Abstract
Chloroplast function depends on the translocation of cytosolically synthesized precursor proteins into the organelle. The recognition and transfer of most precursor proteins across the outer membrane depend on a membrane inserted complex. Two receptor components of this complex, Toc34 and Toc159, are GTPases, which can be phosphorylated by kinases present in the hosting membrane. However, the physiological function of phosphorylation is not yet understood in detail. It is demonstrated that both receptors are phosphorylated within their G-domains. In vitro, the phosphorylation of Toc34 disrupts both homo- and heterodimerization of the G-domains as determined using a phospho-mimicking mutant. In endogenous membranes this mutation or phosphorylation of the wild-type receptor disturbs the association of Toc34, but not of Toc159 with the translocation pore. Therefore, phosphorylation serves as an inhibitor for the association of Toc34 with other components of the complex and phosphorylation can now be discussed as a mechanism to exchange different isoforms of Toc34 within this ensemble.
Keywords: GTPase, membrane complex dynamics, phosphorylation, plastids, protein complex assembly, protein translocation, TOC
Introduction
Plastids are complex organelles undergoing transitions depending on the developmental or metabolic context (Lopez-Juez, 2007), which requires massive changes of the organelle proteome. This process is regulated at multiple levels (Kessler and Schnell, 2006; Oreb et al., 2008) including (i) transcription of nuclear genes encoding plastid destined proteins, (ii) targeting of precursor proteins to the chloroplast, (iii) their recognition and translocation across the chloroplast double membrane, and (iv) protein turnover within the organelle. The translocation process requires functional hetero-oligomeric protein complexes Toc (Translocon at the outer chloroplast envelope membrane) and Tic (Translocon at the inner chloroplast envelope membrane). The Toc core complex is formed by multiple copies of the translocation pore Toc75, the GTP-dependent receptor Toc34 and one tripartite receptor Toc159 (Schleiff et al., 2003a; Kikuchi et al., 2006). The latter protein contains a large membrane-anchored (M) domain, a cytosolic GTPase (G) domain highly homologous to that of Toc34, and an N-terminal acidic (A) domain of unknown function (Chen et al., 2000). Toc34 is anchored in the membrane by a short tail at its carboxy-terminus (Chen and Schnell, 1997). In Arabidopsis, two paralogues of Toc34 exist, namely atToc33 and atToc34, as well as four members of the Toc159 family, atToc159, atToc132, atToc120, and atToc90 (Jackson-Constan and Keegstra, 2001; Oreb et al., 2006). It is proposed that members of both families differentially associate to form translocons specialized for different classes of precursor proteins (Gutensohn et al., 2006; Kessler and Schnell, 2006). According to the current model, atToc33 preferably interacts with atToc159 forming the translocon for photosynthetic proteins whereas atToc34 forms a complex recognizing housekeeping proteins together with atToc132 and/or atToc120. However, according to coimmunoprecipitation experiments (Ivanova et al., 2004), other interaction patterns are also possible. The ability of atToc33 and atToc34 to overtake the function of each other (Jarvis et al., 1998; Constan et al., 2004) in knock-out mutants supports this possibility. It is not yet understood in detail what mechanisms drive the assembly of the Toc complex, but there is evidence that interactions of Toc159 and Toc34 G-domains are important for this process (Kessler and Schnell, 2006; Oreb et al., 2008).
Phosphorylation of the G-domains is discussed as a second circuit for the regulation of complex assembly in this study. For both GTPases Toc159 and Toc34 phosphorylation was demonstrated in vitro (Sveshnikova et al., 2000; Fulgosi and Soll, 2002). In the past, the properties of the phosphorylated Toc34 were explored in some detail (Jelic et al., 2002). Phosphorylation leads to a negative regulation of precursor protein recognition (Sveshnikova et al., 2000) and the GTPase cycle (Jelic et al., 2002, 2003), albeit the phosphorylation sites are located in different areas of the protein from different sources (e.g. atS181, Jelic et al., 2003; psS113, Jelic et al., 2002). Although the phosphorylation sites of atToc33 and psToc34 have been mapped, the in vivo significance and the mechanistic effect are uncertain (Aronsson et al., 2006; Oreb et al., 2007). Here, the aim was to investigate the influence of the post translational modification of the GTPases on the complex formation in endogenous membranes. Evidence is provided here for a regulatory mechanism involving post-translational phosphorylation, which is suggested to change the state of association of the GTPases within the Toc complex.
Materials and methods
Protein production
Mutants of atToc33, atToc33S/A, atToc33S/E (aa 1–251), and psToc159 (aa 658–1048) were generated by conventional PCR (Jelic et al., 2003) and cloned into pET21d (Novagen, Madison, WI, USA) or pGEX6P-1 (GE Healthcare, Freiburg, Germany) to generate His-tagged or GST-fusion proteins. Recombinant proteins were purified by Ni-NTA (Qiagen, Hilden, Germany) or GSH affinity chromatography (GE Healthcare, Freiburg, Germany) according to protocols suggested by the manufacturer.
Determination of the binding constant by size exclusion chromatography
For molecular weight determination, 200 μl of purified protein, typically at concentrations between 0.3 mM and 0.4 mM, was loaded onto a Superdex75 HR 10/300 gel filtration column (GE-Healthcare, Freiburg, Germany), equilibrated with 50 mM TRIS buffered at pH 8, containing 100 mM NaCl, 10 mM imidazole, 10 mM arginine, 5% glycerol, and 1 mM MgCl2. Using different starting concentrations of protein, monomeric and dimeric species were analysed by fitting two independent Gaussian distributions to the elution profile. The dissociation constant was determined according to:
| (1) |
where D is the dimeric fraction and T is the total amount of protein used.
Solid phase binding assay
For all assays only freshly purified protein was used. For determination of nucleotide dependent association, the G-domain of atToc33-His and mutants was preloaded with nucleotides (König et al., 2008) and spotted onto nitrocellulose membranes using a 96 well vacuum manifold (Bethesda Research Laboratories, Bethesda, MD, USA). Nucleotide loading was determined as described in König et al. (2008). Nitrocellulose membranes were saturated with 0.3% low fat milk powder with 0.03% BSA and subsequently incubated with purified GST-atToc33 or GST-psToc159 (35 μg ml−1) preloaded with indicated nucleotides in 20 mM Tricine-KOH at pH 7.6, containing 100 mM NaCl and 1 mM MgCl2. Background binding was controlled by incubation of GST-atToc33 or GST-psToc159 with saturated nitrocellulose membranes without spotted protein and spotted BSA. After two washes (10 min), the bound protein was determined by immunodetection with GST antibodies. Background staining caused by unspecific binding of GST antibodies was determined by immunodetection of membranes with spotted proteins but without an added interaction partner. Intensities were quantified with the AIDA Software (Raytest, Straubenhardt, Germany). The amount of bound protein was corrected for background binding and expressed as a comparison with the maximal binding of wild-type protein. The binding was analysed by:
| (2) |
where S is the concentration of the spotted protein, E the concentration of the added protein (normalized), and K reflects the dissociation constant in a non-normalized situation and an apparent dissociation constant after normalization, as described by König et al. (2008).
In situ analysis of Toc complex assembly
The complementation of the ppi1 mutant with the phospho-mimicking atToc33 construct has been described previously (Aronsson et al., 2006). Wild-type or mutant A. thaliana was grown for 2 weeks on soil in an E-36L growth chamber (Percival Sci., Perry, IA) at 16/8 h light/dark, 21/16 °C, 70% relative humidity. Approximately 5 g leaf material was ground for 2–3 s using a Polytron (Kinematica PT20) with a 13 mm rotor at 40% maximal speed in 25 ml of ice-cold homogenization buffer (0.3 M sorbitol, 5 mM MgCl2, 5 mM EDTA, 20 mM HEPES/KOH, pH 8.0, 10 mM NaHCO3, 50 mM ascorbic acid). The homogenate was filtered through Miracloth and the debris was subjected to homogenization. The procedure was iterated four times before the filtrates were combined and centrifuged at 1000 g for 5 min. Chloroplasts were resuspended in 500 μl homogenization buffer. For coimmunoprecipitation experiments, chloroplasts corresponding to 300 μg protein were used. Outer envelope vesicles (OEVs) (50 μg total protein), isolated as described by Schleiff et al. (2003b), were incubated with 50 μCi γ-32P-ATP/10 μM ATP-mixture in 300 μl 20 mM Tricine-KOH, pH 7.6, 1 mM MgCl2, 0.5 mM MnCl2 for 15 min at RT, and subsequently centrifuged at 100 000 g for 10 min at 4 °C. The pellet was solubilized with 1.5% n-decylmaltoside in 150 μl 20 mM Tricine-KOH, pH 7.6, 150 mM NaCl (buffer A) for 5 min at RT. The sample was centrifuged again, and the supernatant diluted 10 times in buffer A. 750 μl samples were incubated for 1 h at RT under constant agitation with 15 μl of indicated antiserum, before addition of 50 μl Protein A-Sepharose (GE-Healthcare) saturated with 0.1% (w/v) BSA. After incubation for 1 h at RT the matrix was washed three times with 500 μl buffer A+0.2% n-decylmaltoside. Bound protein was eluted by boiling in SDS sample buffer for 2 min and subjected to SDS/PAGE followed by western blotting or autoradiography. OEV from pea (Schleiff et al., 2003a) or envelopes from A. thaliana (Gerdes et al., 2006) were solubilized by addition of 2% decylmaltoside. Phosphorylation of outer envelopes and complex isolation by sucrose density centrifugation was performed as described previously (Sveshnikova et al., 2000; Schleiff et al., 2003a).
Homology modelling
Superimposing the crystal structure of atToc33 onto the monomers of the homeodimer of psToc34 (Sun et al., 2002) with Yasara's SHEBA plugin (Jung and Lee, 2000) yielded the template for further modelling. The heterodimer of the G-domains of atToc159 and atToc33 was built with Modeller (Sali and Blundell, 1993) v8.1. The homology model of the heterodimer was further refined by loop modelling with Modeller (Fiser et al., 2000). An energy minimization of the resulting heterodimer model was performed in Yasara (www.yasara.org) with the Yamber3 force field (Krieger et al., 2004). Afterwards, a short MD simulation was performed in Yasara to further refine the interface of the complex.
Results
The Toc receptors are phosphorylated within their G-domains
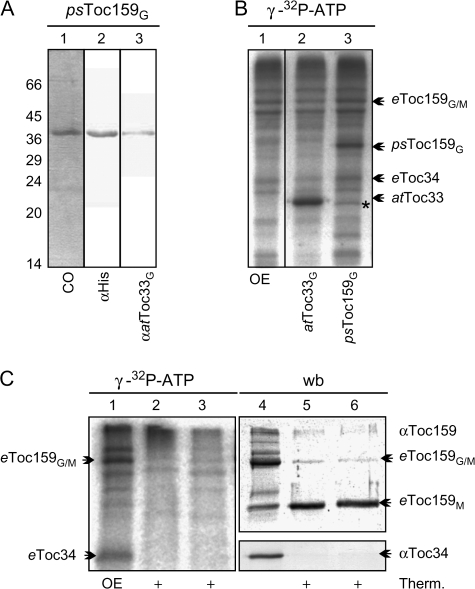
It was observed that both Toc receptors, Toc34 and Toc159, can be phosphorylated (Fulgosi and Soll, 2002) and the phosphorylation site of Toc34 was mapped within its G-domain (Jelic et al., 2002, 2003). However, it had not been clarified whether the tripartite protein Toc159 is phosphorylated in its G-domain as well. Therefore, the heterologously expressed G-domain of psToc159 (Fig. 1A, lanes 1–3) was incubated with isolated OEVs in the presence of γ-32P-ATP as the donor for phosphorylation, because kinase activity has been reported within this membrane (Fulgosi and Soll, 2002). As a positive control, the G-domain of recombinant atToc33 was used. Both proteins are efficiently phosphorylated (Fig. 1B, lanes 2, 3) suggesting that Toc159 might be phosphorylated within its G-domain. To confirm this observation with native proteins, the phosphoprotein pattern of the outer envelope membrane was analysed before and after thermolysin treatment, which is known to degrade the cytosolic A- and G-domains of psToc159 but leaving the membrane-anchored M-domain intact (Becker et al., 2004). Indeed, after proteolysis, as confirmed by the degradation of Toc34 (Fig. 1C, lanes 4–6, western blot) the M-domain of Toc159 remains detectable (Fig. 1C, lanes 5, 6), but is not phosphorylated (Fig. 1C, lanes 2, 3, autoradiogram). Therefore it can be concluded that psToc159, like psToc34, is phosphorylated within the G-domain at the cytosol-facing side of the chloroplast outer membrane.
Fig. 1.
The phosphorylation of Toc159. (A) The G-domain of psToc159 with a C-terminal His-tag was produced in E. coli, purified, subjected to SDS-PAGE, and stained with Coomassie Blue (lane 1) or immunodetected with antibodies against the C-terminal His-tag (lane 2) or against the G-domain of atToc33 (lane 3) because the antibodies against Toc159 present in our laboratory are raised against the M-domain. The migration of the molecular weight marker is indicated on the left. (B) Pea OEVs (lane 1) were incubated with γ-32P-ATP in the presence of atToc33G (lane 2) or psToc159G (lane 3). Proteins were subjected to SDS-PAGE and an autoradiogram is shown. The migration of the endogenous or the added receptor proteins is indicated. (C) Pea OEVs (lanes 1, 4) were phosphorylated as described (lanes 1–6) prior to proteolysis by thermolysine (lanes 2, 5) or thereafter (lanes 3, 6). Proteins were subjected to SDS-PAGE and vizualized by immunodetection with the indicated antibodies (lanes 4–6) or autoradiography (lanes 1–3).
Modification of the phosphorylation site influences the dimerization of Toc GTPases
In order to investigate the effects of phosphorylation, constructs were generated with a mutated phosphorylation site, whereby the serine 181 was substituted either with alanine or glutamate (Aronsson et al., 2006; Oreb et al., 2007). In a previous study, it was demonstrated that the S/E mutant exhibits a reduced capacity to bind and hydrolyse GTP (Oreb et al., 2007), thus resembling the properties of the phosphorylated protein.
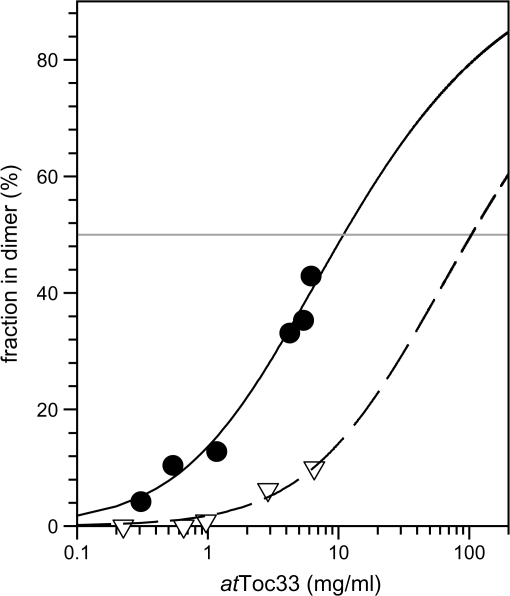
To investigate the effect of the S/E mutation on the dimerization, the concentration dependence of homodimerization was determined by size exclusion chromatography. It was possible to separate the monomeric and dimeric species and determined the dimeric fraction as described in the Materials and methods (Fig. 2). The apparent dissociation constants were calculated according to equation 1. For atToc33, an apparent dissociation constant of 0.4 mM was determined, while the dissociation constant for atToc33S/E is about one order of magnitude higher (Fig. 3). This result suggests that the phospho-mimicking mutant has a lower affinity for self association than the wild type.
Fig. 2.
Homodimerization of atToc33S/E. Indicated concentrations of GDP loaded atToc33 (circles, solid line) or atToc33S/E (triangles, dashed line) were subjected to gel filtration as described in the Materials and methods. The distribution of monomeric and dimeric species was analysed by fitting two Gaussian distributions. The dimer proportion is represented as a function of protein concentration. The apparent KD was calculated according to E1 and lines represent the least square fit analysis to this equation.
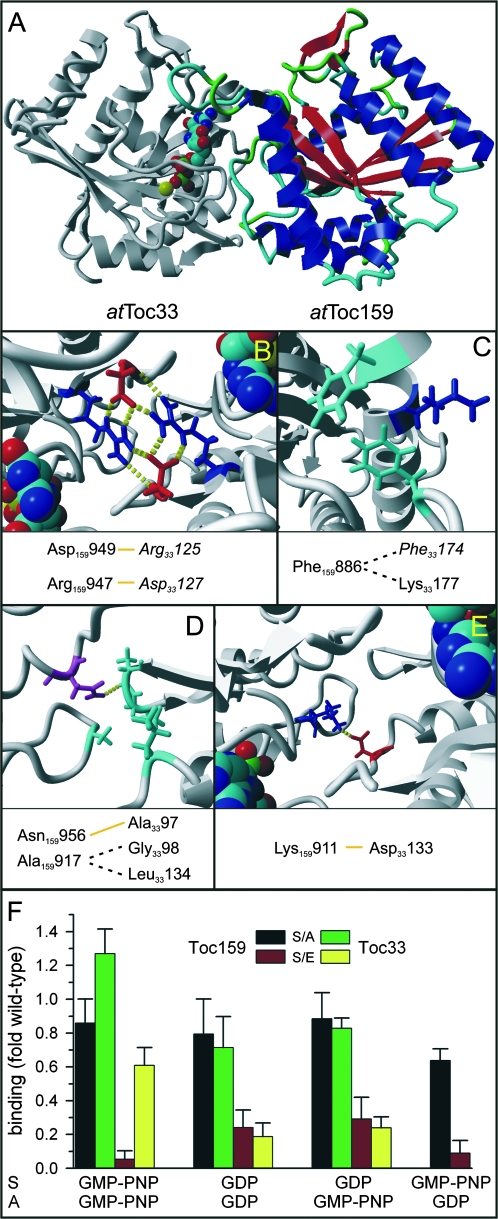
Fig. 3.
Phosphorylation influences heterodimerization of the G-domains of the Toc receptors in vitro. (A) The homology model of the heterodimer generated as described in the Materials and methods section is shown. The model of the G-domain of Toc159 is shown in colour, where structural elements are coloured differently. (B–E) The positions of amino acids previously identified to be involved in homodimerization (in italics) and additional amino acids of atToc33 putatively involved in interactions within the heterodimeric conformation are shown. Yellow lines indicate putative hydrogen bonds and dashed lines putative hydrophobic interactions. Molecular graphics were created with Yasara (www.yasara.org) and Povray (www.povray.org). (F) atToc33-His (for comparison), atToc33S/A-His (black and green bars) or atToc33S/E-His (red and yellow bars) loaded with GMPPNP or GDP were immobilized on nitrocellulose. The membrane was subsequently incubated with GST-atToc33 (green and yellow bars) or GST-psToc159G (black and red bars) loaded with GDP or GMPPNP (The combinations of nucleotides are indicated at the bottom, whereby S denotes the loading state of the spotted protein and A the loading state of the added soluble protein). The amount of bound protein was quantified as described in the Materials and methods and normalized to the ratio of wild-type receptor binding (atToc33/atToc33 or atToc33/psToc159). The average of at least six independent experiments including the three different concentrations is shown.
Given that both homo- and heterodimerization of Toc GTPases is thought to be functionally relevant and a similar interaction interface for both types of associations is possible (Fig. 3A–E; see Discussion), this property was further investigated by a recently described membrane binding assay (König et al., 2008). Here, His-tagged variants were immobilized on a nitrocellulose membrane and incubated with soluble GST-fused Toc proteins. The binding efficiency was subsequently analysed by immunodetection of the GST-moiety and densitometric quantification of the signals. This approach was chosen because it allows the effects of different nucleotide loading states and mutations in a homo- and heterodimeric context to be assessed in parallel.
The affinity of the S/A mutant for dimerization with wild type atToc33 is even slightly enhanced in comparison to the homodimerization of the wild-type receptor when both molecules are charged with GMPPNP (Fig. 3F, green bar, left), but only moderately, if at all, decreased when at least one dimerization partner is in the GDP loaded state (Fig. 3F, green bar, middle). This is also the case for the formation of the heterodimeric complex of the G-domains of Toc159 and atToc33S/A (Fig. 3F, black bar). A far more pronounced effect is observed with the S/E mutant, especially for heterodimerization when atToc33S/E is charged with GMPPNP (Fig. 3F, red bar, left and right). This finding documents that the phospho-mimic has a more drastic effect on heterodimerization than on homodimerization. Furthermore, heterodimerization is mostly affected when atToc33 is in the GMP-PNP state and homodimerization when one of the complex components is in the GDP state.
Phosphorylation modulates the assembly of atToc33 with the Toc core complex
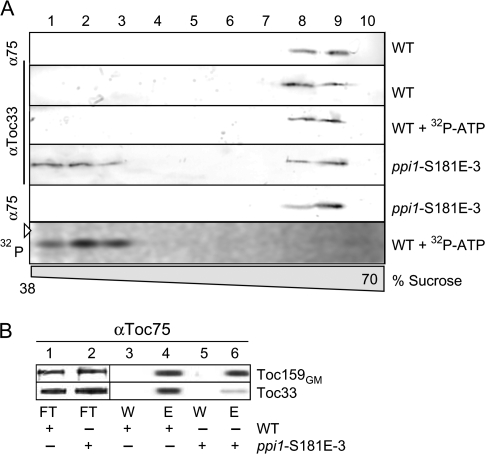
To transfer the in vitro findings into a physiological context, wild-type Arabidopsis plants and ppi1 plants complemented with atToc33S/E (line 3; Aronsson et al., 2006, Oreb et al., 2007) were analysed with respect to the association of atToc33 with the Toc complex. The chloroplasts envelope membranes were isolated, solubilized, subjected to sucrose density centrifugation, and the protein distribution determined by western-blotting using αToc75 and αToc33 antibodies (Fig. 4A). The Toc complex containing both Toc75 and Toc33 is present in high density fractions (Fig. 4A, lanes 8, 9). Fractionating envelopes from ppi1-S181E-3 plants reveals that a significant portion of atToc33S/E is not assembled with the complex and migrates at a lower density (Fig. 4F, lanes 1–3). This sedimentation profile is similar to the phosphorylated form in the wild-type background (compare wt+32P-ATP with western blots) and represents a receptor population not associated with the Toc complex. The data suggest that phosphorylated atToc33 is no longer assembled in the Toc complex in endogenous membranes.
Fig. 4.
Phosphorylation and phospho-mimicking of atToc33 influences the Toc complex integrity. (A) Envelope membranes from wild type (WT) or mutant A. thaliana (ppi1-S181E-3) were solubilized with decylmaltoside after incubation with 32P-ATP (WT+32P-ATP) adjusted with ATP to 1 μM (final) and subjected to a 25–70% sucrose density gradient centrifugation. Fractions of the density region from 38% to 70% were analysed using αToc33 or αToc75 (α75) antibodies. The last panel shows the autoradiogram of the atToc33 blot (αToc33/WT+32P-ATP). The triangle indicates the migration of the 36 kDa molecular weight standard. (B) atToc33 and atToc159 were coimmunoprecipitated using αToc75-antibodies from equivalent amounts of chloroplasts isolated from wt or ppi1-S181E-3 plants. For analysis, 4% of the flow through (FT), 4% of the wash (W), and 50% of the eluate (E) fractions were subjected to SDS–PAGE and immunodectection with αToc159 and αToc33 antibodies, respectively.
To confirm that atToc33S/E indeed has a lower affinity for the core complex, the Toc complex was coimmunoprecipitated by αToc75 antibodies after solubilization of isolated chloroplasts. atToc33, but not atToc33S/E, can efficiently be co-immunoprecipitated with atToc75 as determined by immunodecoration with αToc33 antibodies (Fig. 4B, lanes 2, 4). The reduction becomes most obvious when the precipitation efficiency between Toc159 and Toc33 is compared (Fig. 4B, upper panel versus lower panel). This confirms a specific effect of the S/E exchange and invalidates effects due to up-regulation of atToc33S/E in the transgenic plants (Oreb et al., 2007). While the in vitro data have shown a change in dimerization behaviour of atToc33S/E, these experiments now show that assembly of the native Toc core translocon is also impaired.
The effect of phosphorylation on complex assembly is independent of the positioning of the phosphorylation site of Toc34 in different species
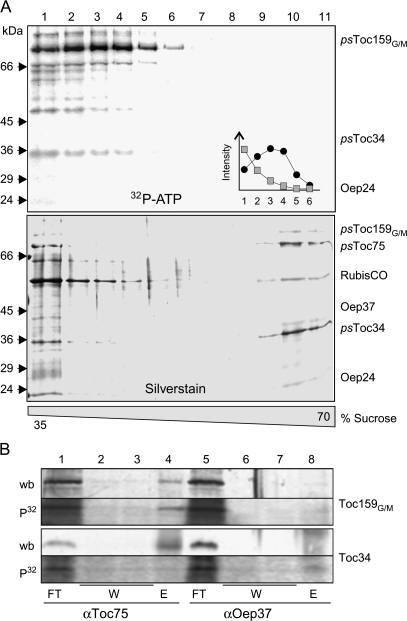
Phosphorylation not only regulates atToc33 (Jelic et al., 2003), but psToc34 is similarly regulated by this post-translational modification (Sveshnikova et al., 2000) even though the position of the phosphorylation sites differs (Jelic et al., 2002, 2003). Thus, it was important to generalize our observations regarding Toc complex assembly using Pisum as an additional system. Hence, complex assembly in purified OEVs from pea chloroplasts, preincubated with radioactive ATP, was analysed by sucrose density centrifugation (Fig. 5A). Again, the phosphorylated, radioactively labelled form of psToc159 and psToc34 is not detectable in high density fractions, containing the assembled Toc complex (Fig. 5A, bottom, silver-stained gel, fractions 9–11). In more detail, the phosphorylated form of psToc34 remains at the top of the gradient (Fig. 5A, fractions 1–4; inset grey squares), whereas phospho-psToc159 migrates further into the gradient (Fig. 5A, fractions 1–6; inset black circles). This could be explained by a remaining association with psToc75, and thus association of Toc complex components was examined by coimmunoprecipitation. Using psToc75-antobodies, both psToc159 and psToc34 can be efficiently coimmunoprecipitated (Fig. 5B, wb, lane 4). While psToc159, when phosphorylated, remains present in the complex, psToc34 is not associated with psToc75 (Fig. 5B, P32, lane 4). The specificity of this assay was controlled by immunoprecipitation using antibodies against the OE protein of 37 kDa, Oep37 (Fig. 5B, lanes 5–8; Schleiff et al., 2003b). This result shows that phosphorylation of the Toc components results in a soluble pool of the small GTPase Toc34.
Fig. 5.
Phosphorylation of psToc34 in OEVs influences the Toc complex integrity. (A) Pea OEVs were solubilized with decylmaltoside after incubation with 32P-ATP adjusted with ATP to 1 μM (final) and subjected to a 25–70% sucrose density gradient centrifugation. Fractions of the density region from 35–70% were collected and subjected to SDS-PAGE followed by silver staining (lower panel) and visualization of radioactivity (upper panel). The inset shows the distribution of phosphorylated psToc34 (squares) or psToc159G/M (circles). (B) Pea OEVs were phosphorylated as described in the Materials and methods followed by immunoprecipitation using αToc75 or αOep37 antibodies. Flow through (10%, FT), wash (10%, W), and precipitate fractions (50%, E) were analysed by western blots (wb) using αToc159 (Toc159GM) or αToc34 antibodies. Phosphorylation was visualized by autoradiography (P32).
Discussion
The significance of the phosphorylation of Toc receptors, even though it has been documented in vitro and in vivo (Sveshnikova et al., 2000; Jelic et al., 2002), is controversial (Aronsson et al., 2006; Oreb et al., 2007). This controversy has three reasons: physiological effects of a phospho-mimicking mutant can only be observed under specific conditions (Aronsson et al., 2006; Oreb et al., 2007), the envelope localized kinases (Fulgosi and Soll, 2002) still await their identification, and the molecular effect of phosphorylation in endogenous membranes remains unclear. In the present study, the last question was targeted and evidence provided that phosphorylation of the G-domains of the Toc receptors (Fig. 1) regulates the association of Toc34 with the Toc core complex. Two underlying mechanisms can be proposed. The first considers the fact that the Toc complex integrity in general (Becker et al., 2004), and the association between Toc34 and Toc159 in particular (Wallas et al., 2003), are dependent on the ability of Toc GTPases to bind GTP. It can be expected to influence all nucleotide-dependent properties, including the integrity of the Toc complex, since phosporylation inhibits binding of GTP. Contradicting such a conclusion is that atToc33S/E is still able to bind GTP (Oreb et al., 2007), but shows a reduced affinity for complex assembly as well. Furthermore, the strongest effect of the phospho-mimicking mutation on heterodimerization was observed when atToc33 is loaded with GMP-PNP, the non-hydrolysable analogue of GTP (Fig. 3).
The second possibility would be a direct modulation of protein–protein interactions by the bulky and negatively charged phosphate at the interface of interacting proteins. The observation that the atToc33 phospho-mimicry mutant has a lower affinity for heterodimerization with atToc159, even in the GDP-loaded state (Fig. 3), supports this notion. Furthermore, on the basis of psToc34 and atToc33 crystal structures (Sun et al., 2002; König et al., 2008) the phosphorylation site of atToc33 can be mapped to the homodimerization interface. In vitro, the ability of the phospho-mimicking atToc33S/E mutant to form homodimers is reduced (Figs 2, 3). Given the high similarity of G-domains of atToc33 and atToc159, the interface for homodimerization can also serve as an interaction site for heterodimerization (Fig. 3). This hypothesis is supported by the observation that in vitro-translated atToc33, which can be assumed to be exclusively monomeric due to its low concentration in the translation mixture, was shown to bind to Toc159 (Smith et al., 2002). Analysing the amino acids assigned as interacting in the homodimer (Sun et al., 2002) in the model of the heterodimer showed that many of these interactions are conserved. For instance, the network of electrostatic interactions by Arg33125 and Asp33127 can be identified (Fig. 3B), as well as the van der Waals interaction of Phe33174 (Fig. 3C), Phe3367 (not shown, with putative interactions with Phe159989, Gln1591001, and Ile1591001) or of Gly3398 and Leu33134 (Fig. 3D). In addition, some new possible contacts could be allocated (Fig. 3C–E), possibly compensating the loss of the interaction of the network involving Arg33130 (Sun et al., 2002), which is not conserved in Toc159. Hence, it can be speculated that (at least in the Arabidopsis system) the phosphate group sterically inhibits the contacts between the G-domains even in the heterodimeric context. This interpretation, however, can not be directly applied to the Pisum orthologues, because the phosphorylation site of psToc34 is distant from the dimerization interface. Hence, in pea, another interaction, for example with the cytosolic domain of the translocation channel Toc75 (Ertel et al., 2005), might be affected. In turn, it is possible that the phosphorylation of the Toc159 G-domain (Fig. 1) disturbs the contacts with Toc34 in pea. The precise determination of the phosphorylation site in Toc159 and the physical mapping of the interaction network within the Toc complex will therefore be necessary to understand the regulation of the system in pea.
In general, regulation of protein complexes by phosphorylation is a widespread regulatory principle, even in protein translocation (Wang and Johnsson, 2005; Glavy et al., 2007). An extensively studied example is the regulation of the light-harvesting complex protein II (LHCPII) by phosphorylation when the activity of photosystem I becomes limiting (Allen, 2005). The phosphorylated LHCPII dissociates from photosystem II and assembles with the photosystem I. The subsequent oxidation of the plastoquinone pool inactivates the kinase and activates the phosphatase, restoring the interaction of LHCPII with photosystem II. Thereby, this cycle fine-tunes the activity of a membrane complex. A similar mechanism can be envisioned for the Toc translocon to switch between different functionalities as proposed and to enable the exchange of receptors. However, our results and the structural context with the phosphorylation site being localized on the dimerization interface suggests that phosphorylation does not serve as a signal for dissociation of the receptor, but rather as an inhibitor of association. Finally, the question about the physiological function of the regulatory circuit of Toc34 will have to be answered, a question which will require the identification and analysis of the Toc-phosphorylating kinase(s).
Acknowledgments
We thank Ivo Tews and Patrick König (Heidelberg, Germany) for critical discussions and Maike Ruprecht (Frankfurt, Germany) for her technical assistance. We thank Paul Jarvis, John Combe (Leicester, UK) and Henrik Aronsson (Gotemborg, Sweden) for providing the transgenic lines. This work was supported by grants to ES from the Volkswagenstiftung and from the DFG (SFB594-B11).
Glossary
Abbreviations
- at/psToc33/34
the homologous GTPases atToc33 from Arabidopsis thaliana and psToc34 from Pisum sativum that have been studied. The first two italicized letters indicate source organism, followed by the GTPase name. Toc34 without denominator refers to both, atToc33 and psToc34
- OE(V)
outer envelope (vesicle)
- ppi1
plastid protein import mutant 1
- LHCPII
light harvesting complex protein 2
References
- Allen JF. Photosynthesis: the processing of redox signals in chloroplasts. Current Biology. 2005;15:R929–R932. doi: 10.1016/j.cub.2005.10.061. [DOI] [PubMed] [Google Scholar]
- Aronsson H, Combe J, Patel R, Jarvis P. In vivo assessment of the significance of phosphorylation of the Arabidopsis chloroplast protein import receptor, atToc33. FEBS Letters. 2006;580:649–655. doi: 10.1016/j.febslet.2005.12.055. [DOI] [PubMed] [Google Scholar]
- Becker T, Jelic M, Vojta A, Radunz A, Soll J, Schleiff E. Preprotein recognition by the Toc complex. EMBO Journal. 2004;23:520–530. doi: 10.1038/sj.emboj.7600089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Schnell DJ. Insertion of the 34 kDa chloroplast protein import component, IAP34, into the chloroplast outer membrane is dependent on its intrinsic GTP-binding capacity. Journal of Biological Chemistry. 1997;272:6614–6620. doi: 10.1074/jbc.272.10.6614. [DOI] [PubMed] [Google Scholar]
- Chen K, Chen X, Schnell DJ. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiology. 2000;122:813–822. doi: 10.1104/pp.122.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D, Patel R, Keegstra K, Jarvis P. An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. The Plant Journal. 2004;38:93–106. doi: 10.1111/j.1365-313X.2004.02024.x. [DOI] [PubMed] [Google Scholar]
- Ertel F, Mirus O, Bredemeier R, Moslavac S, Becker T, Schleiff E. The evolutionarily related beta-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. Journal of Biological Chemistry. 2005;280:28281–28289. doi: 10.1074/jbc.M503035200. [DOI] [PubMed] [Google Scholar]
- Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Science. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulgosi H, Soll J. The chloroplast protein import receptors Toc34 and Toc159 are phosphorylated by distinct protein kinases. Journal of Biological Chemistry. 2002;277:8934–8940. doi: 10.1074/jbc.M110679200. [DOI] [PubMed] [Google Scholar]
- Gerdes L, Bals T, Klostermann E, Karl M, Philippar K, Hünken M, Soll J, Schünemann D. A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. Journal of Biological Chemistry. 2006;281:16632–16642. doi: 10.1074/jbc.M513623200. [DOI] [PubMed] [Google Scholar]
- Glavy JS, Krutchinsky AN, Cristea IM, Berke IC, Boehmer T, Blobel G, Chait BT. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proceedings of the National Academy of Sciences, USA. 2007;104:3811–2816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutensohn M, Fan E, Frielingsdorf S, Hanner P, Hou B, Hust B, Klösgen RB. Toc, Tic, Tat et al.: structure and function of protein transport machineries in chloroplasts. Journal of Plant Physiology. 2006;163:333–347. doi: 10.1016/j.jplph.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Ivanova Y, Smith MD, Chen K, Schnell DJ. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Molecular Biology of the Cell. 2004;15:3379–3379. doi: 10.1091/mbc.E03-12-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Jackson-Constan D, Keegstra K. Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiology. 2001;125:1567–1576. doi: 10.1104/pp.125.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelic M, Soll J, Schleiff E. Two Toc34 homologues with different properties. Biochemistry. 2003;42:5906–5916. doi: 10.1021/bi034001q. [DOI] [PubMed] [Google Scholar]
- Jelic M, Sveshnikova N, Motzkus M, Hörth P, Soll J, Schleiff E. The chloroplast import receptor Toc34 functions as preprotein regulated GTPase. Biological Chemistry. 2002;383:1875–1883. doi: 10.1515/BC.2002.211. [DOI] [PubMed] [Google Scholar]
- Jung J, Lee B. Protein structure alignment using environmental profiles. Protein Engineering. 2000;13:535–543. doi: 10.1093/protein/13.8.535. [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell DJ. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 2006;7:248–257. doi: 10.1111/j.1600-0854.2005.00382.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Hirohashi T, Nakai M. Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant and Cell Physiology. 2006;47:363–371. doi: 10.1093/pcp/pcj002. [DOI] [PubMed] [Google Scholar]
- König P, Oreb M, Höfle A, Kaltofen S, Rippe K, Sinning I, Schleiff E, Tews I. The GTPase cycle of the chloroplast import receptor Toc34: implications from monomeric and dimeric structures. Structure. 2008 doi: 10.1016/j.str.2008.01.008. (in press) [DOI] [PubMed] [Google Scholar]
- Krieger E, Darden T, Nabuurs S, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force field parameterization in crystal space. Proteins. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E. Plastid biogenesis, between light and shadows. Journal of Experimental Botany. 2007;58:11–26. doi: 10.1093/jxb/erl196. [DOI] [PubMed] [Google Scholar]
- Oreb M, Reger K, Schleiff E. Chloroplast protein import: reverse genetic approaches. Current Genome. 2006;7:235–244. [Google Scholar]
- Oreb M, Tews I, Schleiff E. Policing Tic ‘n’ Toc, the doorway to chloroplasts. Trends in Cell Biology. 2008 doi: 10.1016/j.tcb.2007.10.002. (in press) [DOI] [PubMed] [Google Scholar]
- Oreb M, Zoryan M, Vojta A, Maier UG, Eichacker LA, Schleiff E. Phospho-mimicry mutant of atToc33 affects early development of Arabidopsis thaliana. FEBS Letters. 2007;581:5945–5951. doi: 10.1016/j.febslet.2007.11.071. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Eichacker LA, Eckart K, Becker T, Mirus O, Stahl T, Soll J. Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Science. 2003b;12:748–759. doi: 10.1110/ps.0237503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J, Küchler M, Kühlbrandt W, Harrer R. Characterization of the translocon of the outer envelope of chloroplasts. Journal of Cell Biology. 2003a;160:541–551. doi: 10.1083/jcb.200210060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Hiltbrunner A, Kessler F, Schnell DJ. The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. Journal of Cell Biology. 2002;159:833–843. doi: 10.1083/jcb.200208017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YJ, Forouhar F, Li HM, Tu SL, Yeh YH, Kao S, Shr KL, Chou CC, Chen C, Hsiao CD. Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nature Structural Biology. 2002;9:95–100. doi: 10.1038/nsb744. [DOI] [PubMed] [Google Scholar]
- Sveshnikova N, Soll J, Schleiff E. Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proceedings of the National Academy of Sciences, USA. 2000;97:4973–4978. doi: 10.1073/pnas.080491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallas TR, Smith MD, Sanchez-Nieto S, Schnell DJ. The roles of toc34 and toc75 in targeting the toc159 preprotein receptor to chloroplasts. Journal of Biological Chemistry. 2003;278:44289–44297. doi: 10.1074/jbc.M307873200. [DOI] [PubMed] [Google Scholar]
- Wang X, Johnsson N. Protein kinase CK2 phosphorylates Sec63p to stimulate the assembly of the endoplasmic reticulum protein translocation apparatus. Journal of Cell Science. 2005;118:723–732. doi: 10.1242/jcs.01671. [DOI] [PubMed] [Google Scholar]