Abstract
5′-Capping is an early mRNA modification that has important consequences for downstream events in gene expression. We have isolated mammalian cDNAs encoding capping enzyme. They contain the sequence motifs characteristic of the nucleotidyl transferase superfamily. The predicted mouse and human enzymes consist of 597 amino acids and are 95% identical. Mouse cDNA directed synthesis of a guanylylated 68-kDa polypeptide that also contained RNA 5′-triphosphatase activity and catalyzed formation of RNA 5′-terminal GpppG. A haploid strain of Saccharomyces cerevisiae lacking mRNA guanylyltransferase was complemented for growth by the mouse cDNA. Conversion of Lys-294 in the KXDG-conserved motif eliminated both guanylylation and complementation, identifying it as the active site. The K294A mutant retained RNA 5′-triphosphatase activity, which was eliminated by N-terminal truncation. Full-length capping enzyme and an active C-terminal fragment bound to the elongating form and not to the initiating form of polymerase. The results document functional conservation of eukaryotic mRNA guanylyltransferases from yeast to mammals and indicate that the phosphorylated C-terminal domain of RNA polymerase II couples capping to transcription elongation. These results also explain the selective capping of RNA polymerase II transcripts.
Addition of a 5′-terminal cap is an important, early event in mRNA formation (1). This structural hallmark of most eukaryotic mRNAs enhances splicing (2–4), transport (5), translation (6), and stability (7, 8) and is essential for viability (9).
Caps are formed on nascent nuclear pre-mRNAs by conversion of 5′-tri-diphosphate to 5′-diphosphate ends, followed by addition of GMP and methylation (1, 10). The guanylyltransfer reaction characterized in various systems involves formation of an active enzyme intermediate containing GMP covalently attached to lysine (11). In yeast, mRNA capping enzyme consists of separate subunits for RNA 5′-triphosphatase and guanylyltransferase activities (9, 12). cDNA clones coding for mRNA guanylyltransferase in Saccharomyces cerevisiae (9), Schizosaccharomyces pombe (13), and Candida albicans (14) have been sequenced. Each contains the active site lysine in KXDG (13, 15), one of several highly conserved motifs characteristic of a superfamily of nucleotidyl transferases (16). A number of viral capping enzymes also contain these diagnostic sequence motifs, and the recently solved structure of capping enzyme from Chlorella virus PBCV-1 suggests that specific residues in these motifs are important for binding GTP (17). Despite this detail of sequence and structure information, no metazoan capping enzyme previously has been cloned and characterized.
To explore the molecular interactions that result in selective capping of RNA polymerase II (pol II) transcripts in mammalian cells, we have isolated and characterized cDNA clones that code for the human and mouse capping enzymes. Functional studies demonstrated that the mammalian enzyme complements the lethality of a S. cerevisiae mutant (CEG1−) lacking mRNA guanylyltransferase and binds selectively to the elongating form of RNA pol II in which the largest subunit contains a phosphorylated C-terminal domain (CTD).
MATERIALS AND METHODS
Cloning of Mouse and Human Capping Enzyme cDNAs.
To identify genes encoding mammalian mRNA capping enzymes, we used the blast server at the National Center for Biotechnology Information (18) to screen the nonredundant GenBank Database Expressed Sequence Tag (EST) Division by querying with the amino acid sequence of S. pombe mRNA guanylyltransferase, PCE1 (accession no. U16143) (13). Two mouse expressed sequence tags (EST accession nos. AA096583 and AA108985) that displayed significant identity to the C-terminal half of PCE1 were identified and overlapped to predict a partial ORF with homology to PCE1 C-terminal sequence. A blast search of the nonredundant GenBank coding sequences using the mouse partial ORF demonstrated its homology to PCE1 as well as to Caenorhabditis elegans CEL-1 (19) and S. cerevisiae CEG1 (mRNA guanylyltransferase) (9) gene product.
Mouse capping enzyme (MCE) cDNAs were cloned from embryo poly(A)+ RNA (Ambion) by Marathon cDNA amplification according to the manufacturer’s instructions (CLONTECH). Gene-specific primers GSP1 (5′-GGGCGGCTGCTGCACATCTGTCAATG-3′) and GSP2 (5′-CCCAAATGCCTACAACACAGCCATGGC-3′) based on the EST sequences were used for 5′- and 3′-rapid amplification of cDNA ends (RACE) reactions. The resulting single 5′-RACE fragment (1.8 kb) and two 3′-RACE fragments (0.8 kb and 2.3 kb) were cloned into pCR2.1 (Invitrogen) and sequenced by using an Applied Biosystems-373 automated sequencer. The EST also was used to screen a HeLa cDNA library (Stratagene), and a 4.5-kb cDNA insert recovered in pBluescript SK was sequenced as above.
Northern Blot Analysis.
EST AA096583 and human β-actin cDNA (CLONTECH) were used to probe mouse and human multiple tissue and mouse embryo Northern blots as recommended by the manufacturer (CLONTECH).
Plasmids.
All MCE constructs generated by PCR (Advantage KlenTaq polymerase, CLONTECH) or site-specific mutagenesis (QuikChange Kit, Stratagene) first were cloned into pCR2.1 with a BamHI site at the 5′ end. N-terminal truncation mutant Δ210 was further subcloned into the BamHI site of pET-28a (Novagen). A 5′XhoI–3′KpnI fragment coding for wild-type or K294A mutant MCE was isolated from pCR2.1, blunted with Klenow and ligated to SalI-digested, Klenow-treated pG-1 (20) to yield pG-MCE.
Expression of MCE Fragment and Antibody Production.
Based on the combined nucleotide sequence of the mouse ESTs, a DNA fragment encoding a C-terminal region (amino acids 438–597) of the MCE with a 6xHis tag was obtained by PCR by using as template mouse embryo cDNA (Marathon ready, CLONTECH). The purified PCR product was cloned into pCR2.1, and the insert then was excised with NheI and BamHI and cloned in-frame in pET-11a (Novagen) to generate pMCE0.5. Recombinant protein expressed in Escherichia coli BL21 (DE3) transformed with pMCE0.5 was purified on Ni-nitrilotriacetic acid agarose (Qiagen) and used to raise antibodies (αMCE) in rabbits.
In Vitro Translation and Immunoprecipitation.
pCR2.1 plasmids containing wild-type or mutant MCE were used to synthesize proteins in the TNT Coupled Reticulate Lysate System (Promega). Products were immunoprecipitated with αMCE in 25 mM Tris, pH 7.5/100 mM KCl/5 mM MgCl/0.5 mM DTT (buffer B).
MCE Guanylylation and RNA Cap Formation.
Vaccinia virus capping enzyme (1 unit, GIBCO/BRL) or translated and immunoprecipitated proteins prepared as above from 50 μl TNT lysate were incubated for 15 min at 37°C in 20 μl of GTP-labeling buffer consisting of 25 mM Tris at pH 7.5, 5 mM MgCl2, 0.5 mM DTT, α[32P]GTP (10 μCi, 3,000 Ci/mmol, Amersham), and 0.10 μg inorganic pyrophosphatase (Boehringer). Guanylylation of proteins was analyzed by SDS/PAGE followed by autoradiography. T7 polymerase run-off RNA (32-mer) transcribed from BamHI-linearized pGEM1 (Promega) was incubated as indicated in GTP-labeling buffer for 15 min at 37°C. Samples were extracted with phenol/chloroform, precipitated with ethanol, and analyzed by 8% PAGE. For analysis of cap formation, RNA after extraction was passed through a Chroma Spin-10 column (CLONTECH), precipitated with ethanol, resuspended in 8 μl of 50 mM sodium acetate (pH 5.3) containing 4 μg P1 nuclease (Boehringer) and incubated at 37°C for 1 hr. Samples were adjusted to 50 mM Tris (pH 8.0), treated with calf intestine alkaline phosphatase (1 unit, GIBCO/BRL) for 30 min at 37°C (21), spotted on polyethyleneimine-cellulose plates, and analyzed by TLC in 0.4 M ammonium sulfate followed by autoradiography.
RNA 5′-Triphosphatase Assay.
RNA (32-mer) was synthesized as above except that GTP was replaced with γ[32P]GTP (30 μCi, 30 Ci/mmol, DuPont). The 5′ end-labeled RNA was purified by passage through a Chroma Spin-10 column and incubated for 15 min at 37°C with the indicated proteins in GTP-labeling buffer but without α[32P]GTP and inorganic pyrophosphatase (4 × 104 cpm, 15 μl). Reaction products were extracted with phenol/chloroform and analyzed by polyethyleneimine-cellulose TLC in 0.8 M acetic acid containing 0.9 M LiCl followed by autoradiography.
Complementation of a Yeast mRNA Guanylyltransferase Mutant by MCE.
pG-1 (CEN and TRP1; ref. 20) was used to express MCE under control of the constitutive yeast glyceraldehyde-3-phosphate dehydrogenase promoter. The haploid S. cerevisiae strain YBS2 [MATa, leu2, lys2, trp1, ceg1::hisG, pGYCE-360 (CEN, URA3, CEG1)] (22) was transformed with either wild-type or mutant pG-MCE by using the lithium acetate method (23). Trp+ transformants obtained at 30°C were tested for the ability to grow in the presence of 5-fluoroorotic acid, which selects against cells that retain the CEG1-containing URA3 plasmid (24).
Pol II-Capping Enzyme Interaction Studies.
αMCE (1 μg), affinity purified using MCE Δ210 bound to Affi-prep 10 (Bio-Rad) Resin, was incubated with protein A-agarose (Repligen) beads for 30 min at room temperature. The antibody-protein A beads were washed and equilibrated with Hepes buffer, pH 7.8 containing 0.1 M KCl and 0.1% Nonidet P-40 (buffer I) and then incubated with or without partially purified (25) HeLa capping enzyme (120 μg) for 1 hr, rotating at 4°C. This immune complex was washed extensively with buffer I containing 1M KCl and equilibrated with buffer I before incubation for 1 hr, rotating at 4°C with pol II (1.2 μg of DEAE-5PW protein fraction; ref. 26). The immune complexes again were washed extensively, equilibrated with buffer I, and used for Western blot analysis. Blots were developed with αMCE and antibodies against the pol II largest subunit by ECL (Boehringer Mannheim).
Immunoprecipitation of 6xHis-tagged MCE Δ210 was performed using mAbs against hexa-histidine tag (CLONTECH). One microgram of antibodies was incubated with protein G-agarose beads (Boehringer Mannheim) for 30 min at room temperature, and the resulting complexes were washed and equilibrated with buffer I, incubated with purified MCE Δ210 (0.25 μg), and processed as above.
RESULTS
Nucleotidyl Transferase Superfamily Sequence Motifs Are Conserved in Human and Mouse mRNA Capping Enzymes.
One human and two mouse cDNAs encoding mRNA guanylyltransferase were isolated. Each contains a single ORF of 1,794 bp that begins with the first ATG in an initiator consensus sequence (27) (Fig. 1A). The 3′ untranslated region sequence of the shorter mouse clone is repeated in the longer one, which also has a second AATAAA polyadenylation signal. Complete sequences of the predicted polypeptides are shown in Fig. 1B. The mouse and human predicted proteins are 95% identical and consist of 597 amino acids with calculated molecular masses that are in close agreement with the 68 kDa reported previously for mammalian capping enzymes (10, 28).
Figure 1.
MCE and human capping enzyme cDNA clones. (A) Schematic diagram of the ORFs (1,794 bp, solid bars), relative sizes of the untranslated regions, and the sequences and positions of initiation sites and polyadenylation signals. (B) Predicted amino acid sequences of the mouse and human enzymes. Residues that are identical (95% of total) are omitted from the human sequence.
mRNA capping proceeds by removal of the γ-phosphate from 5′-triphosphate termini followed by GMP transfer from guanylylated capping enzyme intermediate to the resulting 5′-diphosphate ends. In yeast, these reactions are catalyzed by an 80-kDa RNA 5′-triphosphatase and a 52-kDa mRNA guanylyltransferase, whereas in mammals both activities apparently are present as distinct domains of a single polypeptide (10). Sequence alignment of the C-terminal regions of the mammalian clones and C. elegans CEL-1 gene (19) with yeast guanylyltransferases indicates that they contain the conserved sequence motifs that identify members of the nucleotidyl transferase superfamily (16) (Fig. 2). These include a KXDG motif (bracket), which corresponds to the active site, guanylylated lysine in a variety of viral and cellular capping enzymes.
Figure 2.
Sequence alignments of cellular mRNA guanylyltransferases. The deduced amino sequences of the C-terminal regions of the mouse and human enzymes (residues 232–597) are compared with C. elegans CEL-1 (residues 238–573; ref. 19) and to the nearly full-length S. pombe (13) and S. cerevisiae (9) guanylyltransferase subunits of capping enzyme. Dark background indicates regions of sequence identity; the active site motif, including Lys-294, in the mammalian enzymes is bracketed.
Capping Enzyme Expression In Vivo and In Vitro.
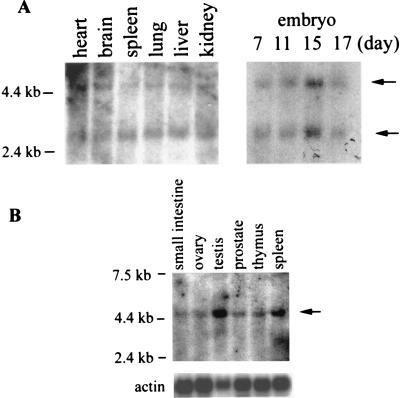
Northern analyses of poly(A)+ RNA from mouse embryos and several adult tissues revealed two specific transcripts (Fig. 3A, arrows), consistent with the two mouse cDNA clones that differed in the sizes of the 3′-untranslated regions. Human tissues contained a single transcript (Fig. 3B).
Figure 3.
Expression of capping enzyme transcripts in vivo. (A) Poly(A)+ RNAs prepared from mouse adult tissues (Left) and embryos at the indicated days of gestation (Right) were probed with a mouse EST as described in Materials and Methods. (B) Human adult tissue poly(A)+ RNAs were probed with the same EST and a human β-actin cDNA. Positions of marker RNAs and hybridized transcripts (arrows) are indicated.
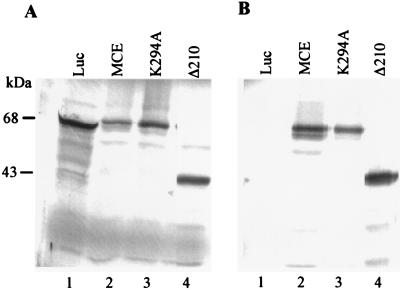
MCE cDNAs corresponding to the full-length wild type or a K294A mutant protein, or a C-terminal fragment beginning with amino acid 211 (Δ210), were inserted into a T7 expression vector and used in a coupled transcription/translation system. Products of the expected sizes were detected by [35S]methionine labeling and SDS/PAGE (Fig. 4A). A cDNA encoding luciferase (Luc) served as a control. The MCEs, but not Luc, were immunoprecipitated with αMCE (Fig. 4B).
Figure 4.
In vitro synthesis and specific immunoprecipitation of MCEs. (A) 35S-methionine-labeled proteins were synthesized in vitro by using luciferase (Luc) cDNA (lane 1) or cDNAs for wild type (lane 2), mutant (lane 3), and C-terminal fragment (Δ210, lane 4) of MCE. Products were analyzed by SDS/PAGE and autoradiography. (B) Products shown in A were immunoprecipitated with αMCE and analyzed by SDS/PAGE and autoradiography.
Guanylyltransferase Activity and Identification of the Active Site.
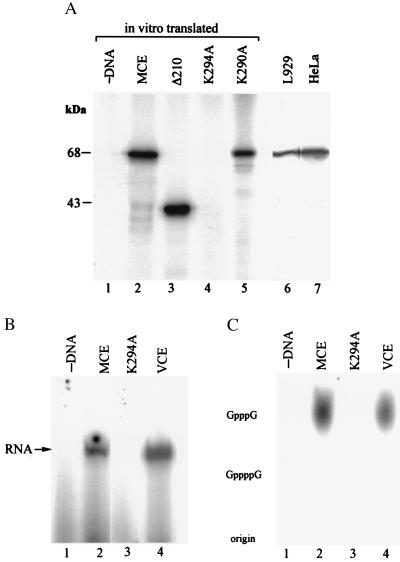
To test for covalent attachment of GMP to the polypeptide encoded by the isolated mouse cDNA, proteins made in vitro were immunoprecipitated with αMCE, incubated with α[32P]GTP and analyzed by SDS/PAGE. As shown in Fig. 5A, the full-length wild-type and K290A mutant proteins, as well as the C-terminal fragment, were guanylylated. Immunoprecipitates of samples from HeLa and mouse L cells also yielded a 32P-labeled polypeptide that comigrated with the full-length guanylylated in vitro products. To verify that Lys-294 in the KXDG motif is the site of guanylylation, the Lys was mutated to Ala. Although similar amounts of the K294A mutant and wild-type proteins were synthesized (Fig. 4A), the K294A mutant protein (unlike mutant K290A) was not guanylylated (Fig. 5A).
Figure 5.
Guanylyltransferase activity of MCEs. (A) In vitro translated and immunoprecipitated proteins were incubated with α[32P]GTP as described in Materials and Methods and analyzed by SDS/PAGE and autoradiography. Lane 1, no added DNA; lanes 2–5, products synthesized with cDNA templates for full-length MCE, C-terminal fragment (Δ210), and mutants K294A and K290A, respectively. Mouse L cell nuclear extract (lane 6) and partially purified HeLa cell capping enzyme (lane 7) were immunoprecipitated with αMCE and then incubated with α[32P]GTP. Proteins were analyzed by SDS/PAGE and autoradiography. (B) 5′-Terminal guanylylation of T7 transcripts was assayed by incubating the RNA with α[32P]GTP and immunoprecipitates of in vitro synthesized wild-type (lane 2) or K294A mutant (lane 3) MCE or vaccinia virus capping enzyme (VCE, lane 4). (C) Cap formation on the radiolabeled RNAs shown in B was verified by digestion with P1 nuclease followed by alkaline phosphatase and TLC analysis of the digests with authentic GpppG and GppppG as markers.
Capping enzymes synthesized in vitro were immunoprecipitated and tested for cap formation on T7 transcripts (presumably containing 5′-terminal pppG). The wild-type enzyme (Fig. 5B, lane 2), like purified vaccinia virus capping enzyme (Fig. 5B, lane 4), catalyzed transfer of radiolabeled GMP from α[32P]GTP to the RNA, whereas the K294A mutant (lane 3) was inactive. 32P-labeled RNA samples were treated with P1 nuclease and phosphatase (CIAP) under conditions that convert capped RNAs to Pi and caps (29), and the digests were analyzed by TLC together with authentic GpppG and GppppG as markers. As shown in Fig. 5C, mouse and vaccinia virus capping enzymes both produced GpppG caps on the T7 transcripts. In the presence of S-adenosylmethionine, vaccinia, but not MCE, produced methylated caps (data not shown). The results suggest that recombinant mammalian capping enzyme contains RNA 5′-triphosphatase in addition to guanylyltransferase activity.
MCE Contains RNA 5′-Triphosphatase.
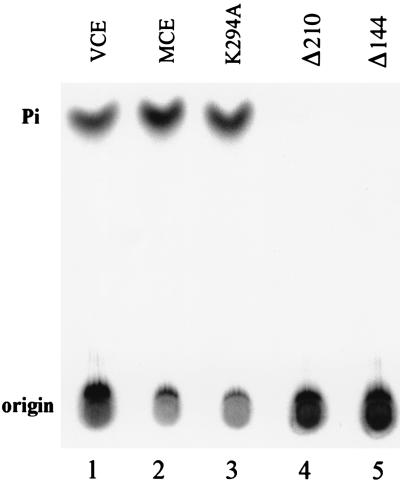
Mouse and human capping enzymes and the related C. elegans CEL-1 ORF have N-terminal extensions that are absent from yeast mRNA guanylyltransferases and contain a stretch of 15 identical amino acids, GVHCTHGFNRTGFLI. It includes (I/V)HCxxGxxR(S/T)G, a motif characteristic of protein tyrosine phosphatases (30, 31). However, CEL-1 encodes RNA 5′-triphosphatase rather than protein phosphatase activity (19). Treatment of γ[32P]GTP, 5′-end-labeled T7 transcripts with wild-type (Fig. 6, lane 2) or mutant K294A (Fig. 6, lane 3) MCE generated 32Pi, as did incubation with vaccinia capping enzyme (Fig. 6, lane 1). N-terminal truncation of 144 (Fig. 6, lane 5) or 210 (Fig. 6, lane 4) amino acids resulted in C-terminal fragments that were without RNA 5′-triphosphatase but retained guanylyltransferase (Fig. 5A, lane 3 and data not shown). Thus, the N-terminal region of mammalian capping enzyme is required for RNA 5′-triphosphatase but not for mRNA guanylyltransferase activity.
Figure 6.
MCE contains RNA 5′-triphosphatase. RNA containing 5′-terminal γ[32P]GTP, prepared as described in Materials and Methods, was incubated with in vitro-synthesized, immunoprecipitated MCE wild type (lane 2), mutant K294A (lane 3), N-terminal truncation mutants Δ210 (lane 4), and Δ144 (lane 5) or vaccinia virus capping enzyme (lane 1). Samples were analyzed for release of labeled Pi by TLC and autoradiography.
Mammalian Capping Enzyme Complements S. cerevisiae ΔCEG1 Mutant.
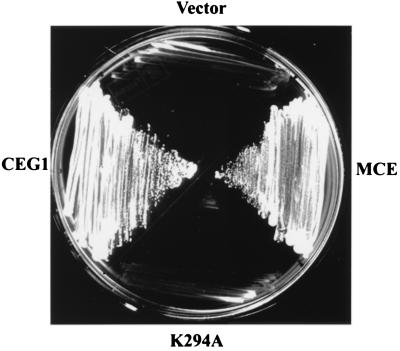
YBS2, a haploid S. cerevisiae strain in which the chromosomal CEG1 (mRNA guanylyltransferase) gene is deleted, is kept viable by the presence of an extrachromosomal allele supplied on a URA3 plasmid. A plasmid shuffle strategy (24) was used to determine if the mouse MCE gene can supply the essential mRNA guanylyltransferase function. Plasmids pG-MCE and pG-MCE(K294A) that constitutively express wild-type and mutant alleles of MCE and carry a TRP1 marker were introduced into YBS2 cells. Trp+ transformants then were plated on medium containing 5-fluoroorotic acid. As shown in Fig. 7, wild-type MCE sustained growth of YBS2 on counterselective medium, similar to control cells containing pGYCE358 (CEG1 and TRP1). By contrast, the K294A active site mutant that was devoid of mRNA guanylyltransferase activity did not complement YBS2 for growth on 5-fluoroorotic acid. Cells containing the empty parental vector pG-1 also failed to grow on 5-fluoroorotic acid. These results demonstrate that the mouse MCE gene fully complements S. cerevisiae ΔCEG1 mutant for growth.
Figure 7.
Complementation of S. cerevisiae lacking mRNA guanylyltransferase. Growth was measured on selective media using CEG1− cells transformed with empty parental pG-1 (vector), pG-1 containing wild-type (MCE) or mutant (K294A) MCE or with a CEG1-containing URA3 plasmid (CEG1) as detailed in Materials and Methods.
Selective Binding of Full-Length and C-Terminal Fragment of Mammalian Capping Enzyme to the Elongating Form of pol II.
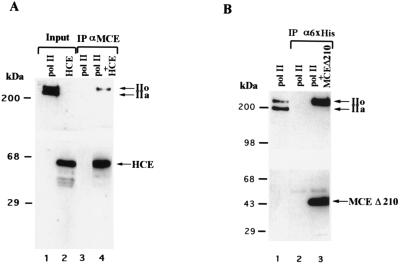
Because addition of the cap to mRNA occurs early during transcription (32–34), we investigated whether capping enzyme is part of the human pol II holoenzyme (35). To our surprise, we found that it is not (data not shown), and we therefore tested for the direct interaction of capping enzyme with pol II. Immunocomplexes containing human capping enzyme were incubated with purified human RNA pol II that contains approximately equal amounts of the initiating (nonphosphorylated CTD) and elongating (phosphorylated CTD) forms of the enzyme (Fig. 8A, lane 1). Western blot analysis demonstrated that αMCE specifically immunoprecipitated the elongating form of pol II (Fig. 8A, lane 4). Selective immunoprecipitation of the phosphorylated form of pol II was not because of a nonspecific interaction of the antibodies as it was dependent on the presence of capping enzyme (Fig. 8A, lane 3).
Figure 8.
Capping enzyme directly interacts with the phosphorylated form of RNA pol II. (A) A HeLa cell protein fraction containing approximately equal amounts of the phosphorylated (pol IIo) and unphosphorylated (pol IIa) forms of pol II (lane 1) was mixed with αMCE-protein A beads in the presence or absence of affinity-purified HeLa capping enzyme (HCE, lane 2). Immunoprecipitates (lane 3 and 4) were extensively washed and loaded onto a 5–20% gradient SDS-polyacrylamide gel with the input (10%, lanes 1 and 2) for Western blot analysis. Immunoblots were developed for the CTD of the largest subunit of pol II (Upper) or capping enzyme (Lower). (B) The carboxy-terminal domain of capping enzyme is sufficient to mediate direct and selective interaction with the phosphorylated form of pol II. The HeLa protein fraction containing the two forms of pol II (lane 1) was mixed with antihexahistidine tag antibody (α6xHis)-protein G beads in the presence or absence of purified, recombinant hexa-histidine tagged carboxy-terminal domain of MCE Δ210. Immunoprecipitates (lanes 2 and 3) were extensively washed and loaded onto a 5–20% gradient SDS-polyacrylamide gel with the input (10%, lane 1) for Western blot analysis. Immunoblots were developed for the CTD of the largest subunit of pol II (Upper) or MCE (Lower).
The C-terminal guanylyltransferase domain of capping enzyme was sufficient for interaction with pol II. Consistent with the results in Fig. 8A, the phosphorylated form of pol II was specifically coimmunoprecipitated with 6xHis-tagged MCE Δ210 in the presence of anti-hexahistidine antibody (Fig. 8B, lane 3), and this was dependent on the presence of capping enzyme (Fig. 8B, lane 2). We conclude that the C-terminal guanylyltransferase domain of capping enzyme directly interacts with the elongation competent, phosphorylated form of RNA pol II.
DISCUSSION
The functional importance of mRNA 5′-capping in the control of eukaryotic gene expression has been demonstrated at several levels, including pre-mRNA synthesis and splicing in the nucleus, mRNA transport to the cytoplasm, ribosome binding during translation initiation, and transcript turnover. The presence of a 5′-terminal cap impacts on each of these stages in the lifetime of cellular mRNAs, and mRNA capping is essential for viability in S. cerevisiae. In addition, most viruses that replicate in the cytoplasm have evolved mechanisms to assure that the viral mRNAs are capped.
In mammalian cells, caps are added exclusively to newly initiated RNA pol II transcripts. Presumably, this specificity is based on the selective association of capping enzyme with one or more subunits of pol II. The results presented in Fig. 8 directly demonstrate an interaction between capping enzyme and pol II and suggest that CTD phosphorylation regulates this interaction. Previous studies have demonstrated that CTD phosphorylation regulates pol II function during the transcription cycle, most likely by selecting interacting factors (36, 37). The form of pol II that associates with transcription initiation complexes contains an unphosphorylated CTD, whereas the pol II engaged in elongation contains a phosphorylated CTD (36). The unphosphorylated form of pol II selectively interacts with the TATA-binding protein (38), TFIIE (39), and presumably other general transcription initiation factors (40), as well as with a large number of other initiation factors that regulate transcription efficiency and are components of a large polymerase complex, the so-called pol II holoenzyme (35, 41–43).
Recent studies have shown that pol II also interacts with factors involved in RNA processing (44, 45), and it has been hypothesized that these interactions are established after CTD phosphorylation (44, 46). Our results demonstrating a specific interaction between capping enzyme and the phosphorylated form of pol II further support this hypothesis. The studies of Hagler and Shuman (34), using the vaccinia virus transcription system, demonstrated that the cap structure is added immediately after the 5′-end of the nascent RNA is exposed (after formation of 30 phosphodiester bonds). Lis and coworkers (33), using the Drosophila HSP70 gene, arrived at a similar conclusion. In intact isolated nuclei, the uninduced HSP70 gene contains an RNA pol II engaged in transcription but paused approximately 25–30 nucleotides downstream of the transcription start site. The CTD of the paused RNA pol II is unphosphorylated. Early after heat shock induction, the pol II enters into a productive elongation mode that is accompanied by phosphorylation of the CTD (47). Analysis of the formation of the cap at the Drosophila heat shock locus established that the paused RNAs are mostly uncapped, but become capped after the formation of approximately 30 phosphodiester bonds (33). Based on our results establishing that capping enzyme interacts preferentially with pol II containing a phosphorylated CTD, we propose that concomitant to entry into the elongation mode, pol II interacts with capping enzyme and the nascent RNA becomes capped. This event, together with CTD phosphorylation, results in a signal for the association of the RNA processing machinery with the transcribing (elongating) machinery. In this model, 3′ extension of 5′-uncapped and exonuclease-sensitive transcripts is avoided, possibly another example of CTD-mediated mRNA quality assurance (48, 49).
It is perhaps noteworthy in light of capping enzyme binding to the elongating form of pol II that the presence of a 5′-cap is a prerequisite for 3′-processing of histone mRNA (50) and possibly other transcripts (51). In addition, termination of influenza virus mRNA synthesis is cap-dependent (52), and vaccinia virus mRNA termination requires the presence of viral capping enzyme (53). Recent studies indicate that the human capping enzyme gene is on chromosome 6 (R.P., unpublished results), and further analyses may uncover linkages between this key enzyme and hereditary disorders as recently described for transcription factors (54, 55).
Acknowledgments
We thank Dr. Beate Schwer for pGYCE-358 and S. cerevisiae strain YBS2, Yingxia Wen for purified 6xHis-tagged MCE Δ210, and Dr. John Lis for helpful comments. This research was supported by grants from the Howard Hughes Medical Institute and the National Institutes of Health (GM 37120 to D.R.), the New Jersey Commission on Cancer Research (797-033 to R.P.), and the New Jersey Commission on Science and Technology.
ABBREVIATIONS
- pol II
polymerase II
- CTD
C-terminal domain
- EST
expressed sequence tag
- MCE
mouse capping enzyme
Footnotes
References
- 1.Shatkin A J. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Konarska M M, Padgett R A, Sharp P A. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 3.Krainer A R, Maniatis T, Ruskin B, Green M R. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 4.Edery I, Sonenberg N. Proc Natl Acad Sci USA. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 6.Shatkin A J. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 7.Furuichi Y, LaFiandra A, Shatkin A J. Nature (London) 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 8.Shimotohno K, Kodama Y, Hashimoto J, Miura K I. Proc Natl Acad Sci USA. 1977;74:2734–2738. doi: 10.1073/pnas.74.7.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 10.Mizumoto K, Kaziro Y. Prog Nucleic Acid Res Mol Biol. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- 11.Shuman S. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Shatkin A J. Nucleic Acids Res. 1984;12:2303–2315. doi: 10.1093/nar/12.5.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuman S, Liu Y, Schwer B. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada-Okabe T, Shimmi O, Doi R, Mizumoto K, Arisawa M, Yamada-Okabe H. Microbiology. 1996;142:2515–2523. doi: 10.1099/00221287-142-9-2515. [DOI] [PubMed] [Google Scholar]
- 15.Fresco L D, Buratowski S. Proc Natl Acad Sci USA. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shuman S, Schwer B. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 17.Håkansson K, Doherty A J, Shuman S, Wigley D B. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Takagi T, Moore C R, Diehn F, Buratowski S. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 20.Schena M, Picard D, Yamamoto K R. Methods Enzymol. 1991;194:389–392. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Shuman S. J Virol. 1996;70:6162–6168. doi: 10.1128/jvi.70.9.6162-6168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwer B, Shuman S. Proc Natl Acad Sci USA. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker D M, Guarente L. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 24.Sikorski R S, Boeke J D. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 25.Reinberg D, Roeder R G. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 26.Lu H, Flores O, Weinmann R, Reinberg D. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Furuichi Y, Shatkin A J. Mol Cell Biol. 1982;2:993–1001. doi: 10.1128/mcb.2.8.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin A J. J Biol Chem. 1976;251:5043–5053. [PubMed] [Google Scholar]
- 30.Denu J M, Stuckey J A, Saper M A, Dixon J E. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 31.Fauman E B, Saper M A. Trends Biochem Sci. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- 32.Coppola J A, Field A S, Luse D S. Proc Natl Acad Sci USA. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasumssen E B, Lis J T. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagler J, Shuman S. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 35.Maldonado E, Shlekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 36.Dahmus M E. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 37.Zawel L, Reinberg D. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 38.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 39.Maxon M E, Goodrich J A, Tjian R. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 40.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 41.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y-J, Björklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 43.Chao D M, Gadbols E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 44.Greenleaf A L. Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- 45.Kuchin S, Yeghiayan P, Carlson M. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brien T, Hardin S, Greenleaf A, Lis J T. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 48.Steinmetz E J. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 49.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 50.Georgiev O, Moss J, Birnstiel M L. Nucleic Acids Res. 1984;12:8539–8551. doi: 10.1093/nar/12.22.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sisodia S S, Sollner-Webb B, Cleveland D W. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beaton A R, Krug R M. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shuman S, Broyles S S, Moss B. J Biol Chem. 1987;262:12372–12380. [PubMed] [Google Scholar]
- 54.Hanawalt P C. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 55.Latchman D S. N Engl J Med. 1996;334:28–33. doi: 10.1056/NEJM199601043340108. [DOI] [PubMed] [Google Scholar]