Abstract
The lipophilic biopolyester suberin forms important boundaries to protect the plant from its surrounding environment or to separate different tissues within the plant. In roots, suberin can be found in the cell walls of the endodermis and the hypodermis or periderm. Apoplastic barriers composed of suberin accomplish the challenge to restrict water and nutrient loss and prevent the invasion of pathogens. Despite the physiological importance of suberin and the knowledge of the suberin composition of many plants, very little is known about its biosynthesis and the genes involved. Here, a detailed analysis of the Arabidopsis aliphatic suberin in roots at different developmental stages is presented. This study demonstrates some variability in suberin amount and composition along the root axis and indicates the importance of ω-hydroxylation for suberin biosynthesis. Using reverse genetics, the cytochrome P450 fatty acid ω-hydroxylase CYP86A1 (At5g58860) has been identified as a key enzyme for aliphatic root suberin biosynthesis in Arabidopsis. The corresponding horst mutants show a substantial reduction in ω-hydroxyacids with a chain length <C20, demonstrating that CYP86A1 functions as a hydroxylase of root suberized tissue. Detailed expression studies revealed a strong root specificity and a localized expression in the root endodermis. Transgenic expression of CYP86A1 fused to GFP distributed CYP86A1 to the endoplasmic reticulum, indicating that suberin monomer biosynthesis takes place in this sub-cellular compartment before intermediates are exported in the apoplast.
Keywords: Cell wall, cytochrome P450, root, suberin, ω-hydroxyacids
Introduction
A fundamental development for plants to colonize the terrestrial habitat was the evolution of protective surface tissues deposited in the cell walls, the polyesters cutin and suberin. These aliphatic biopolymers prevent the uncontrolled loss of water and nutrients from plants and infection by pathogens. Cutin covers all aerial parts of the plant in a primary developmental stage, whereas suberin depositions are more variable. Suberin can be found in the aerial part of a plant in the cell walls of bark tissues, bundle sheath cells of grasses, conifer needles, and seeds (reviewed in Kolattukudy, 1981, 2001; Bernards, 2002). Subterrestrial suberin is located in the cell walls of the root endodermis and periderm of all angiosperm species, including Arabidopsis (Ma and Peterson, 2003; Franke et al., 2005) and in rhizodermal and hypodermal cell walls of primary roots (Perumalla et al., 1990). For periderms of many species, it was shown that the apoplastic deposition of suberin acts as a barrier that strongly reduces the movement of water and dissolved nutrients and ions (Schönherr and Ziegler, 1980; Vogt et al., 1983; Groh et al., 2002). Similarly, suberized root cell walls prevent the movement of water and ions. A good example is the suberized Casparian strip in the endodermis of the root (Schreiber et al., 1994) that enables this inner plant tissue to control water and ion uptake into the stele (Sattelmacher et al., 1998; Ma and Peterson, 2003). When suberin is formed in outer tissues like the periderm of the root, it also acts as an antimicrobial barrier against pathogens (Lulai and Corsini, 1998). Additional suberin depositions can occur in response to wounding and pathogen attacks (Agrios, 1997) and in response to unfavourable environmental conditions such as drought and salt stress (North and Nobel, 1994; Reinhardt and Rost, 1995; Steudle and Peterson, 1998). This increased suberization, and similarly the induction of a suberized exodermis in aeroponically grown corn roots, led to an increased resistance to water movement in roots (Zimmermann et al., 2000). This indicated that ‘root hydraulics’ can be modulated by the deposition of the aliphatic polymer suberin.
To understand the property of suberin to act as an apoplastic barrier in multiple ways, the composition of suberin must be considered. The macromolecule suberin is composed of a polyaromatic domain and a polyaliphatic domain (Kolattukudy, 2001; Bernards, 2002). The polyaromatic domain, which is derived from the phenylpropanoid pathway, is restricted to the primary cell wall and is covalently attached to cell wall carbohydrate units. The polyaliphatic domain is a three-dimensional polyester network mainly composed of oxygenated long-chain fatty acids, partially cross-linked by glycerol and with integrated aromatic compounds. The aliphatic domain is supposed to be the main reason for the physiological important water-sealing properties of suberin and was therefore the focus of the present studies. Aliphatic compounds in Arabidopsis root suberin are mainly ω-hydroxyacids and α,ω-diacids of carbon chain-length C16–C24, with lower amounts of very long-chain fatty acids (VLCFA) (C16–C24), primary alcohols (C16–C20), and 2-hydroxyacid (C24) (Franke et al., 2005; Beisson et al., 2007) comparable to the suberin composition of other species (Holloway, 1983; Matzke and Riederer, 1991, Schreiber et al., 1999).
Based on carbon flux studies in wound-healing potato tuber tissue (Yang and Bernards, 2006) and the chemical composition of suberin from Arabidopsis and other species (Franke and Schreiber, 2007) two main metabolic pathways have been suggested to be characteristic for aliphatic suberin biosynthesis. One is the elongation of C16 and C18 fatty acid precursors to C20–C32 VLCFA (in Arabidopsis up to C24); the other pathway consists of ω-oxygenation reactions needed for the transformation into fatty acid derivatives such as ω-hydroxyacids and α,ω-diacids. Fatty acid elongation (FAE) is a four-step reaction sequence involving endoplasmic reticulum (ER)-localized multienzyme complexes that are also required in the production of VLCFA for seed storage lipids (James et al., 1995), waxes (Millar et al., 1999; Todd et al., 1999), and membrane lipids (Zheng et al., 2005). By contrast, fatty acid ω-oxygenation in plants has mostly been implicated in cell wall polyester formation, such as cutin synthesis, although hydroxy fatty acids have recently been reported to be involved in fertilization and signalling processes (Kandel et al., 2006).
The oxygenation of fatty acids can be performed by NADPH-dependent cytochrome-P450 monooxygenases (P450), catalysing the insertion of one of the atoms from molecular oxygen into a substrate (Werck-Reichhart and Feyereisen, 2000). In plants this has been demonstrated in biochemical studies showing that P450-containing microsomes from Vicia sativa and Pisum are capable of catalysing the formation of ω-hydroxyacids (Soliday and Kolattukudy, 1977; Benveniste et al., 1982; Pinot et al., 1992, 1993). Subsequent molecular approaches led to the isolation and cloning of CYP94A1 from Vicia sativa the first characterized plant ω-hydroxylase (Tijet et al., 1998). Shortly after, CYP86A1, cloned from Arabidopsis, was found to catalyse the ω-hydroxylation of saturated and unsaturated fatty acids in microsomal preparations from yeast, heterologously expressing the encoded P450 (Benveniste et al., 1998). Furthermore CYP86A8 from Arabidopsis and several CYP94 family members were biochemically characterized as ω-hydroxylases (Wellesen et al., 2001; Benveniste et al., 2006), leading to the ontological classification of P450 genes from the CYP86 clan, especially the CYP86 and CYP94 subfamilies, as fatty acid ω-hydroxylases.
The most prominent substance class in Arabidopsis aliphatic root suberin are ω-hydroxyacids. Although to date many P450 ω-hydroxylases have been functionally characterized, the biological process they act in, is mostly unknown and no P450 gene that is directly involved in the synthesis of these predominant suberin monomers could be identified. Instead, the recent characterization of mutants in a putative glycerol-3-phosphate acyltransferase (GPAT5) identified the first gene involved in suberin biosynthesis, probably involved in the formation of fatty acid-containing acylglycerol precursors for suberin deposition (Beisson et al., 2007). In the past three decades, fatty acid ω-hydroxylases have mostly been implicated in cutin biosynthesis. This was supported by the isolation of Arabidopsis mutants in CYP86A2 and CYP86A8, both characterized by defects in cuticle formation (Wellesen et al., 2001; Xiao et al., 2004). Similar to CYP86A1, fatty acid ω-hydroxylase activity was demonstrated biochemically for CYP86A8, whereas the involvement of CYP86A2 in ω-hydroxylation was indicated by the strong reduction in ω-oxygenated fatty acids in stem cutin of corresponding mutants. Based on these investigations, members of the CYP86A subfamily are also potential candidates for enzymes involved in the ω-hydroxylation of aliphatic suberin monomers. This is supported by P450-specific expression studies revealing root expression for some of the members of the CYP86A subfamily. Of these, CYP86A1 is specifically expressed in roots (Duan and Schuler, 2005). In addition, transcriptom analysis of developing phelloderm in the suberin model Quercus suber identified a P450 with closest similarity to the Arabidopsis homologue CYP86A1 and was suggested to be involved in suberin formation (Soler et al., 2007). Altogether, CYP86A1 seemed to be a good candidate for an involvement in suberin biosynthesis.
Here the characterization of T-DNA insertion mutants in the Arabidopsis CYP86A1 gene is reported. The in vivo role of CYP86A1 as a hydroxylase of root suberized tissue (HORST) is demonstrated. The horst mutants exhibit a highly reduced aliphatic root suberin. Furthermore the monomer composition of the aliphatic root suberin is altered in a way that is consistent with the biochemical in vitro characterization of CYP86A1 as an ω-hydroxylase.
Materials and methods
Plant material and growth conditions
All plants used are in the Arabidopsis thaliana L. Heynh. ecotype Columbia background. Plants were either cultivated in Floradur potting mix (Floraguard, Germany), or on agar sucrose medium [1.5% (w/v) sucrose, 0.7% (w/v) agar agar] containing MS salts (Murashige and Skoog, 1962) adjusted to pH 5.7 using KOH. When grown on agar sucrose medium, seeds where surface-sterilized by shaking for 15 min in sterilization-solution [50% (v/v) EtOH, 1% (v/v) NaHClO], rinsed three times with 100% EtOH, and dried. Plants were grown in a growth chamber at 22 °C with a photoperiod of 16 h/8 h light/dark cycle and 100 μE m−2 s−1 light intensity.
Mutant isolation
The T-DNA insertion mutant lines SALK_107454 and SALK_104083 (Alonso et al., 2003) were obtained from the Nottingham Arabidopsis Stock Centre. For the genotype screening to isolate homozygous mutant alleles, the T-DNA left border primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) has been used together with the gene-specific primers 5′-AAGAACCAGCTCAAGGCCACC-3′ (forward) and 5′-AGCAAAAAGCCTAAACCGGGA-3′ (reverse) for line SALK_107454 and 5′-AACGAGTTTCTTGAGCCTCAAG-3′ (forward) and 5′-ACCAGGATTTCAAATACGTCG-3′ (reverse) for line SALK_104083. Homozygous T-DNA insertion mutants were verified by RT-PCR analysis (see below) of 10-d-old seedlings cultivated on MS-agar and examined for the presence or absence of a gene transcript of CYP86A1 (At5g58860).
Generation of plant transformation vectors and transgenic Arabidopsis
For cloning and generation of expression constructs, the Gateway® Cloning Technology (Hartley et al., 2000) was used according to the manufacture's suggestions (Invitrogen, Karlsruhe, Germany). After PCR amplification of the desired fragment, the amplicon was cloned by homologous recombination using Gateway® BP Clonase™ II Enzyme Mix and the pDONR/Zeo entry vector (Invitrogen). The generated entry clones were recombined with selected binary destination vectors (pMDC) provided by Curtis and Grossniklaus (2003). The correctness of all vectors was confirmed by sequencing. For cloning of the putative CYP86A1-promoter, the 1426 bp upstream region of At5g58860 was amplified from genomic DNA using the Gateway-compatible primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACGTGTTGATTATGTTGATGATGCTGAG-3′ (forward; LS248) and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGTTTAGGCTTTTTGCTTTTGTTCTGT-3′ (reverse) and inserted in the pDONR/Zeo entry vector yielding plasmid p176. LR recombination of p176 with destination vector pMDC162 yielded the promoter reporter gene (β-glucoronidase; GUS) fusion PromCYP86A1:GUS for expression studies. Similarly, the coding sequence of At5g58860 was amplified from cDNA with the primer pair 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACAGAACAAAAGCAAAAAGCCTAAACC-3′ (forward) and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGCAAGCACCTCACCACGAG-3′ (reverse; LS298) to generate the entry clone p161. The C-terminal fusion to GFP (CYP86A1:GFP) was obtained by recombination of p161 with destination vector pMCD84. The 3.4 kb genomic DNA fragment for complementation analysis was generated by PCR using primers LS248 and LS298 amplifying the promoter of At5g58860 together with the genomic sequence. After cloning in entry vector p166, the genomic fragment was inserted in pMDC99 for mutant complementation. Transgenic plants were generated by introduction of the plant expression constructs into Agrobacterium tumefaciens (strain GV3101) and subsequent floral dip transformation as described previously (Clough and Bent, 1998).
Semi-quantitative RT-PCR analysis
The RNA for organ-specific expression studies was extracted from 5-week-old soil-grown Arabidopsis plants using hot phenol according to De Vries et al. (1988). For expression studies confirming the knock-out or complementation, the total RNA was isolated from 10-d-old seedlings with TRI-Reagent (Molecular Research Center, Cincinnati, OH, USA) according to the instructions of the manufacturer. The RT-PCR was performed using the SuperScript™ III One-Step RT-PCR System with Platinum® Taq DNA Polymerase (Invitrogen) with each reaction containing 200 ng RNA. ACTIN was used as a control. The primer sequences and predicted amplicon sizes were 5′-ACAGAACAAAAGCAAAAAGCCTAAACC-3′ (forward) and 5′-TGCAAGCACCTCACCACGAG-3′ (reverse) for CYP86A1 (1572 bp) and 5′-GTGATGATGCCCCGAGAGC-3′ (forward) and 5′-GACCCGCAAGATCAAGACGA-3′ (reverse) for ACTIN (At5g09810) (480 bp). RT-PCR conditions were 30 min at 50 °C followed by 25–40 cycles of 30 s at 94 °C, 30 s at 57 °C and 90 s at 68 °C, and finally 5 min at 68 °C.
GUS-staining procedure
Transgenic T1 and T2 plants containing the PromCYP86A1:GUS construct were selected on agar sucrose medium containing hygromycin (50 μg ml−1). Hygromycin-resistant seedlings were either transferred to Floradur potting mix (Floraguard) and grown to maturity, or directly stained. All samples were stained according to De Block and Debrouwer (1992) with 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc) (1 mM) in staining buffer solution [10−1 M sodium phosphate buffer (pH 7.0), 10−2 M EDTA, 0.5 mM FeK3(CN)6, 0.1% (v/v) Triton X-100] at 37 °C for 2–18 h. To stop staining and remove chlorophyll in above-ground organs, samples were transferred to 70% ethanol with two or three changes. Specimens were examined using an Olympus SZ61 binocular (Olympus, Japan) or fixed for light microscopy.
Transient expression of GFP-fusions in Nicotiana benthamiana
Agrobacterium tumefaciens strain GV3101 containing the CYP86A1:GFP construct under the control of the CaMV35S-promoter (35S) were grown in 2 ml LB medium containing the appropriate antibiotics. Following overnight growth at 28 °C, bacteria were pelleted from 500 μl culture by centrifugation and resuspended in 2 ml infiltration medium (20 mM citric acid, 2% sucrose, 200 μM Acetosyringon; adjusted to pH 5.2) according to Wydro et al. (2006) with minor modifications. Bacteria were resuspended in this medium. Cultures were incubated at room temperature for 3 h before infiltration. Bacterial suspensions were infiltrated into young but fully expanded leaves of 6- to 8-week-old N. benthamiana plants using a needle-less syringe. After infiltration, plants were placed at 20 °C with a photoperiod of 16 h/8 h light/dark. After 40–48 h, GFP fluorescence was monitored by confocal laser scanning microscopy. As a positive control for co-localization studies, a similar 35S-driven fusion construct, HDEL:DsRED, possesing the ER resident protein retention signal HDEL (Haseloff et al., 1997) fused to the Discosoma sp. red fluorescing protein (DsRed; Jach et al., 2001) was co-infiltrated in N. benthamiana leaves.
Transfected N. benthamiana leaves were examined with an Olympus FV 1000 confocal microscope (Olympus, Germany) using a ×63 water-immersion objective. GFP fluorescence was imaged using excitation with the 488 nm line of an argon/krypton laser and a 530 nm band pass emission filter. Serial confocal optical sections were taken at different step sizes. Projections of serial confocal sections were done using Olympus FV100 software.
Light microscopic and histochemical techniques
Whole root bundles or root parts were washed with water and fixed in a mixture of formaldehyde (3.7%; v/v) in phosphate buffer saline (10−2 M NaPO4, 0.137 M NaCl, 27×10−4 M KCl, adjusted to pH 7.4) for at least 24 h. The specimens were embedded in frozen section medium (Neg-50; Richard-Allan Scientific, USA) and frozen at –25 °C. Sections (20 μm thick) were prepared using a cryo microtome (Cryostat H 500). Sections were transferred to microscope slides, embedded in glycerol:water (1:1; v/v) and examined with an Axioplan microscope (Zeiss, Germany) with bright field illumination. Sudan staining was performed according to Brundrett et al. (1991) with Sudan red 7B (0.1%; w/v) in 50% (v/v) PEG-400, 45% (v/v) glycerol, 5% (v/v) H2O at room temperature for 1 h. After staining, sections were rinsed with 1% SDS, washed thoroughly with water and mounted in glycerol:water (1:1; v/v) before microscopic examination.
Analytical techniques
Isolation and depolymerization of suberized root cell wall material:
As described previously (Franke et al., 2005) one to five roots of 5-week-old plants were carefully dug out from the potting mix, washed with water and incubated in polysaccharide hydrolases [1% v/v cellulase (Celluclast, Novo Nordisc, Denmark), 1% v/v pectinase (Trenolin, Germany), 10−3 M NaN3 in 10−2 M citric buffer pH 3] for 2 weeks with one exchange of enzyme solution. The remaining cell wall materials were washed with borate buffer (10−2 M, pH 9) and distilled water. Finally samples were copiously extracted with chloroform:methanol (1:1; v/v), dried, weighed, and used for depolymerization reaction. The samples were depolymerized by transesterification with 2 ml 1 M MeOH/HCl (Supelco, USA) for 2 h at 80 °C. After addition of 2 ml saturated NaCl/H2O and 10 μg dotriacontane as internal standard, aliphatic monomers were gradually extracted (three times with 1 ml) in hexane. The combined organic phase was evaporated in a steam of nitrogen to a volume of ∼100 μl before analysis using gas-liquid chromatography (GC) and mass spectrometry (MS).
GC and GC-MS analysis:
All samples were treated with bis-(N,N,-trimethylsilyl)-tri-fluoroacetamide (BSTFA; Macherey-Nagel, Germany) for 40 min at 70 °C to convert free hydroxyl and carboxyl groups into their corresponding trimethylsilyl (TMS) ethers and esters. Monomers were identified on the basis of their electron-impact MS spectra (70 eV, m/z 50–700) after capillary GC (DB-1, 30 m, 0.32 mm, 0.1 μm) on an Agilent 6890N gas chromatograph combined with an Agilent 5973N quadrupole mass-selective detector (Agilent Technologies, Germany). The depolymerization products were separated by on-column-injection at 50 °C, 2 min at 50 °C, 10 °C/min to 150 °C, 1 min at 150 °C, 3 °C/min to 310 °C, 15 min at 310 °C. Quantitative determination of the components was carried out with an identical GC system coupled with a flame ionization detector based on the internal standard. All analyses are presented as means ±standard deviation of three to five replicates.
Leaf wax extraction and polyester analysis from leaf and seed coat
Leaf wax and leaf polyester analysis was basically performed as described by Kurdyukov et al. (2006). For wax extraction, 6–10 rosette leaves from 35-d-old plants were harvested, individually dipped in chloroform for 10 s and scanned to determine the leaf surface area using imaging software. The chloroform extract was spiked with 10 μg of tetracosan (Fluka, Germany) as internal standard and evaporated under a steam of nitrogen to a volume of ∼100 μl prior to GC and GC-MS analysis. Following BSTFA derivatization, wax extracts were separated by the GC system described above applying the following temperature gradient: on-column-injection at 50 °C, 2 min at 50 °C, 40 °C/min to 200 °C, 2 min at 200 °C, 3 °C/min to 310 °C, 30 min at 310 °C.
Leaf polyesters were analysed from totally extracted, delipidated leaves. After collecting and scanning 15–20 rosette leaves from 5-week-old plants, leaves were completely extracted in chloroform:methanol (1:1; v/v). The solvent was changed five times within 2 weeks. Samples were dried and used for depolymerization reaction as outlined above (Franke et al., 2005).
Adapted from the protocols of Molina et al. (2006) for seed coat polyester analysis, at least 15 μg of fully developed and dried seeds were crushed in liquid nitrogen using a mortar and pestle. The resulting powder was exhaustively extracted in chloroform:methanol (1:1; v/v) for 2 weeks with five changes of solvent. After drying, the residue was used for depolymerization reaction.
Results
ω-Hydroxyacid suberin monomers increase during root development
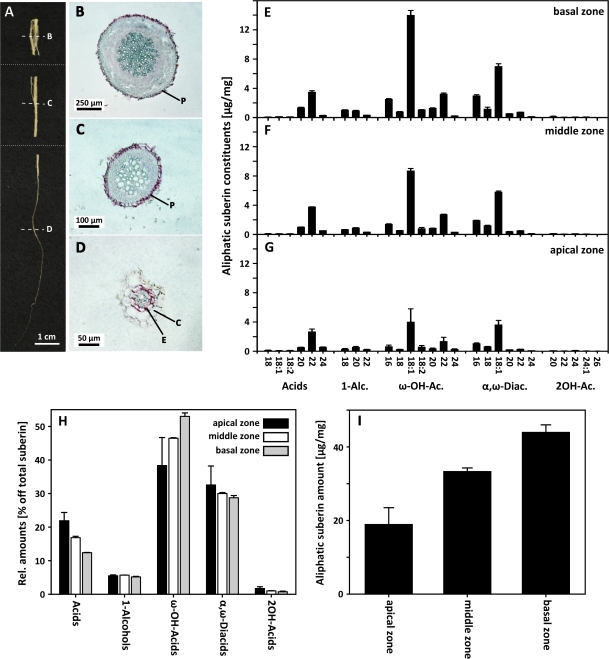
To gain more insights into the biochemical reactions involved in the formation of aliphatic root suberin, suberin compositional changes in the course of root development were studied in more detail. Roots of 5-week-old, fully developed Arabidopsis plants were dissected in three zones along the root axis for histochemical and chemical analysis (Fig. 1A). The basal zone ranged from the root base 15 mm towards the tip. The middle zone contained the tissue of the next consecutive 25 mm and the apical root zone is represented by the remaining distal tissue including the root tip. Representative cross-sections were microscopically examined and histochemically analysed using Sudan 7B, a lipophilic dye commonly used to stain suberin depositions. The basal root zone is mainly represented by roots in a secondary developmental stage, characterized by suberin depositions in the cell walls of the outermost peridermal cell layer (Fig. 1B). Similar suberized periderms were observed in cross-sections from the middle root zone (Fig. 1C). The apical root zone was dominated by roots in a primary developmental stage characterized by the presence of an endodermis with apoplastic suberin depositions (Fig. 1D).
Fig. 1.
Tissue distribution and suberin composition in different developmental stages of the Arabidopsis root. (A) Arabidopsis root sectioned in the three zones investigated. The positions of the cross-sections from B, C, and D are indicated by dashed lines. (B–D) Bright field microscopic picture of representative cross-sections of the Arabidopsis root stained with the lipophilic dye Sudan 7B. The red-stained suberin is deposited in the periderm of the roots in a secondary developmental stage (B, C) or in the endodermal cell walls of the roots in a primary developmental stage (D). (E–I) Aliphatic suberin composition and amount in the different developmental stages of the Arabidopsis root. (E–G) Suberin monomer composition in μg mg−1 dry weight in the basal (E), middle (F), and apical (G) root zone. (H) Substance class distribution in the three root zones in relative amounts. (I) Total aliphatic suberin amount in μg mg−1 dry weight in the three root zones. Values 1-Alc., 1-Alcohols; ω-OH-Ac., ω-hydroxyacids; α,ω-Diac., α,ω-diacids; 2OH-Ac., 2-hydroxyacid are given as mean ±SD of three replicates, each containing 2–8 mg isolated suberized root cell wall material. P, Periderm; E, endodermis; C, cortex.
To analyse the quantity and composition of the aliphatic root suberin, root tissue of the different root zones was subjected to depolymerization and subsequently analysed using GC and GC-MS. The same monomers were detected in the suberin hydrolysate of all three root zones: unsubstituted fatty acids ranging in chain length from C18 to C24, primary alcohols (C18–C22), ω-hydroxyacids (C16–C24), α,ω-diacids (C16–C24), and 2-hydroxyacids (C20–C26) (Fig. 1E–G). The most prominent substance classes in all root zones are ω-hydroxyacids (37–52%) and α,ω-diacids (28–31%) (Fig. 1H). Along the root axis all aliphatic suberin monomers increase from the tip to the base. The total aliphatic suberin amount in the basal root part is more than twice the amount in the apical part (Fig. 1I). In the substance class composition, the relative amounts of α,ω-diacids and alcohols did not change. However, significant changes in the relative amounts of ω-hydroxyacids and fatty acids were detected among the different root zones. Comparing the apical and the basal root zones, the relative amount of ω-hydroxyacids increased >40% in the basal zone. By contrast the relative amount of fatty acids is significantly reduced in this zone; 12% compared with 21% in the apical zone. A reduction from the tip to the base was also observed in the minor compounds of the 2-hydroxyacid class.
horst knock-out mutants have a highly modified and reduced root suberin
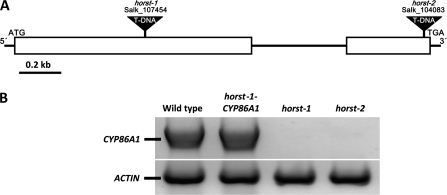
The abundance of ω-hydroxyacids and α,ω-diacids in aliphatic root suberin in Arabidopsis and the significant increase in ω-hydroxyacids as suberin formation goes on during root development indicates that ω-hydroxylases are required for suberin monomer biosynthesis. Of the known fatty acid ω-hydroxylases, CYP86A1 is preferentially expressed in roots. Therefore CYP86A1 seemed to be a good candidate for an involvement in suberin biosynthesis. To investigate the in vivo function of CYP86A1, a reverse genetics approach was chosen. Two allelic T-DNA insertion mutant lines in CYP86A1 were obtained from the Nottingham Arabidopsis Stock Centre and further characterized. Since it was supposed that CYP86A1 encodes a ω-hydroxylase in suberin formation the mutant lines were named horst-1 (SALK_107454) and horst-2 (SALK_104083) for hydroxylase of root suberized tissue. PCR amplification of T-DNA flanking genomic sequences showed that horst-1 carries the T-DNA insertion in the first exon of CYP86A1, whereas in horst-2 the T-DNA insertion is located in the second exon (Fig. 2A). The T3 progeny of the SALK insertion lines were genotyped using PCR, and homozygous T4 horst-1 and horst-2 plants were used to determine the CYP86A1 transcript levels by semi-quantitative RT-PCR (Fig. 2B). In contrast to wild type, no CYP86A1 transcript could be detected in total RNA from horst-1 and horst-2 seedlings. No obvious phenotypes could be observed when horst-1 and horst-2 mutant lines were grown together and compared with wild-type plants.
Fig. 2.
CYP86A1 locus and mutant verification. (A) Structure of the CYP86A1 gene and designated locations of the horst alleles. (B) RT-PCR analysis of CYP86A1 expression in 10-d-old seedlings of wild-type, horst-1, horst-2, and horst1-CYP86A1 complemented plants.
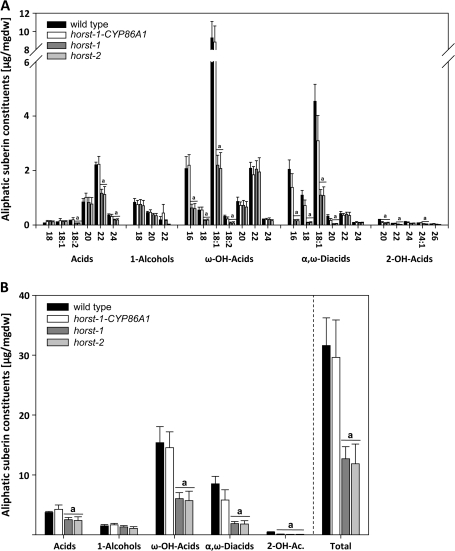
To investigate a potential involvement of CYP86A1 in suberin biosynthesis, a qualitative and quantitative root suberin analysis of horst mutants was performed by GC and GC-MS and compared with the corresponding wild type. horst-1 and horst-2 aliphatic suberin exhibited significant reductions in the amount of specific monomers of all compound classes except alcohols (Fig. 3A, B). The most pronounced reductions were detected in ω-hydroxyacids of chain length C16 and C18 and α,ω-diacids with chain length C16–C20 (Fig. 3A). However, significant reductions could also be detected in C22 and C24 fatty acids and significant but less pronounced reductions can be detected in 2-hydroxyacids of horst-1 and horst-2 suberin. In total, horst-1 and horst-2 aliphatic root suberin amount is reduced by 60.86% (±7.96) and 63.4% (±10.61), respectively, compared with the wild type (Fig. 3B).
Fig. 3.
Aliphatic suberin composition in wild type and horst mutant plants. (A) Suberin monomer composition. (B) Total amount of each substance class and total aliphatic suberin amount. Suberin analysis of 5-week-old plants in μg mg−1 dry weight ±SD of four or five replicates, each containing one or two roots. Statistically significant changes are indicated at ≥99%.
CYP86A1 is required for the biosynthesis of suberin in Arabidopsis roots
To verify that the suberin phenotypes determined in horst mutant lines are attributed to the knockout of CYP86A1, horst-1 plants were transformed with a wild-type genomic fragment comprising the 1.4 kb 5′ region upstream of the predicted CYP86A1 start codon and the CYP86A1 encoding genomic sequence. RT-PCR analysis on total RNA from hygromycin-resistant seedlings (horst-1-CYP86A1) confirmed the successful reintegration and expression of wild-type CYP86A1. Wild-type levels of CYP86A1 intron-excised transcript were detected in horst-1-CYP86A1 transgenic plants (Fig. 2B). Root suberin analysis of the complemented CYP86A1-transformed horst-1 plants revealed a quantitative and qualitative aliphatic suberin composition similar to the wild type (Fig. 3).
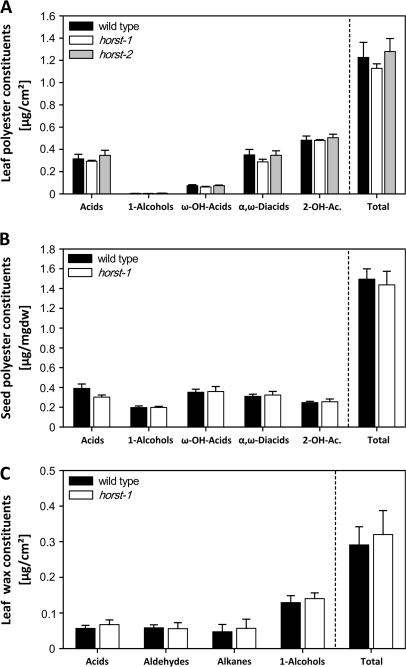
Since ω-hydroxyacids and α,ω-diacids, the most strongly affected compounds in the horst mutant suberin, are also abundant monomers in the biopolyesters of leaves and seeds, these tissues were also analysed for compositional changes in their long-chain aliphatic compounds. No significant differences could be detected in the leaf and seed polyester composition of the wild-type and horst-1 mutant plants (Fig. 4A, B). Other significantly reduced monomers in horst suberin are unsubstituted fatty acids. However, the amount of long-chain fatty acids as part of leaf waxes and other fatty acid-derived wax monomers is not modified in horst-1 mutant plants (Fig. 4C).
Fig. 4.
Cutin, seed coat, and wax analysis in wild type and horst mutant plants. (A) Leaf cutin substance class composition and total amount in μg cm−2 ±SD of three replicates, each containing 15–20 leaves. (B) Seed coat substance class composition and total amount in μg mg−1 dry weight ±SD of five replicates, each containing at least 16 μg seeds. (C) Leaf wax substance class composition and total amount in μg cm−2 ±SD of five replicates, each containing seven leaves.
The suberin biosynthetic gene CYP86A1 is expressed in roots
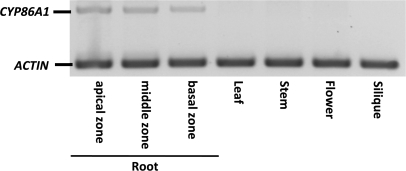
To investigate the expression pattern of CYP86A1 in more detail on the organ and tissue level, experiments using RT-PCR and GUS reporter gene constructs were performed. RT-PCR analysis on total RNA preparations from different organs resulted in no detectable CYP86A1 transcript levels in above-ground organs such as leaves, stems, flowers, and siliques (Fig. 5). In roots, high expression levels for CYP86A1 were detected in all three root zones from the tip to the base.
Fig. 5.
Organ-specific expression pattern of CYP86A1. Expression determined by RT-PCR of CYP86A1 transcript levels in three different root zones, leaves, stems, flowers, and siliques of 5-week-old fully developed Arabidopsis plants. In each reaction, 0.2 μg of total RNA was used. ACTIN was used as a control.
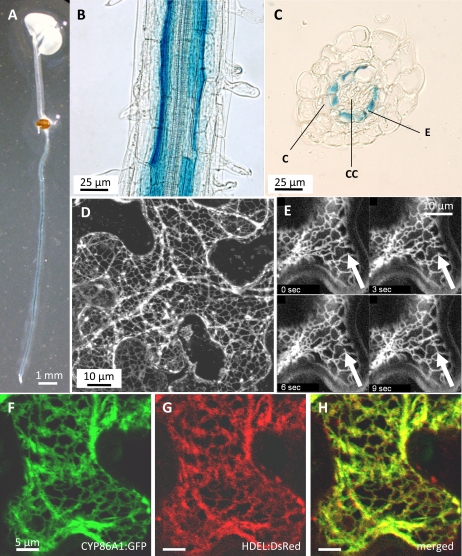
For expression analysis of CYP86A1 on the tissue level the 1.4 kb putative promoter region upstream of the predicted CYP86A1 start codon was fused in frame to the β-glucuronidase (GUS) gene and used to transform wild-type Arabidopsis. Several independent PromCYP86A1:GUS transgenic plant lines were investigated. GUS staining was observed in the root of all PromCYP86A1:GUS seedlings, while GUS activity was not found in the hypocotyl or cotyledons (Fig. 6A). Microscopic examination of the intact seedling roots showed GUS staining in the centre of the root, inside cortical tissue (Fig. 6B). In cross-sections of GUS-stained PromCYP86A1:GUS roots, GUS protein activity could be located to the endodermis (Fig. 6C). Similar to seedlings, no GUS staining could be observed under these conditions in above-ground organs like leaves or stems of mature plants.
Fig. 6.
Tissue expression pattern and sub-cellular protein localization. (A–C) GUS staining in Arabidopsis transformed with a PromCYP86A1:GUS construct: (A) in a 7-d-old seedling GUS staining is only observed in the root; (B) close-up view of the intact seedling root with a GUS staining limited to the centre of the root; (C) cross-section through the seedling root showing GUS staining restricted to the endodermis. (D–H) Confocal microscope pictures of transiently expressed green fluorescent protein (GFP) and red fluorescent protein (DsRED) tagged peptides in epidermis of tobacco (N. benthamiana) leaves: (D) projection of serial confocal optical sections through an epidermis cell expressing a CYP86A1:GFP fusion protein, showing reticulate ER-like structures (brightness indicates GFP fluorescence); (E) close-up view showing the vitality of the cell by movement of the ER-like structures over the time indicated by an arrow (also see Video S1 in Supplementary material available at JXB online); (F–H) co-localization of CYP86A1:GFP (green, F) and HDEL:DsRed (red, G) in ER network, shown in the merged image (yellow, H). CC, Central cylinder; E, endodermis; C, cortex.
The CYP86A1:GFP protein is targeted to the ER
In order to examine the sub-cellular protein localization of CYP86A1, the green-fluorescent-protein (GFP) was fused to the 3′ end of the coding sequence of CYP86A1. Employment of the native promoter to drive the expression of this construct in transgenic Arabidopsis plants resulted in a weak GFP fluorescence in roots, only slightly above background (data not shown). The weak fluorescence signal did not allow the GFP localization inside the root tissue with a resolution high enough for sub-cellular localization in endodermal tissue. Therefore, we focused on heterologous expression and transiently expressed this construct (CYP86A1:GFP) under the control of the CaMV35S-promoter (35S) in the epidermis of Nicotiana benthamiana leaves. Using confocal laser scanning microscopy, intense GFP fluorescence was observed in reticulate structures characteristic for ER (Fig. 6D). Continuous observation of the GFP-tagged CYP86A1 revealed the movement of these membranous structures, typical of ER in vital cells (Fig. 6E; Video S1 in Supplementary material available at JXB online). Co-expression with an ER marker construct containing the HDEL retention signal for ER fused to DsRed (HDEL:DsRed) revealed it co-localized with CYP86A1:GFP (Fig. 6F–H).
Discussion
Root suberin analysis in many species has usually been applied to enzymatically isolated endodermal and hypodermal/peridermal cell walls (Schreiber et al., 1999, 2007). However, dissection and isolation of suberized tissue from the frail Arabidopsis root is not feasible. Therefore previous analyses of Arabidopsis root suberin have been carried out with whole root samples (Franke et al., 2005; Beisson et al., 2007). These analyses represented a ‘mixed’ suberin composition as the whole roots comprised both root tissues in a primary and secondary developmental stage. The allocation of fully developed Arabidopsis roots into three segments along the root axis, enabled the generation of root tissue for comparative compositional suberin analysis in root zones of different developmental stages. As demonstrated in the histochemical analysis of representative cross-sections (Fig. 1B, D), the analysis of the basal root zone sample represents a suberin predominantly derived from peridermal cell walls of a root in a secondary developmental stage, whereas suberin in the apical zone is representative of endodermal suberin from roots in a primary developmental stage. The middle zone mostly comprises roots in a secondary developmental stage (Fig. 1C). This differentiation pattern and the peridermal origin are consistent with previous solely microscopic studies of 4-week-old Arabidopsis roots. Baum et al. (2002) observed first periclinal divisions of the pericycle 35 mm from the tip and a phellogen was detected 20 mm from the base. The development of a periderm and therefore the transition from a primary to a secondary developmental stage probably takes place between the distal part of the middle root zone and the proximal region of the apical root zone.
The chemical composition determined for the aliphatic suberin of the three root zones is comparable to previous analysis in Arabidopsis roots (Franke et al., 2005; Beisson et al., 2007). Independent of the zonal origin of the root samples, the same aliphatic suberin monomers could be identified in all three root zones (Fig. 1E–G). However, pronounced quantitative and qualitative differences could be detected in the suberin of the three root zones. The total amount of aliphatic suberin in the basal root part is more than twice the amount in the apical root part (Fig. 1,I). This is the result of a substantial increase in root diameter from the tip to the base. In addition, the proportion of suberized tissue layers increases during root development: in the primary developmental stage suberin depositions occur only in an inner cell layer, the endodermis. By contrast the suberized tissue of the root in the secondary developmental stage, the periderm, forms the outermost cell layer of the root. Compared with the whole root surface the endodermis represents an inner ring of a lower circumference compared with the area covered by the periderm.
The most obvious compositional changes along the root zones were observed in ω-hydroxyacids and primary fatty acids (Fig. 1H). From the tip to the base the amount of ω-hydroxyacids, characteristic suberin monomers, increases substantially, and this increase correlates with a decrease in primary fatty acids. Similarly, in suberin of endodermal and rhizodermal/hypodermal cell walls of corn roots, primary fatty acids decrease in sub-apical root zones whereas ω-hydroxyacids increase (Zeier et al., 1999). Although these changes were most pronounced between the very apical zone (0–80 mm from the tip), characterized by Casparian strip suberin, and the basal zones, characterized by suberin lamellae around the cell wall, the increase in ω-hydroxyacids on the expense of primary fatty acids continued over the next two root zones (80–160 mm and 160–240 mm from the tip, respectively) towards the base. The compositional changes observed in suberin along the axis of corn and Arabidopsis roots point out the importance of ω-hydroxylation in the suberization of cell walls in general and might indicate an increase in the activity of fatty acid ω-hydroxylation during development.
Due to the two opposite functional groups, ω-hydroxyacids are chemically ideally suited to form a polyester. Whether the increase in ω-hydroxyacids is associated with physiological or functional properties of suberized barriers in an endodermis or in a periderm still remains to be established.
The majority of fatty acid hydroxylation reactions in plants is catalysed by cytochrome P450 of the CYP86 clan, especially the CYP86 and CYP94 families (Duan and Schuler, 2005; Kandel et al., 2006). Although many CYP86 and CYP94 have been characterized biochemically, their physiological function in a biological process is mostly unknown. horst-1 and horst-2, two allelic Arabidopsis mutants carrying a T-DNA insertion in At5g58860 were used to study the in vivo function of CYP86A1. As a consequence of the transcriptional knock-out of CYP86A1 major suberin monomers are significantly reduced resulting in a >60% reduced amount of total aliphatic suberin (Fig. 3B). Wild-type levels of both total aliphatic root suberin amount and amounts of single suberin components were restored by complementation with a genomic DNA fragment spanning the CYP86A1 locus-including promoter region (Fig. 3A, B). Taken together, the analysis of the aliphatic root suberin composition of the two allelic horst-1 and horst-2 mutants and complemented plants provides the genetic evidence for the involvement of CYP86A1 in suberin biosynthesis. This is further supported by very similar suberin monomer profiles from two other allelic cyp86A1 insertion mutants, reported by Li et al. (2007) during the preparation of this paper.
Recent P450 protein sequence analysis has set CYP86A1 on a single clade within the CYP86A subfamily in Arabidopsis (Nelson et al., 2004; Duan and Schuler, 2005), probably indicative of unique functional properties. Therefore, the detailed monomer composition obtained by GC-MS of horst mutant suberin is also strongly indicative for the substrate specificity of CYP86A1 in vivo, although some overlap in catalytic properties with other ω-hydroxylases probably exists (see below). The strong reduction in suberin ω-hydroxyacids <C20 in horst plants compared with the wild-type plants is essentially in agreement with the in vitro catalytic activity of CYP86A1. Microsomal preparations from yeast expressing CYP86A1, actively metabolized C16>C18:1>C18:2>C14>C12 fatty acids to the corresponding ω-hydroxyacids with highest activities towards C16 and C18:1 fatty acids (Benveniste et al., 1998). Similarly, CYP86A1 from baculovirus-infected insect cells has been shown to bind these substrates (Rupasinghe et al., 2007). However, the CYP86A1-mutant suberin analysis not only confirms these studies, it also provides new information, as substrates longer than C18 have not been tested in in vitro studies. The fact that the content of C20, C22, and C24 ω-hydroxyacids is not affected by the horst mutation indicates a strong chain-length specificity of CYP86A1 catalysed ω-hydroxylation. Most likely fatty acids longer than C20 do not act as substrates for CYP86A1 in vivo. Inconsistent with the inability of heterologously expressed CYP86A1 to efficiently bind or use stearic acid (C18:0) as a substrate in vitro (Benveniste et al., 1998; Rupasinghe et al., 2007), the horst aliphatic root suberin is also significantly reduced in saturated C18 ω-hydroxyacid. Although some deficiencies of the heterologous in vitro systems cannot be excluded (e.g. endogenous yeast reactions compete for the stearic acid), this might indicate that in situ additional factors affect CYP86A1 substrate specificity, enabling the conversion of C18 fatty acid to C18 ω-hydroxyacid. An alternative explanation would be that in wild-type plants an elongation of ω-hydroxyacids from C16 to C18 could occur, but only the CoA-activation, required for fatty acid elongation (FAE), has been demonstrated for C16 ω-hydroxyacid (Schnurr et al., 2004).
As C16 and C18 ω-hydroxyacids are not entirely omitted in the horst mutant's suberin, other ω-hydroxylases must be encoded in the genome which provide a functional redundancy, at least partially compensating the knock-out of CYP86A1. In plants, fatty acid ω-hydroxylation activity has been demonstrated for P450 of the CYP78, CYP92, CYP96, and many members of the CYP86 and CYP94 families (Kandel et al., 2006). In Arabidopsis, four members of the CYP94 family (CYP94B1, CYP94B2, CYP94B3, and CYP94C1) were shown to be ω-hydroxylases after heterologous expression in yeast (Benveniste et al., 2006). Similar to CYP86A8 (Wellesen, et al., 2001), all 4 CYP94 ω-hydroxylated C12:0, C14:0, C16:0, and C18:1 fatty acids. Recently, the biochemically less-characterized CYP86A members in Arabidopsis, CYP86A2, CYP86A4, and CYP86A7, have been shown to ω-hydroxylate C18:1 fatty acid (Rupasinghe et al., 2007). Of these P450 with ω-hydroxylase activity, the corresponding genes for CYP94B1, CYP94B3, CYP86A2, CYP86A4, and CYP86A8 have been reported to be also expressed in roots (Wellesen et al., 2001; Duan and Schuler, 2005; Schuler et al., 2006). Although not preferentially expressed in suberizing tissue, residual root expression in one or more of these ω-hydroxylases could result in ω-hydroxylation activity partially compensating the lack of CYP86A1 activity.
In addition to the strong reduction in ω-hydroxyacids <C20, specific suberin monomers in the compound classes of α,ω-diacids, acids and 2-hydroxyacids are also significantly reduced in the horst mutant suberin (Fig. 3). According to the biochemical pathways suggested for ω-oxidation in apoplastic polyester biosynthesis, α,ω-diacid formation can be performed in two different ways (Franke et al., 2005; Kurdyukov et al., 2006). One requires two sequential dehydrogenase reactions on ω-hydroxyacids as initial substrates. These reactions were initially demonstrated in suberizing potato tuber tissue (Agrawal and Kolattukudy, 1978) and are supposed to involve HOTHEAD-like oxydoreductases. A mutation in HOTHEAD (HTH) leads to an increase in ω-hydroxyacids in cutin polyester (Kurdyukov et al., 2006). Therefore the strong reduction in C16–C18 α,ω-diacids is most likely the result of a depletion in ω-hydroxyacids, the substrate for the dehydrogenase activity of HTH-like oxidoreductase. In this context, it is interesting to note that the HTH gene is also expressed in roots. In a second pathway to α,ω-diacids, CYP86A1 could act as a multifunctional ω-hydroxylase, similar to CYP94A5 and CYP94C1 which catalyse the multi-step oxidation of fatty acids to the corresponding ω-hydroxyacid and α,ω-diacids (Le Bouquin et al., 2001; Kandel et al., 2007). However, for CYP86A1 this multifunctional activity has not been demonstrated in vitro under conditions successfully applied to other multifunctional ω-hydroxylases.
Interestingly, the supposed substrates of the C16 and C18 ω-hydroxylation catalysed by CYP86A1, mainly medium chain fatty acids, do not accumulate in the mutant suberin. Instead long-chain fatty acids and 2-hydroxy fatty acids, aliphatic suberin monomers that are not direct or downstream products of fatty acid ω-oxidation, are affected by the mutation in CYP86A1 (Fig. 3). This indicates significant interaction between metabolic branches in aliphatic suberin biosynthesis and maybe other lipid biosynthetic pathways. So far undiscovered, complex regulatory mechanisms in suberin biosynthetic pathways lead to more or less specific secondary effects on suberin composition by ‘down-regulation’ of upstream reactions in suberin biosynthetic pathways. A very specific effect can be observed on the chain length distribution of fatty acids. C22 and C24 fatty acids are significantly reduced in the horst suberin, indicating that the expression or activity of enzymes involved in C20 FAE is affected by the CYP86A1 knock out. FAE has been suggested as a major metabolic activity in aliphatic suberin biosynthesis in potato and Arabidopsis (Franke et al., 2005; Yang and Bernards, 2006). Of the 21 Arabidopsis β-ketoacyl-CoA synthases (FAE-KCS), which catalyse the rate-limiting and product-determining step in FAE (Millar and Kunst, 1997), at least six FAE-KCS have been shown to have the catalytic properties to provide C22 or C24 fatty acids (Trenkamp et al., 2004; Blacklock et al., 2006; Paul et al., 2006). Studying these FAE-KCS in the horst mutants could provide a first insight in a coordinated regulation or ‘cross talk’ between FAE and fatty acid hydroxylation.
Similarly, other metabolic fates, e.g. 2-hydroxylation, seem to be secondarily affected by an unknown inhibition mechanism. The reduction in C20 α,ω-diacids also seems to be a secondary effect as the supposed precursor, C20 ω-hydroxyacid, is not reduced in horst suberin. Maybe, HTH-like oxidoreductases, metabolizing C20 ω-hydroxyacid, are inhibited or depressed in the horst mutant.
The suberin analysis of the horst mutant and complemented lines demonstrated that CYP86A1 is required for suberin biosynthesis. In agreement with the tissue distribution of suberin depositions in roots in a primary developmental stage (Fig. 1D), GUS activity in plants transformed with a PromCYP86A1:GUS construct was preferentially observed in the endodermis (Fig. 6C), indicating the role of CYP86A1 in early deposition of suberin. In peridermal tissue of secondary roots, CYP86A1-promoter driven GUS expression could not be detected under the applied conditions (data not shown). However, consistent with the predominance of ω-hydroxyacids in apical, middle, and basal root zone suberin, RT-PCR analysis revealed that CYP86A1 is expressed throughout root development (Fig. 5). Furthermore, CYP86A1 is also specifically root expressed with no detectable expression levels in above-ground organs. These results are in agreement with predictions for gene expression (Zimmermann et al., 2004) and previous whole-organ expression studies (Duan and Schuler, 2005). Consistent with the lack of expression in above-ground organs, aliphatic cell wall depositions such as wax or ω-hydroxyacid-containing polyesters in above-ground organs, such as cutin and the seed coat, are not altered in the substance class composition (Fig. 4). This emphasizes the specific involvement of CYP86A1 in root suberization.
At the sub-cellular level, visualization of the CYP86A1–GFP fusion protein (Fig. 6D, E) strongly indicates an ER localization of CYP86A1. Although, heterologous overexpression may eventually lead to artificial ER retention, the localization of CYP86A1 to the ER was supported by co-localization studies with an ER reporter construct (HDEL:DsRed protein), when simultaneously expressed in N. benthamiana epidermis cells (Fig. 6F–H). An ER localization of the fatty acid ω-hydroxylase CYP86A1 is also in agreement with studies from decades ago showing that P450-containing plant microsomes can catalyse the production of ω-hydroxyacids (Benveniste et al., 1982; Pinot et al., 1992; 1993). Current gene ontology predicts CYP86A1 to have an N-terminal hydrophobic transmembrane domain and is targeted to the endomembrane system (Schwacke et al. 2003). Furthermore, the ER localization of the GFP-tagged CYP86A1 refines biochemical studies that assigned CYP86A1 activity to endomembrane compartments due to the successful employment of microsomal preparations from CYP86A1-expressing yeast to determine the catalytic properties of CYP86A1 (Benveniste et al., 1998). In this context, it is interesting to note that FAE, the other key reaction in aliphatic suberin biosynthesis, has been demonstrated in microsomes of suberizing corn root tissue (Schreiber et al., 2005). The FAE activity in different root zones correlated with the demand in suberin constituents. In addition, similar GFP-tagging approaches successfully determined the ER localization of reaction steps in FAE (Kunst and Samuels, 2003; Zheng et al., 2005). Together, these results suggest that core reactions of the suberin biosynthetic machinery take place at the ER. It remains to be determined if the ER-produced aliphatic suberin constituents are exported to the apoplast as monomers or preformed esters.
In summary, CYP86A1 has been identified as a key enzyme for aliphatic suberin biosynthesis in Arabidopsis roots. Corresponding mutants exhibiting a 60% reduction in aliphatic suberin do not show any phenotypic variation, indicating that small amounts of aliphatic suberin are sufficient to provide the physiological function, at least under the optimized laboratory culture conditions. Further experiments on the behaviour of horst to different abiotic and biotic stresses might shed light on the physiological consequence of an altered suberin substance class composition and total aliphatic suberin amount during the plant life cycle. Mutants with reduced and modified suberin content are now available to test the barrier properties in relation to the chemical composition when tests have been developed for Arabidopsis.
Supplementary material
Video S1: Dynamics of CYP86A1:GFP in ER-like reticulate structures
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre (Loughborough, UK) and the SALK Institute Genomic Analysis Laboratory (La Jolla, CA, USA) for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. We are also grateful for support from the Department of Plant Cell Biology, IZMB, University of Bonn: Claudia Heym and Boris Voigt for constructing the HDEL:DsRed plasmid and Dr Jozef Šamaij for fruitful discussions. This work was supported by the Deutsche Forschungsgemeinschaft (DFG).
References
- Agrawal VP, Kolattukudy PE. Purification and characterization of a wound-induced ω-hydroxyfatty acid:NADP oxidoreductase from potato tuber disks (Solanum tuberosum L) Archives of Biochemistry and Biophysics. 1978;191:452–465. doi: 10.1016/0003-9861(78)90384-3. [DOI] [PubMed] [Google Scholar]
- Agrios GN. Plant pathology. San Diego: Academic Press; 1997. [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Baum SF, Dubrovsky JG, Rost TL. Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. American Journal of Botany. 2002;89:908–920. doi: 10.3732/ajb.89.6.908. [DOI] [PubMed] [Google Scholar]
- Beisson F, Yonghua L, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. The Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste I, Saito T, Wang Y, Kandel S, Huang H, Pinot F, Kahn R, Salaün JP, Shimoji M. Evolutionary relationship and substrate specificity of Arabidopsis thaliana fatty acid hydroxylase. Plant Sciences. 2006;170:326–338. [Google Scholar]
- Benveniste I, Salaün JP, Simon A, Reichhart D, Durst F. Cytochrome P-450- dependent ω-hydroxylation of lauric acid by microsomes from pea seedlings. Plant Physiology. 1982;70:122–126. doi: 10.1104/pp.70.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste I, Tijet N, Adas F, Philipps G, Salaün JP, Durst F. CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochemical and Biophysical Research Communications. 1998;243:688–693. doi: 10.1006/bbrc.1998.8156. [DOI] [PubMed] [Google Scholar]
- Bernards MA. Demystifying suberin. Canadian Journal of Botany. 2002;80:227–240. [Google Scholar]
- Blacklock BJ, Jaworski JG. Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochemical and Biophysical Research Communications. 2006;346:583–590. doi: 10.1016/j.bbrc.2006.05.162. [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Kendrick B, Peterson CA. Efficient lipid staining in plant-material with Sudan Red 7B or Fluorol Yellow-088 in polyethylene glycol-glycerol. Biotechnic and Histochemistry. 1991;66:111–116. doi: 10.3109/10520299109110562. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in plants. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M, Debrouwer D. In-situ enzyme histochemistry on plastic-embedded plant material: the development of an artefact-free β-glucoronidase assay. The Plant Journal. 1992;2:261–266. [Google Scholar]
- De Vries S, Hoge H, Bisseling T. Isolation of total and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoot RA, editors. Plant molecular biology. Dordrecht: Kluwer Academic Publishers; 1988. pp. 1–5. [Google Scholar]
- Duan H, Schuler MA. Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiology. 2005;137:1067–1081. doi: 10.1104/pp.104.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. Apoplastic polyesters in Arabidopsis surface tissues: a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Franke R, Schreiber L. Suberin – a biopolyester forming apoplastic plant interfaces. Current Opinion in Plant Biology. 2007;10:252–259. doi: 10.1016/j.pbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Groh B, Hubner C, Lendzian KJ. Water and oxygen permeance of phellems isolated from trees: the role of waxes and lenticels. Planta. 2002;215:794–801. doi: 10.1007/s00425-002-0811-8. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Research. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway PJ. Some variations in the composition of suberin from the cork layers of higher plants. Phytochemistry. 1983;22:495–502. [Google Scholar]
- Jach G, Binot E, Frings S, Luxa K, Schell J. Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. The Plant Journal. 2001;28:483–491. doi: 10.1046/j.1365-313x.2001.01153.x. [DOI] [PubMed] [Google Scholar]
- James DW, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. The Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel S, Sauveplane V, Compagnon V, Franke R, Millet Y, Schreiber L, Werck-Reichhart D, Pinot F. Characterization of a methyl jasmonate and wounding-responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS Journal. 2007;274:5116–5127. doi: 10.1111/j.1742-4658.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- Kandel S, Sauveplane V, Olry A, Diss L, Benveniste I, Pinot F. Cytochrome P450-dependent fatty acids hydroxylases in plants. Phytochemistry Reviews. 2006;5:359–372. [Google Scholar]
- Kolattukudy PE. Structure, biosynthesis, and biodegradation of cutin and suberin. Annual Review of Plant Physiology and Plant Molecular Biology. 1981;32:539–567. [Google Scholar]
- Kolattukudy PE. Suberin from plants. In: Doi Y, Steinbuechel A, editors. Biopolymers I: Biological systems and biotechnological production 3. Münster: Wiley–VCH; 2001. pp. 41–68. [Google Scholar]
- Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Progress in Lipid Research. 2003;42:51–80. doi: 10.1016/s0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bär S, Franke R, Efremova N, Tiedjen K, Schreiber L, Saedler H, Yephremov A. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006;224:315–329. doi: 10.1007/s00425-005-0215-7. [DOI] [PubMed] [Google Scholar]
- Le Bouquin R, Skrabs M, Kahn R, Benveniste I, Salaün JP, Schreiber L, Durst F, Pinot F. CYP94A5, a new cytochrome P450 from Nicotiana tabacum is able to catalyze the oxidation of fatty acids to the ω-alcohol and to the corresponding diacid. European Journal of Biochemistry. 2001;268:3083–3090. doi: 10.1046/j.1432-1327.2001.02207.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proceedings of the National Academy of Sciences, USA. 2007;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulai EC, Corsini DL. Differential deposition of suberin phenolic and aliphatic domains and their roles in resistance to infection during potato tuber (Solanum tuberosum L.) wound-healing. Physiological and Molecular Plant Pathology. 1998;53:209–222. [Google Scholar]
- Ma FS, Peterson CA. Current insights into the development, structure, and chemistry of the endodermis and exodermis of roots. Canadian Journal of Botany. 2003;81:405–421. [Google Scholar]
- Matzke K, Riederer M. A comparative study into the chemical constitution of cutins and suberins from Picea abies (L.) Karst., Quercus robur L., and Fagus sylvatica L. Planta. 1991;185:233–245. doi: 10.1007/BF00194066. [DOI] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. The Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. The Plant Journal. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M. The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry. 2006;67:2597–2610. doi: 10.1016/j.phytochem.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. Comparative genomics of rice and Arabidopsis: analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiology. 2004;135:756–772. doi: 10.1104/pp.104.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North GB, Nobel PS. Changes in root hydraulic conductivity for two tropical epiphytic cacti as soil-moisture varies. American Journal of Botany. 1994;81:46–53. [Google Scholar]
- Paul S, Gable K, Beaudoin F, Cahoon E, Jaworski J, Napier JA, Dunn TM. Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. Journal of Biological Chemistry. 2006;281:9018–9029. doi: 10.1074/jbc.M507723200. [DOI] [PubMed] [Google Scholar]
- Perumalla CJ, Peterson CA, Enstone DE. A survey of angiosperm species to detect hypodermal casparian bands. I. Roots with a uniseriate hypodermis and epidermis. Botanical Journal of the Linnean Society. 1990;103:93–112. [Google Scholar]
- Pinot F, Bosch H, Alayrac C, Mioskowski C, Vendais A, Durst F, Salaün JP. ω-Hydroxylation of oleic acid in Vicia sativa microsomes. Plant Physiology. 1993;102:1313–1318. doi: 10.1104/pp.102.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot F, Salaün JP, Bosch H, Lesot A, Mioskowsky C, Durst F. ω-Hydroxylation of Z9-octadecenoic, Z9,10-epoxystearic and 9,10-dihydroxystearic acids by microsomal cytochrome P450 systems from Vicia sativa. Biochemical and Biophysical Research Communications. 1992;184:183–193. doi: 10.1016/0006-291x(92)91176-q. [DOI] [PubMed] [Google Scholar]
- Reinhardt DH, Rost TL. Salinity accelerates endodermal development and induces an exodermis in cotton seedling roots. Environmental and Experimental Botany. 1995;35:563–574. [Google Scholar]
- Rupasinghe SG, Duan H, Schuler MA. Molecular definitions of fatty acid hydroxylases in Arabidopsis thaliana. PROTEINS: Structure, Function, and Bioinformatics. 2007;68:279–293. doi: 10.1002/prot.21335. [DOI] [PubMed] [Google Scholar]
- Sattelmacher B, Muhling KH, Pennewiss K. The apoplast – its significance for the nutrition of higher plants. Zeitschrift fur Pflanzenernährung und Bodenkunde. 1998;161:485–498. [Google Scholar]
- Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. The Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr J, Ziegler H. Water permeability of Betula periderm. Planta. 1980;147:345–354. doi: 10.1007/BF00379844. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Breiner HW, Riederer M, Duggelin M, Guggenheim R. The casparian strip of Clivia miniata Reg. roots – isolation, fine structure and chemical nature. Botanica Acta. 1994;107:353–361. [Google Scholar]
- Schreiber L, Franke R, Hartmann K. Chemical composition of apoplastic transport barriers in roots: quantification of suberin depositions in endodermal and hypodermal root cell walls. In: Sattelmacher B, Horst WJ, editors. The apoplast of higher plants: compartment of storage, transport and reactions. Heidelberg: Springer; 2007. pp. 109–118. [Google Scholar]
- Schreiber L, Franke R, Lessire R. Biochemical characterisation of elongase activity in corn (Zea mays L.) roots. Phytochemistry. 2005;66:131–138. doi: 10.1016/j.phytochem.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Hartmann K, Skrabs M, Zeier J. Apoplastic barriers in roots: chemical composition of endodermal and hypodermal cell walls. Journal of Experimental Botany. 1999;50:1267–1280. [Google Scholar]
- Schuler MA, Duan H, Bilgin M, Ali S. Arabidopsis cytochrome P450s through the looking glass: a window on plant biochemistry. Phytochemistry Reviews. 2006;5:205–237. [Google Scholar]
- Schwacke R, Schneider A, Van Der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiology. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Serra O, Molinas M, Huguet G, Fluch S, Figueras M. A genomic approach to suberin biosynthesis and cork differentiation. Plant Physiology. 2007;144:419–431. doi: 10.1104/pp.106.094227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliday CL, Kolattukudy PE. Biosynthesis of cutin: ω-hydroxylation of fatty acids by a microsomal preparation from germinating Vicia faba. Plant Physiology. 1977;59:1116–1121. doi: 10.1104/pp.59.6.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steudle E, Peterson CA. How does water get through roots? Journal of Experimental Botany. 1998;49:775–788. [Google Scholar]
- Tijet N, Helvig C, Pinot F, Le Bouquin R, Lesot A, Durst F, Salaün JP, Benveniste I. Functional expression in yeast and characterization of a clofibrate-inducible plant cytochrome P-450 (CYP94A1) involved in cutin monomers synthesis. The Biochemical Journal. 1998;332:583–589. doi: 10.1042/bj3320583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. The Plant Journal. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- Trenkamp S Martin W, Tietjen K. Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proceedings of the National Academy of Sciences, USA. 2004;101:11903–11908. doi: 10.1073/pnas.0404600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt E, Schönherr J, Schmidt HW. Water permeability of periderm membranes isolated enzymatically from potato tubers (Solanum tuberosum L.) Planta. 1983;158:294–301. doi: 10.1007/BF00397330. [DOI] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proceedings of the National Academy of Sciences, USA. 2001;98:9694–9699. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R. Cytochrome P450: a success story. Genome Biology. 2000;1 doi: 10.1186/gb-2000-1-6-reviews3003. reviews3003.1-3003.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydro M, Kozubek E, Lehman P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotioana benthamiana. Acta Biochimica Polonica. 2006;53:289–298. [PubMed] [Google Scholar]
- Xiao FM, Goodwin SM, Xiao YM, Sun ZY, Baker D, Tang XY, Jenks MA, Zhou JM. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO Journal. 2004;23:2903–2913. doi: 10.1038/sj.emboj.7600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-L, Bernards MA. Wound induced metabolism in potato (Solanum tuberosum) tubers: biosynthesis of aliphatic domain monomers. Plant Signaling and Behavior. 2006;1:59–66. doi: 10.4161/psb.1.2.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J, Ruel K, Ryser U, Schreiber L. Chemical analysis and immunolocalisation of lignin and suberin in endodermal and hypodermal/rhizodermal cell walls of developing maize (Zea mays L.) primary roots. Planta. 1999;209:1–12. doi: 10.1007/s004250050601. [DOI] [PubMed] [Google Scholar]
- Zheng HQ, Rowland O, Kunst L. Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. The Plant Cell. 2005;17:1467–1481. doi: 10.1105/tpc.104.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann HM, Hartmann K, Schreiber L, Steudle E. Chemical composition of apoplastic transport barriers in relation to radial hydraulic conductivity of corn roots (Zea mays L.) Planta. 2000;210:302–311. doi: 10.1007/PL00008138. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.