition of an EcoRI site in loxPc (Watt et al., 2004). We electroporated Gata4+/loxPneo ES cells with a Cre expression plasmid, pHDMCCre8, grew the cells for 3 days without selection, and then cultured in 2μM gancyclovir for 5 days to select for deletion of the Neo-tk cassette. Deletion of the Neo-tk cassette could occur by recombination between loxPa and loxPb (see Fig. 1 of Watt et al (Watt et al., 2004)), deleting only the Neo-tk cassette and leaving behind a single loxPa/b site and the loxPc site flanking exons 3–5. This produces the Gata4 loxP allele with an 11kb SacI fragment and a 2kb EcoRI fragment. Alternatively, recombination between loxP a and loxP c deletes the neo-tk cassette and 2kb of genomic DNA leaving behind a single loxP a/c site. This produces a Gata4– allele with a 9kb SacI fragment (not shown) and a 2kb EcoRI fragment (Fig. 1a,c) (Watt et al., 2004). Gata4−/− ES cells were generated by targeting Gata4+/− ES cells with same targeting vector followed by transient expression of Cre as described above. We confirmed the genotype of Gata4−/− ES cells by Southern blot (Fig. 1c) and loss of GATA4 expression by RT-PCR analyses of embryoid bodies (Fig. 1e) and immunohistochemistry on Gata4−/− ES cell–derived embryos (Watt et al., 2004). We next targeted the Gata6 gene in Gata4−/−ES cells using the targeting vector described by Morrisey et al (Morrisey et al., 1998) and used by us previously to generate Gata6−/− ES cells (Zhao et al., 2005). This vector contains a Pgk–Neo cassette, which replaces exons encoding both zinc fingers and results in a Gata6 null allele (Morrisey et al., 1998). We electroporated Gata4−/− ES cells with the targeting vector and collected colonies that were resistant to growth in G418 (350μg/ml). We next identified Gata6+/− ES cells by genomic Southern blot analyses using a probe that flanks exon 5 and identifies a 13kb wild type Gata6 BamH1 fragment and an 8kb BamH1 fragment in the targeted allele (Fig. 1b, c). We next cultured Gata4−/−Gata6+/− ES cells in culture medium supplemented with an elevated concentration (1.5mg/ml) of G418, as described (Zhao et al., 2005), and identified Gata4−/−Gata6−/− ES cells by Southern blot analyses (Fig. 1c). We produced embryoid bodies after treating ES cells with Noggin and withdrawing leukemia inhibitory factor following the procedure described by Yuasa et al (Yuasa et al., 2005). Embryoid bodies were collected at day 9 after removal of LIF from three independent experiments. We produced embryos directly from ES cells by tetraploid embryo complementation as described elsewhere (Nagy and Rossant, 1993) and considered noon on the day we identified a vaginal plug in surrogate mothers as E0.5.
Figure 1. Loss of both GATA4 and GATA6 disrupts cardiac myocyte gene expression in ES cell embryoid bodies.
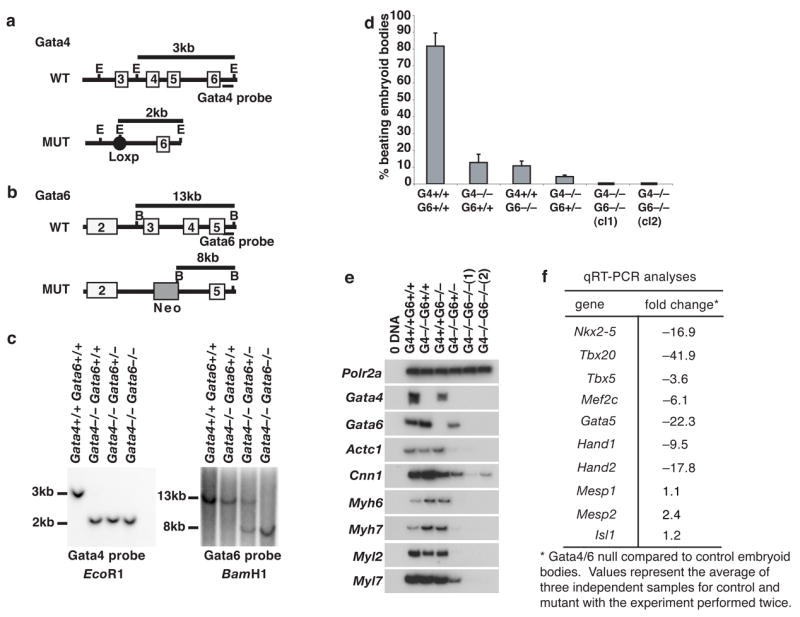
a,b, Schematic overview of the strategy used to generate Gata4−/− Gata6−/− ES cells. Genomic structure of wild type (WT) and mutated (MUT) GATA4 (a) and GATA6 (b) alleles with exons presented as open boxes, the loxP site as a closed circle, and the Pgk-Neo cassette as a shaded box. Position of Southern blot probes and sizes of relevant genomic DNA fragments following digestion with EcoRI(E) or BamHI(B) are indicated. c, Southern blot confirming the genotype of ES cell lines with DNA fragment sizes shown in kb. d, Graph showing percentage of contractile embryoid bodies generated from control (R1) and experimental Gata4−/−Gata6+/+, Gata4+/+ Gata6−/−, Gata4−/− Gata6+/−, Gata4−/− Gata6−/− (clones 1 and 2) ES cells in three independent experiments (error bars). e, RT-PCR analysis of steady-state cardiac mRNA levels in control and GATA knockout embryoid bodies. Polr2a; RNA Polymerase II, Actc1; cardiac alpha actin, Cnn1;Calponin, Myh6;cardiac alpha myosin heavy chain, Myh7;cardiac beta myosin heavy chain, Myl2;cardiac myosin light chain 2v, Myl7;myosin light chain 2a. f, Table showing changes, determined by real-time quantitative RT-PCR analyses, in abundance of mRNAs encoding cardiac transcription factors.
Oligonucleotide arrays, RT-PCR and real-time qRT-PCR
We collected total RNA from three independent control and Gata4−/−Gata6−/− embryoid body preparations using an RNeasy purification kit (Qiagen). Probes were prepared following Affymetrix protocols. Each sample was hybridized to an individual Affymetrix GeneChip Mouse Genome 2.0 array and data were analyzed using DCHIP Ver. 1.3 software. We carried out semi-quantitative RT-PCR as described previously (Zhao et al., 2005). Real time quantitative RT-PCR was performed using SYBR green incorporation with reactions run on a BioRad iCycler™ following the manufacturer’s protocol using empirically optimized primer pairs. All oligonucleotide sequences are available on request, with the exception of proprietary oligonucleotides purchased from Superarray Bioscience Corp that were optimized for qRT-PCR amplification of Mesp1 (cat#PPM24667A) or Mesp2 (cat# PPM27883A).
Immunohistochemistry, antibodies, histochemistry and in situ hybridization
We collected embryos, fixed them with 4% paraformaldehyde, and stored them in 70% ethanol. We processed embryos for paraffin sections as described previously (Watt et al., 2004; Zhao et al., 2005). We performed immunohistochemistry on either whole embryos or sections following microwave antigen retrieval as we discussed elsewhere (Watt et al., 2004; Zhao et al., 2005). We used the following primary antibodies: anti-smooth muscle actin (Sigma A-2547; 1:800); MF20, anti-myosin heavy chain (Developmental Hybridoma Bank; 1:1000), FoxA1 (C-20, Santa Cruz, sc-6553; 1:400), sarcomeric actin (Sigma A-2172; 1:800), HNF4α (C-19, Santa Cruz, sc-6556; 1:500), CD31/PECAM (CD31, BD Pharmingen #553370; 1:50). Whole mount in situ hybridization was performed using digoxigenin–labeled probes generated by in vitro transcription (Roche) following standard procedures.
RESULTS
GATA4 and GATA6 are essential for differentiation of cardiac myocytes from ES cells
To test the proposal that GATA4 and GATA6 had redundant yet essential roles in controlling cardiac myocyte differentiation, we generated Gata4−/− Gata6+/+ (Watt et al., 2004), Gata4+/+ Gata6−/− (Zhao et al., 2005), Gata4−/− Gata6+/−, and Gata4−/− Gata6−/− ES cells by gene targeting (Fig. 1a–c) (details of targeting are presented in the methods section). ES cells of each genotype were then induced to form cardiac myocytes following the protocol described by Yuasa et al (Yuasa et al., 2005), and the number of beating embryoid bodies, which presumably reflects cardiac myocyte differentiation, was determined by visual inspection. As described previously (Yuasa et al., 2005) this protocol resulted in the efficient generation of beating cells from wild type R1 ES cells with beating observed in >80% of embryoid bodies (Fig. 1d). When either Gata4 or Gata6 was disrupted the number of beating embryoid bodies was dramatically reduced, with only 10% of the embryoid bodies containing contractile cells (Fig. 1d). This reduction in the number of beating cells is similar to that previously reported for loss of GATA4 in ES cell embryoid bodies and may reflect a loss of extraembryonic endoderm necessary for maturation rather than a cell autonomous effect (Narita et al., 1997a). No significant difference was observed in the number of beating embryoid bodies when cells lacked either GATA4 or GATA6. However, if one allele of Gata6 was disrupted in the Gata4−/− ES cells then the number of beating embryoid bodies decreased further to 5% (Fig. 1d). Most notably, no beating cells were identified in embryoid bodies lacking both GATA4 and GATA6 (Fig 1d).
If the loss of beating cells reflects a failure in cardiac myocyte differentiation we predicted that cardiac gene expression should be disrupted in Gata4−/−;Gata6−/− embryoid bodies. RT-PCR was therefore performed on control (Gata4+/+ Gata6+/+), Gata4−/−, Gata6−/−, Gata4−/− Gata6+/−, and Gata4−/− Gata6−/− embryoid bodies to determine the abundance of a subset of mRNAs encoding proteins whose expression is characteristic of cardiac myocyte differentiation (Fig. 1e). These included cardiac alpha actin, calponin, and alpha and beta myosin heavy and light chains (encoded by Actc1, Cnn1, Myh6, Myh7, Myl2, Myl7, respectively). Previous in vivo analyses had demonstrated that loss of either GATA4 or GATA6 had relatively little impact on expression of these key cardiac myocyte mRNAs (Watt et al., 2004; Zhao et al., 2005). Consistent with these analyses and despite the observation that contractility was reduced in both Gata4−/− and Gata6−/−embryoid bodies, cardiac myocyte gene expression was unaffected by loss of either GATA4 or GATA6 alone when compared with control embryoid bodies (Fig. 1e). However, we measured a large reduction in the levels of these cardiac myocyte mRNAs in embryoid bodies generated from Gata4−/− Gata6+/− ES cells, and they were virtually undetectable in embryoid bodies generated from Gata4−/− Gata6−/− ES cells (Fig. 1e). These changes in cardiac mRNA levels were confirmed by oligonucleotide array analyses (not shown).
A complex network of cardiac transcription factors is believed to define the differentiated state of cardiac myocytes (Olson and Schneider, 2003; Srivastava and Olson, 2000), and so we next determined the level of mRNAs encoding such transcription factors using qRT-PCR. Figure 1f shows that the level of Nkx2.5, Tbx20, Tbx5, Mef2c, Gata5, Hand1, and Hand2 mRNAs was substantially reduced in the absence of both GATA4 and 6 compared to wild type cells. Of note, however, the level of Isl1, Mesp1, and Mesp2 mRNAs, which are characteristically expressed in cardiac progenitor cells (Cai et al., 2003; Kitajima et al., 2000; Moretti et al., 2006), was not reduced. Cumulatively, these data confirm that GATA4 and GATA6 have redundant functions in controlling cardiac myocyte differentiation and furthermore suggest that they are dispensable for formation of cardiac progenitor cells.
GATA4 and GATA6 are redundant but essential for cardiac development in mouse embryos
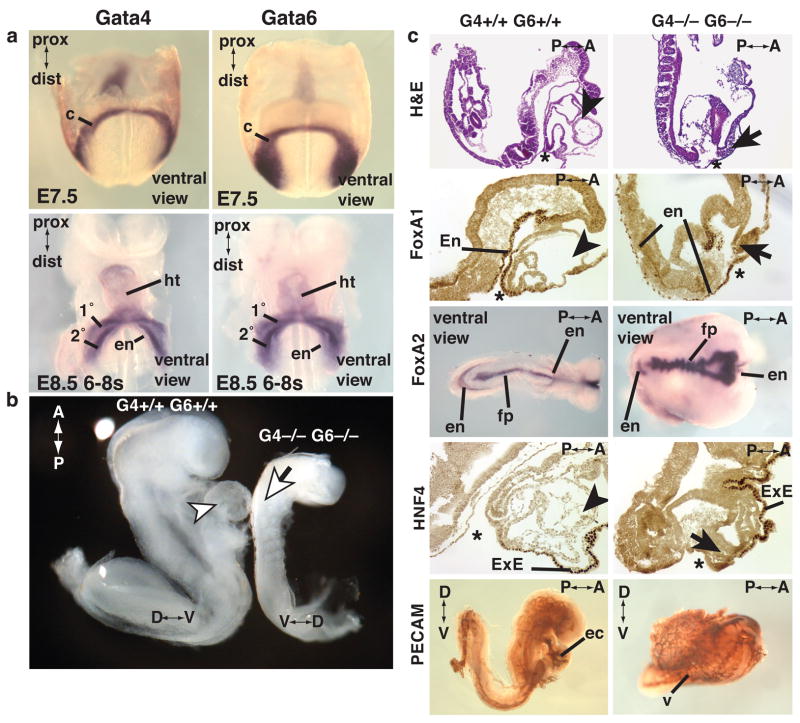
The possibility of functional complementation requires that both GATA factors be expressed during the onset of cardiac development in overlapping cell compartments. We therefore performed in situ hybridization analyses to detect Gata4 and Gata6 mRNAs in developmentally matched embryos. Figure 2a shows that both Gata4 and Gata6 mRNAs could be detected in overlapping patterns in cardiac progenitor cells that are present in the cardiac crescent at E7.5 (early headfold stage), as well as in the nascent heart tube, cardiac outflow and inflow tracts at E8.5 (6–8 somites), which is consistent with previous reports (Dodou et al., 2004; Morrisey et al., 1996; Waldo et al., 2001). We next compared development of the heart in embryos generated from either control or Gata4−/− Gata6−/− ES cells by tetraploid embryo complementation in order to definitively establish whether these factors were essential for cardiac myocyte differentiation in vivo. Figure 2b shows that by E8.5 control wild type ES cell–derived embryos had formed a distinctive heart tube that protruded from the ventral face of the embryo (arrowhead). In contrast, the heart appeared to be completely absent from embryos (n>97) generated from Gata4−/− Gata6−/− ES cells (Fig. 2b). Acardia was found to occur in embryos generated from three independently targeted Gata4−/− Gata6−/− ES cell lines and was 100% penetrant. The absence of the heart in GATA4/6 null embryos did not appear to be a consequence of a general embryonic arrest or a defect in gastrulation because histological sections revealed that Gata4−/− Gata6−/− embryos retained many features characteristic of this developmental stage (Fig. 2c). This included the generation of the expected number of somite pairs (8–12 somite pairs per embryo), the formation of a headfold and the development of the neural tube (Fig. 2c, H&E). The presence of condensed somitic mesoderm confirmed that formation, migration and patterning of the paraxial mesoderm was intact (Fig. 2c, H&E). In addition, the presence of definitive endoderm lining the entire ventral aspect of both control and experimental embryos was confirmed using immunohistochemistry to detect FoxA1 (Fig. 2c, FoxA1) and in situ hybridization to detect FoxA2 (Fig. 2c, FoxA2), although it was noted that morphogenesis of the foregut was deficient in the mutant embryos. The possibility that the FoxA1/A2 expressing endoderm was in fact extra embryonic endoderm was excluded because this endoderm failed to express HNF4α (Fig. 2c, HNF4), which is present exclusively in the extra embryonic endoderm at this developmental stage (Duncan et al., 1994; Taraviras et al., 1994). Detection of Foxa2 mRNA also revealed that the floor plate of the neural tube had been generated in the mutant embryos, although the domain of Foxa2 expression appeared to be expanded compared with controls. Finally, like control embryos, Gata4−/− Gata6−/− embryos were found to generate blood vessels that expressed the endothelial cell marker CD31 (Fig. 2c, PECAM). Together, these data demonstrate that loss of both GATA4 and GATA6 has a relatively specific effect on the development of the cardiogenic mesoderm and does not cause a general deficiency in gastrulation or mesodermal migration.
Figure 2. Loss of both GATA4 and GATA6 precludes development of the heart.
a, Distribution of Gata4 and Gata6 mRNAs detected in E7.5, early headfold stage, and E8.5, 6–8 somite stage, embryos by in situ hybridization. b, Micrograph showing typical E8.5 embryos generated from either control (G4+/+ G6+/+) or GATA4/6 doubly null (G4−/− G6−/−) ES cells from which the yolk sac has been removed. c, Sections of control or GATA4/6 null embryos stained with hematoxylin and eosin (H&E), by immunohistochemistry for expression of FoxA1, HNF4α, or Pecam (CD31), and by in situ hybridization for Foxa2 mRNA. The presence of cardiac tissue in control embryos is indicated with an arrowhead, the expected location of the heart in mutant embryos with an arrow, and the anterior intestinal portal by an asterisk (*). c, cardiac crescent; 1°,presumptive primary heart field; 2° presumptive second heart field; en, definitive endoderm; ht, primary heart tube; fp, neural tube floorplate; ec, endocardium; ExE, extra embryonic endoderm; v, blood vasculature. The positions of the proximal-distal (prox-dist), anterior-posterior (A–P) and dorsal-ventral (D–V) axes are shown where appropriate.
We next performed immunohistochemistry to detect markers of cardiac myocytes to determine whether GATA4 and GATA6 were required for the onset of myocyte differentiation or whether the absence of the heart was primarily due to a block in morphogenesis of differentiated cardiac cells. Figure 3 shows that smooth muscle actin (SMA), alpha myosin heavy chain (αMHC), and alpha cardiac actin (α-sarcomeric actin) could be detected in the differentiating cardiac myocytes forming the heart tube as well as in the yolk sacs of E8.5 (8–12 somites) embryos derived from control ES cells. In contrast, expression of these proteins was undetectable in Gata4−/− Gata6−/− embryos, although staining was easily identifiable in the yolk sac (Fig. 3). We also examined expression of the co-activator Smarcd3 (encoding Baf60c) as an example of a gene expressed in myocytes that is not a structural component of myofibrils. Smarcd3 mRNA was found to be robustly expressed in the cardiac crescent in control embryos, as described previously (Lickert et al., 2004), but was absent from the corresponding region of Gata4−/− Gata6−/− embryos (Fig. 3, Smarcd3). At about E7.5 (early headfold), cardiac progenitors have formed the cardiac crescent that overlays the anterior intestinal portal, and expression of myocyte markers can first be identified in a subset of cells within the crescent that have committed to a cardiac myocyte cell-fate (Olson and Schneider, 2003). While we identified nascent cardiac myocytes in control embryos using immunohistochemistry to detect smooth muscle actin (Fig. 3, bottom panel, arrowhead), no smooth muscle actin positive cells, outside of the yolk sac, could be detected in Gata4−/− Gata6−/− embryos (Fig. 3, Sma).
Figure 3. Differentiated cardiac myocytes are undetectable in GATA4/GATA6 null embryos.
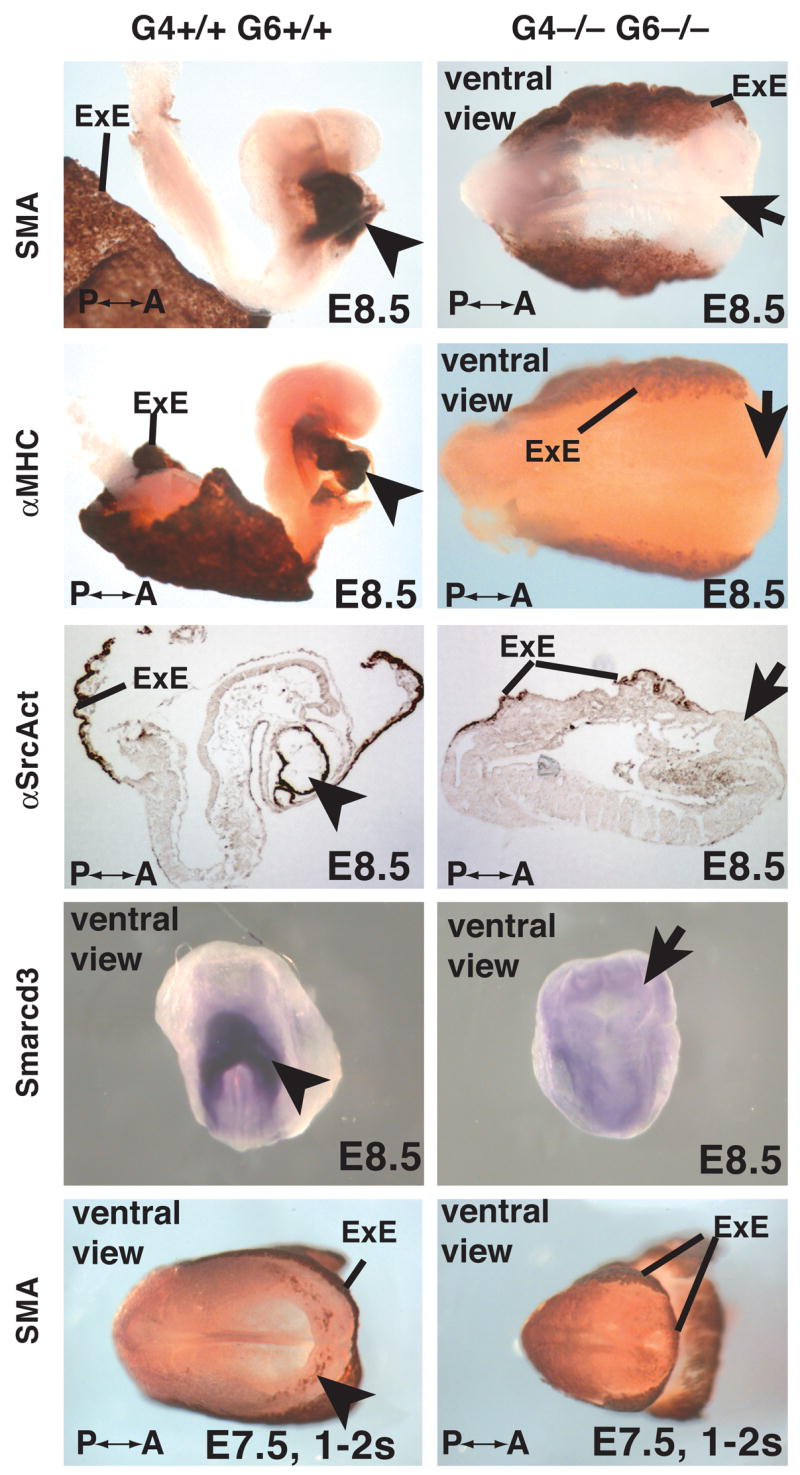
Micrographs showing control (G4+/+ G6+/+) and mutant (G4−/− G6−/−) whole embryos or sections of embryos at E8.5 (8–10 somites) or E7.5 (1–2 somites) stained by immunohistochemistry (brown staining) to reveal expression of smooth muscle actin (SMA), myosin heavy chain (MHC), sarcomeric actin (αSrcAct), or by in situ hybridization to identify Smarcd3 mRNA. Cardiac myocytes are indicated with an arrowhead in control embryos and the expected position of such cells with an arrow in mutant embryos. Anterior-posterior axis (A-P) and extra-embryonic endoderm (ExE) is shown where appropriate.
Although transcription factors expressed in cardiac progenitor cells could be detected in Gata4−/−Gata6−/− embryoid bodies, we considered the possibility that the absence of differentiated cardiac myocytes could result from a failure in formation of progenitor cells in vivo. We therefore examined the expression of mRNAs encoding cardiac progenitor cell markers Isl1, Nkx2.5, and Tbx5. Isl1 mRNA, which is predominantly expressed in progenitors within the second heart field (Cai et al., 2003), could be detected in both control and mutant embryos at both E8.5 and E7.5 (Fig. 4, Isl1). Similarly, Nkx2.5 mRNA, which is expressed in both primary and second heart fields, was also detected in both control and mutant embryos at E8.5 (Fig. 4, Nkx2.5). Both Isl1 and Nkx2.5 mRNA were found in tissue sections to be most abundant in two lateral domains of mesoderm that mark the cardiac progenitors (Fig. 4). In contrast to those markers expressed in the second heart field, expression of Tbx5 mRNA, which is predominantly expressed in the primary heart field progenitor cells and was detectable in control embryos, was undetectable in Gata4−/−Gata6−/− embryos (Fig. 4, Tbx5). These data imply that while GATA4 and GATA6 act to control the onset of cardiac myocyte differentiation they are dispensable for formation of second heart field progenitor cells.
Figure 4. GATA4 and GATA6 are dispensable for formation of second heart field cardiac progenitor cells.
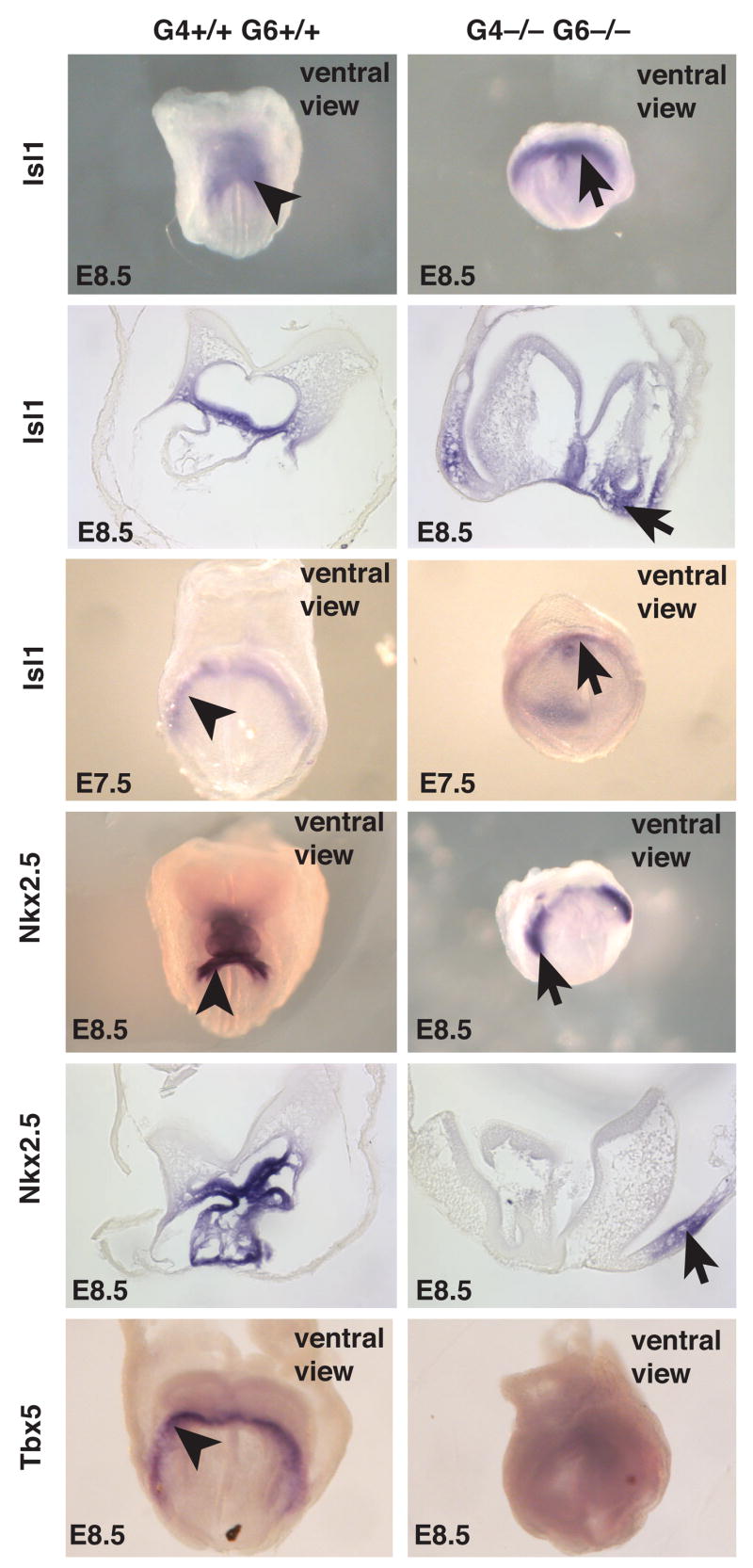
Micrographs showing the expression of Isl1 in E8.5 (8–10 somites) and E7.5 (early headfold) embryos, and Nkx2.5 and Tbx5 in E8.5 embryos (8–10 somites) by in situ hybridization. Arrowheads and arrows show the presence of presumptive cardiac progenitor cells in control and mutant embryos, respectively. Sections through Isl1 and Nkx2.5–stained embryos confirm the presence of these mRNAs in the mesoderm. All panels show ventral views of embryos with the proximal-distal axis positioned from top to bottom.
DISCUSSION
In summary, we conclude that GATA4 and GATA6 have overlapping but essential roles in ensuring that progenitors follow a cardiac myocyte cell fate during mammalian development. The mechanism through which this occurs is likely to be complex, and clarity will require further study. However, our data demonstrate that, while formation of the second heart field progenitor cells and their anterior migration to the cardiac crescent is independent of GATA4 or GATA6 function, the differentiation of these progenitors into cardiac myocytes as well as formation of the primary heart field progenitors is blocked by the absence of both GATA4 and GATA6. Such a conclusion is also supported by our analyses of cardiac myocyte differentiation from ES cells in which expression of markers of cardiac myocytes and transcription factors expressed in the primary heart field are dramatically reduced in the absence of GATA4 and GATA6, whereas markers of the second heart field, such as Isl1, or early undefined cardiac progenitor cells, such as Mesp1 and Mesp2, are unaffected by loss of GATA4 and GATA6. In addition to cardiac myocytes, GATA4 and GATA6 are both expressed in the early definitive endoderm, and so the question of whether the dependence on GATA4 and GATA6 for differentiation of cardiac myocytes is strictly cell autonomous has yet to be resolved. Based on the above analyses of Gata4−/−Gata6−/− embryos, we are confident that the definitive endoderm is generated in the absence of GATA4 and GATA6; however, we cannot definitively exclude the possibility that deficiencies in endoderm morphogenesis or differentiation contribute to the absence of cardiac myocytes in these embryos. Aggregation chimeras generated between ROSA26 morulae and Gata4−/−Gata6−/− ES cells did not answer this possibility because in such chimeras both the endoderm and developing heart were populated by wild type cells (not shown). Nevertheless, a significant body of evidence supports the view that although the endoderm is required for later myocardial contraction (Jacobson and Sater, 1988), it is dispensable for the onset of cardiac myocyte differentiation. For example, expression of cardiac differentiation markers in chick lateral plate mesoderm occurs normally following the removal of the definitive endoderm (Gannon and Bader, 1995), and Zebrafish casanova mutants, which suffer from a complete absence of definitive endoderm, exhibit cardia bifida but still generate cardiac mycocytes (Alexander et al., 1999). A cell autonomous role for GATA control of cardiac myocyte differentiation is also supported by the observation that the GATA factors, whose own expression responds to BMP signaling (Schultheiss et al., 1997), can directly control transcription of genes encoding numerous cardiac transcription factors and structural proteins, including MEF2c (Dodou et al., 2004) and NKX2.5 (Reecy et al., 1999). Such findings, along with our current analyses, support a model whereby, in response to BMP signaling, the GATA factors act as a molecular lynchpin to ensure robust expression of the network of transcription factors that orchestrates expression of cardiac myocyte genes during progenitor cell differentiation.
Acknowledgments
We thank R. Misra for RT-PCR primers, E. Morrisey for supplying the Gata6 targeting vector, B. Brunneau for Smarcd3 and Tbx5 and J. Lough for Nkx2.5 and Isl1 in situ hybridization probes, and A.F. Parlow (NHPP) for PMSG. Funding for this project was provided by an American Heart Association fellowship to A.J.W. and National Institutes of Health (NIDDK) grants to S.A.D., as well as gifts from the Marcus Family, Jack and Pheobe Lewis, and the Sophia Wolf Quadracci Memorial Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215:343–57. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–42. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Bader D. Initiation of cardiac differentiation occurs in the absence of anterior endoderm. Development. 1995;121:2439–50. doi: 10.1242/dev.121.8.2439. [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–14. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev Biol. 2007;312:613–22. doi: 10.1016/j.ydbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson AG, Sater AK. Features of embryonic induction. Development. 1988;104:341–59. doi: 10.1242/dev.104.3.341. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–26. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–32. [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–60. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–72. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Tymitz KM, Richardson JA, Olson EN. Abnormalities of the genitourinary tract in female mice lacking GATA5. Mol Cell Biol. 2000;20:5256–60. doi: 10.1128/mcb.20.14.5256-5260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–65. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–22. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Parmacek MS. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J Biol Chem. 1997;272:8515–24. doi: 10.1074/jbc.272.13.8515. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–90. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J. Production of completely ES cell-derived fetuses. In: Joyner A, editor. Gene targeting: A practical approach. Oxford Unviersity Press; Oxford, UK: 1993. pp. 147–179. [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Cardiomyocyte differentiation by GATA-4-deficient embryonic stem cells. Development. 1997a;124:3755–64. doi: 10.1242/dev.124.19.3755. [DOI] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev Biol. 1997b;189:270–4. doi: 10.1006/dbio.1997.8684. [DOI] [PubMed] [Google Scholar]
- Nemer G, Nemer M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev Biol. 2003;254:131–48. doi: 10.1016/s0012-1606(02)00026-x. [DOI] [PubMed] [Google Scholar]
- Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev. 2003;17:1937–56. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. Embo J. 2003;22:4260–73. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–35. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development. 1999;126:839–49. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–95. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–62. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–6. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Taraviras S, Monaghan AP, Schutz G, Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–88. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–8. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, Okano H, Fukuda K. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23:607–11. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25:2622–31. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]