Abstract
Deleted in Breast Cancer-1 (DBC1) and its paralog CARP-1 are large multi-domain proteins, with a nuclear or perinuclear localization, and a role in promoting apoptosis upon processing by caspases. Recent studies on human DBC1 show that it is a specific inhibitor of the sirtuin-type deacetylase, Sirt1, which deacetylates histones and p53. Using sensitive sequence profile searches and HMM-HMM comparisons we show that the central conserved globular domain present in the DBC1 and it homologs from diverse eukaryotes is a catalytically inactive version of the Nudix hydrolase (MutT) domain. Given that Nudix domains are known to bind nucleoside diphosphate sugars and NAD, we predict that this domain in DBC1 and its homologs binds NAD metabolites such as ADP-ribose. Hence, we propose that DBC1 and its homologs are likely to regulate the activity of SIRT1 or related deacetylases by sensing the soluble products or substrates of the NAD-dependent deacetylation reaction. The complex domain architectures of the members of the DBC1 family, which include fusions to the RNA-binding S1-like domain, the DNA-binding SAP domain and EF-hand domains, suggest that they are likely to function as integrators of distinct regulatory signals including chromatin protein modification, soluble compounds in NAD metabolism, apoptotic stimuli and RNA recognition.
Introduction
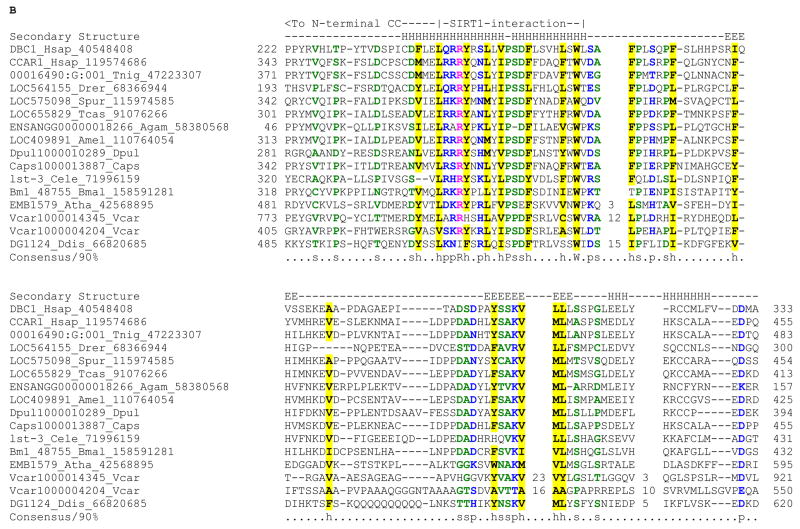
DBC1 was originally identified as a gene homozygously deleted in breast cancer and some other tumors 1. It encodes a large protein, which has been shown to act in a pro-apoptotic fashion upon processing by caspases 2. Its close paralog in vertebrates CARP-1 (CCAR1) is also a pro-apoptotic gene that up-regulates p21WAF1/CIP1 and down-regulates cyclin B1 3. DBC1 and CARP-1 proteins share an N-terminal coiled-coil (zipper) region and multiple conserved globular domains, including a C-terminal inactive EF-hand module that is unlikely to bind Ca2+ ions 2. Homologous proteins with conservation spanning the above regions shared by DBC1 and CARP-1 (the DBC1 family) are also found in plants and the slime mold Dictyostelium, suggesting that the family emerged prior to the divergence of the crown group of eukaryotes. Both DBC1 and CARP-1 have been shown to localize to the nucleus or the nuclear envelope, but they might also localize to the cytoplasm especially during apoptosis 2, 3. However, their exact biochemical roles remained unclear until two recent concomitant reports on human DBC1. These studies showed that DBC1 is a specific inhibitor of the enzyme SIRT1, which mediates NAD-dependent deacetylation of proteins such as histones and p53 4, 5. Thus, by inhibiting the anti-apoptotic SIRT1, DBC1 is able to promote apoptotic pathways via p53 hyperacetylation. It might also counter the repressive action of SIRT1 mediated lysine deacetylation of histones 4, 5. While the conserved coiled-coil region in DBC1 has been shown to be required for its interaction with SIRT1 (Fig. 1), the roles of the conserved globular domains present in the DBC1 family are unclear. Given that these domains are conserved throughout the eukaryotic crown group we were interested in investigating if they might throw light on the conserved regulatory processes mediated by the DBC1 family of proteins.
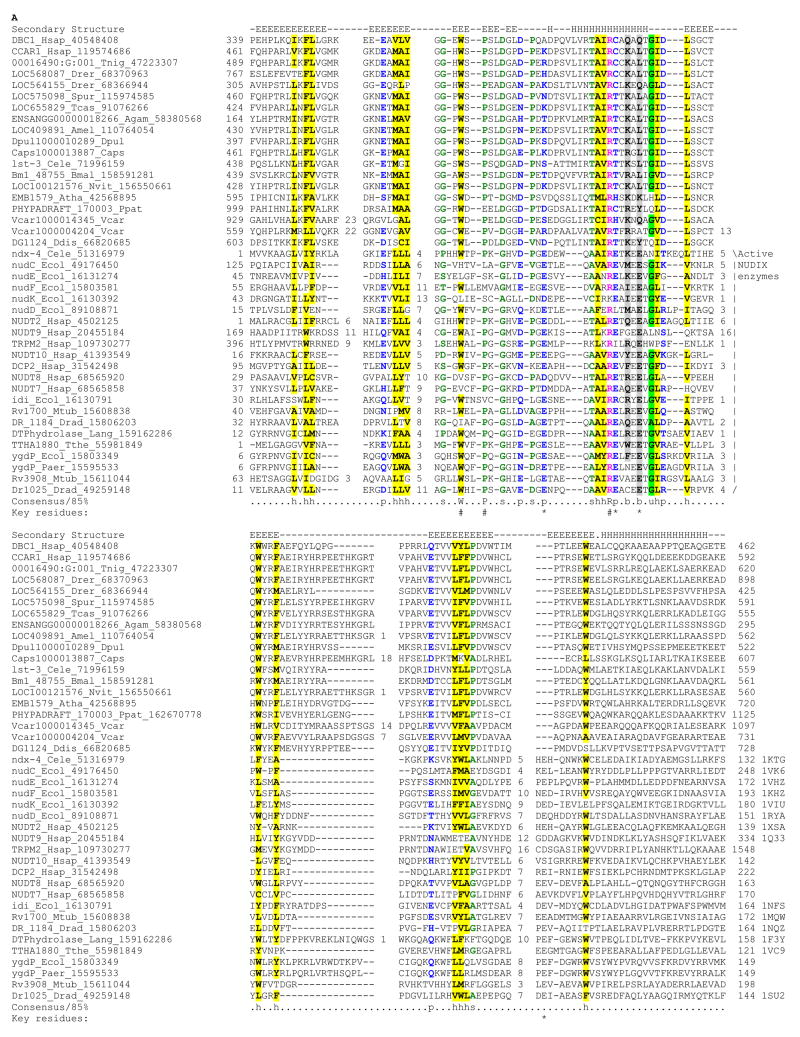
Fig. 1. A) Multiple alignment of Nudix domains of the DBC1 family with known representatives.
Multiple sequence alignment of the Nudix domain was constructed using Kalign after parsing high-scoring pairs from PSI-BLAST search results. The alignment was refined based on the pairwise alignments produced by the profile-profile searches with the HHpred program against the Nudix profile. The secondary structure from the crystal structures is shown above the alignment with E representing a strand and H a helix. The 85% consensus shown below the alignment was derived using the following amino acid classes: hydrophobic (h: ALICVMYFW, yellow shading); small (s: ACDGNPSTV, green); polar (p: CDEHKNQRST, blue) and its charged subset (c: DEHKR, pink), and big (b: FILMQRWYEK; grey shading). The limits of the domains are indicated by the residue positions, on each end of the sequence. The numbers within the alignment are non-conserved inserts that have not been shown. The key positions involved in stabilizing the substrate-binding cleft and constituting the active site motif of the active version are respectively marked with “#” and “*” symbols. The sequences are denoted by their gene name followed by the species abbreviation and GenBank Identifier (gi). The PDB ids, when, available are shown to the right end of the alignment. The species abbreviations are: Agam : Anopheles gambiae; Amel : Apis mellifera; Atha : Arabidopsis thaliana; Bmal : Brugia malayi; Caps : Capitella sp; Cbri : Caenorhabditis briggsae; Cele : Caenorhabditis elegans; Ddis : Dictyostelium discoideum; Dpul : Daphnia pulex; Drad : Deinococcus radiodurans; Drer : Danio rerio; Ecol : Escherichia coli; Hsap : Homo sapiens; Lang : Lupinus angustifolius; Mtub : Mycobacterium tuberculosis; Nvit : Nasonia vitripennis; Paer : Pseudomonas aeruginosa; Ppat : Physcomitrella patens; Spur : Strongylocentrotus purpuratus; Tcas : Tribolium castaneum; Tnig : Tetraodon nigroviridis; Tthe : Thermus thermophilus; Vcar : Volvox carteri
B) Multiple alignment of the N-terminal extension in DBC1
Multiple sequence alignment of the conserved N-terminal extension of the inactive Nudix domains in the DBC1 family was constructed as described above. The helical region that interacts with the SIRT1 in DBC1 is marked. The extended coiled coil region continues N-terminal to this region.
In this study, using sensitive sequence profile analysis methods and structural comparisons, we show that the largest central conserved globular domain present in the DBC1 family is an inactive version of the Nudix hydrolase (MutT) fold, and is likely to bind NAD derivatives such as ADP-ribose. Based on this we predict that DBC1 and its homologs are likely to regulate the activity of SIRT1 or related deacetylases by sensing the soluble products or substrates of the NAD-dependent deacetylation reaction.
Results and Discussion
Identification of an inactive version of the Nudix (MutT) domain in the DBC1 family
To computationally investigate the possible biochemical roles of DBC1 and CARP-1 we sought to systematically identify the conserved domains found in them. Due to the presence of large non-globular and compositionally biased regions in these proteins we first used the SEG program to objectively identify their globular domains (Materials and Methods). To determine the relationships of these predicted globular regions, we then queried them against a library of sensitive position-specific-score matrices (PSSMs) and hidden Markov models (HMM) of previously characterized domains 6. As a result, it became clear that DBC1, CARP-1 and all other members of the family shared two previously uncharacterized globular regions in addition to the C-terminal inactive EF-hand modules 7. All members of the DBC1 family from animals, and one of the two paralogs from the chlorophyte Volvox also contain an N-terminal S1-like RNA-binding domain (Fig. 2). Further, CARP-1 (but not DBC1) and all invertebrate members of the family possess a DNA-binding SAP domain inserted between the two uncharacterized globular domains shared by all members of the DBC1 family (Fig. 2).
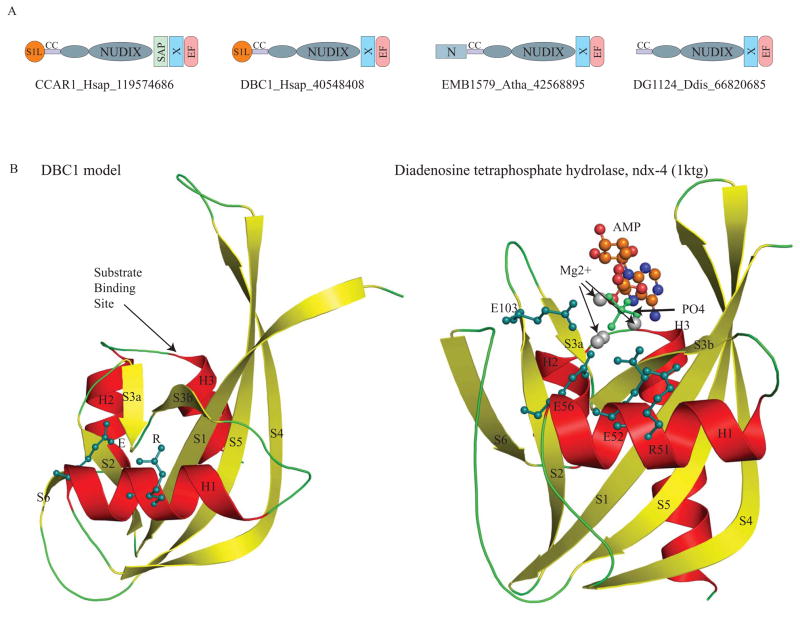
Fig. 2. A. Domain architectures of the DBC1 family.
The domain architectures found in the DBC1 family are shown. Domain architectures are labeled with a representative gene name, the species abbreviation, and the Genebank identifier (GI) number separated by underscores. The N-terminal extension of the Nudix domain is shown as a small ellipse. Domain abbreviations are: S1L – S1-like OB fold domain; EF – EF hand; CC – coiled coil region; and N – N terminal domain specific to plant members of the DBC1 family.
B. Model of the DBC1 Nudix domain compared with the crystal structure of the catalytically active version ndx-4 (pdb: 1ktgA).
The DBC1 model was constructed using DR1025, a nucleotide pyrophosphatase (PDB: 1SU2), DR1184, a CoA pyrophosphatase (PDB: 1NQZ), both from Deinococcus and ndx-4, a diadenosine tetraphosphate hydrolase(PDB: 1KTG) from Caenorhabditis, as templates. The crystal structure of ndx-4, a diadenosine tetraphosphate hydrolase (1KTG), is shown for comparison. The conserved active-site residues, the substrate binding site, Magnesium ions, phosphate ion and the substrate ADP are shown.
The larger of the two predicted uncharacterized globular domains shared by the DBC1 family is located just C-terminal to the conserved coiled-coil region, which, in DBC1, was shown to be required for the interaction of with SIRT1 4, 5 (Fig. 1). We searched the non-redundant database using a PSSM derived from an alignment that included all representatives of this predicted domain from the DBC1 family with the PSI-BLAST program. This search recovered several members of the Nudix hydrolase superfamily prior to convergence (e.g. diadenosine tetraphosphatase, gi: 4502125; NUDT8, gi: 32469515; Pyrobaculum Pcal_0237, gi: 126458862; expect-value=10−2). To further test the validity of this possible relationship we performed a HMM-HMM comparison with HHpred program, using the HMM of the above domain of the DBC1 family in a search against a panel of HMMs derived using the globular domains in the PDB database as seeds (Materials and Methods). The top hits in this search were the Nudix hydrolases with highly significant p-values (p=10−3–10−9). A reciprocal PSI-BLAST search with a PSSM prepared from an alignment of the Nudix domain including diverse members of the superfamily against a database of predicted proteins from all completely sequenced eukaryotic genomes also recovered members of the DBC1 family (e-value 10−2–10−4). Prediction of the secondary structure with the Jpred program by using a combination of residue frequencies in a column, HMM and PSSM derived from the alignment of this domain of the DBC1 family showed that the conserved core contained four conserved strands and a helix between strand 2 and strand 3 (Fig. 1). This secondary structure corresponds exactly to the β-grasp fold found in the Nudix hydrolases 8, which taken together with the results of the sequence profile analysis suggests that this conserved domain in the DBC1 family adopts the same fold as the Nudix hydrolases.
Nudix hydrolases hydrolyze diverse nucleoside diphosphates and contain a characteristic active site defined by a Gx5Ex7Rx4E motif on the conserved helix of their fold 9, 10 (Fig. 1). The acidic residues in this motif primarily participate in chelating Mg2+ ions and as a general base in catalysis 11 (Fig. 1). The conserved arginine, while not directly participating in substrate contact, interacts with surrounding residues and is thereby critical for maintaining the substrate-binding pocket. Interestingly, the multiple alignment of the above-detected Nudix domain in the DBC1 family with other catalytically active members of the superfamily showed that the former only conserved the arginine, but not the other key acidic residues in the active site motif (Fig 1, 2). This observation indicates the Nudix domain is likely to be catalytically inactive in all members of the DBC1 family. However, the strict conservation of the arginine belonging to the active-site motif suggested it might stabilize the binding pocket typical of the Nudix superfamily even in the DBC1 family. To further investigate this possibility, we used the crystal structures of diadenosine tetraphosphatase (PBD: 1su2 and 1ktg) 12 and an ADP-ribose pyrophosphatase (PBD: 1nqz) 13 as templates to construct a structural model for the inactive Nudix domain of DBC1. It must be stressed that such homology based model for distantly related sequences are only rough approximations of the actual structure. Nevertheless, in the absence of the actual structure it can be used in conjunction with the conservation pattern to explore certain aspects, such as the general features of the binding pocket in the DBC1 family. Importantly, the conservation of a key tryptophan, small residue (usually proline) suggested that the inactive DBC1 Nudix domain preserves the characteristic “outflow” which interrupts the second strand of the β-grasp fold and forms the base of the substrate binding pocket (Fig. 1, 2) 8. Further the model suggests that all the key residues lining the predicted active-site pocket are of comparable size to those observed in the active Nudix hydrolases, and are typically polar or positive charged. Hence, the Nudix domain of the DBC1 family, while catalytically inactive, would still be able to accommodate soluble ligands with a nucleoside diphosphate moiety.
Functional implications for the predicted ligand-binding Nudix domain in the DBC1 family
The above discovery of a potential nucleoside diphosphate-binding Nudix domain in the DBC1 family has considerable functional implications in light of the recent experimental evidence for DBC1 being a SIRT1 inhibitor. The sirtuins, like Sirt1, catalyze deacetylation of acetyl-lysine in histones and other proteins by using NAD, which in the process is converted to O-acetyl ADP-ribose (OAAR), while releasing nicotinamide 14. Both ADP sugars and NAD are well-known substrates of nudix hydrolases: for example, in animals NUDT9 is a specific ADP ribose hydrolase, whereas NudC from proteobacteria shows a substrate preference for NAD 10, 11. Given this precedence, we suggest that the inactive Nudix domain found in the DBC1 family is likely to bind either the substrate (NAD) or the product (OAAR) of the SIRT1 enzyme. In particular by binding the ADP-ribose derivatives produced by the SIRT1 reaction DBC1 could sense the presence of Sirtuin deacetylase activity and specifically down-regulate it. This suggestion is supported by close proximity of the SIRT1 -binding coiled-coil region and the inactive Nudix domain (Fig. 1). In addition to the sirtuins, action of other NAD-dependent enzymes, such as poly-ADP-ribose polymerase which are also involved in apoptosis, might also alter concentrations of NAD metabolites sensed by the DBC1 family 15. In this context, it is of interest to note that the calcium channel TRPM2 (NUDT9L1) also contains an intracellular Nudix domain with a partially disrupted active site 16. This Nudix domain has low turnover and mediates channel-opening through ADP-ribose-binding and its closing through gradual hydrolysis of the bound ADP-ribose. Presence of the conserved inactive EF-hand module in the DBC1 proteins suggests that, although they might not bind Ca2+, they might hetero-dimerize with other functional EF-hand proteins activated by the Ca2+ influx due to TRPM2-opening in response to high ADP-ribose concentration arising from elevated NAD metabolism.
Further, the presence of S1-like RNA-binding domains in several members of the DBC1 family suggests that they might additionally interact with RNA. Given that different S1-like domain proteins interact with both miRNAs 17 and mRNAs 18, it would be of interest to investigate if the regulation chromatin modification by the DBC1 family is also regulated by the presence of transcripts or miRNAs. Similarly, the presence of a SAP domain in some members of the DBC1 family, like CARP-1, suggests that these versions are likely to localize to scaffold or matrix attachment sites 19. Hence, in vertebrates, where both DBC1 and CARP-1 are present, there might be a compartmentalization of their roles -- the former functioning in the nuclear interior and the latter functioning in the nuclear periphery. Sequence profile searches initiated with different starting queries corresponding to the second uncharacterized globular domain in the DBC1 showed that it was an α-helical domain restricted to the DBC1 family. It might mediate an additional conserved interaction of this family with as yet unknown partners.
Conclusions
The recent finding that DBC1 interacts with the histone and p53 deacetylase Sirt1 has provided the first hints regarding the actual biochemical functions of these conserved multi-domain proteins. We present evidence that the DBC1 family contains a catalytically inactive, ligand-binding version of the Nudix domain located immediately C-terminal to the coiled-coil region which interacts with Sirt1. We hence speculate that the DBC1 family is likely to sense soluble products of NAD metabolism, such as ADP-sugars produced by the action of sirtuins, thereby regulating these deacetylases. This would imply that the Nudix domains of the DBC1 family are functionally analogous to the Macro domains found in histone macroH2A1, certain certain sirtuins and Swi2/Snf2 ATPases 6. Certain Macro domains are enzymes which convert ADPR-1″ monophosphate to ADPR 20, whereas others are catalytically inactive high-affinity ADPR-binding domains 15. The latter version of the Macro domain in macroH2A1 binds OAADPR formed by Sirt1 action and might thereby regulate local heterochromatin structure 21. The complex domain architectures of the DBC1 proteins suggest that they are likely to act in linking a variety of distinct regulatory process including chromatin protein deacetylation, NAD metabolism, apoptotic signaling and RNA metabolism. Given the involvement of members of the DBC1 family in tumorigenesis and apoptosis, the prediction of a soluble ligand-binding domain in the makes them attractive as potential targets for therapeutic intervention with small molecules.
Material and Methods
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the PSI-BLAST programs 22. Profile searches using the PSI-BLAST program were conducted either with a single sequence or a sequence with a PSSM used as the query, with a profile inclusion expectation (E) value threshold of 0.01, and were iterated until convergence 22. For all compositionally biased queries the correction using composition-based statistics was used in the PSI-BLAST searches 23. Multiple alignments were constructed using the Kalign program 24, followed by manual correction based on the PSI-BLAST results. The multiple alignment was used to create a HMM using the Hmmbuild program of the HMMER package 25. It was then optimized with Hmmcaliberate and the resulting profile was used to search a database of completely sequenced genomes using the Hmmsearch program of the HMMER package 25. HMM-HMM comparison searches were performed using the HHpred program 26, 27. The JPRED program 28 and the COILS program were used to predict secondary structure. Globular domains were predicted using the SEG program with the following parameters: window size 40, trigger complexity=3.4; extension complexity=3.75 29.
The Swiss-PDB viewer 30 and Pymol programs 31 were used to carry out manipulations of PDB files. The model was generated using SWISS-MODEL 32 (http://swissmodel.expasy.org/). Briefly, this process consisted of threading the human DBC1 Nudix domain on to the known structures of Nudix domains (templates) using the multiple sequence alignment and the alignment window of the SWISS-PDB viewer program. This alignment of the DBC1 Nudix domain to the template structures was then submitted for modeling to the SwissModel server. Energy minimization of the modeled domain was carried out using the GROMOS 43B1 force field incorporated in SwissModel.
Acknowledgments
The work by the authors was supported by the intramural funds of the National Library of Medicine, NIH.
References
- 1.Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci U S A. 2002;99:13647–52. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundararajan R, Chen G, Mukherjee C, White E. Caspase-dependent processing activates the proapoptotic activity of deleted in breast cancer-1 during tumor necrosis factor-alpha-mediated death signaling. Oncogene. 2005;24:4908–20. doi: 10.1038/sj.onc.1208681. [DOI] [PubMed] [Google Scholar]
- 3.Rishi AK, Zhang L, Yu Y, Jiang Y, Nautiyal J, Wali A, Fontana JA, Levi E, Majumdar AP. Cell cycle- and apoptosis-regulatory protein-1 is involved in apoptosis signaling by epidermal growth factor receptor. J Biol Chem. 2006;281:13188–98. doi: 10.1074/jbc.M512279200. [DOI] [PubMed] [Google Scholar]
- 4.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 5.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Lewit-Bentley A, Rety S. EF-hand calcium-binding proteins. Curr Opin Struct Biol. 2000;10:637–43. doi: 10.1016/s0959-440x(00)00142-1. [DOI] [PubMed] [Google Scholar]
- 8.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin EV. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–43. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys. 2005;433:129–43. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Ranatunga W, Hill EE, Mooster JL, Holbrook EL, Schulze-Gahmen U, Xu W, Bessman MJ, Brenner SE, Holbrook SR. Structural studies of the Nudix hydrolase DR1025 from Deinococcus radiodurans and its ligand complexes. J Mol Biol. 2004;339:103–16. doi: 10.1016/j.jmb.2004.01.065. [DOI] [PubMed] [Google Scholar]
- 13.Kang LW, Gabelli SB, Bianchet MA, Xu WL, Bessman MJ, Amzel LM. Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family. J Bacteriol. 2003;185:4110–8. doi: 10.1128/JB.185.14.4110-4118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–40. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–20. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naziroglu M. New molecular mechanisms on the activation of TRPM2 channels by oxidative stress and ADP-ribose. Neurochem Res. 2007;32:1990–2001. doi: 10.1007/s11064-007-9386-x. [DOI] [PubMed] [Google Scholar]
- 17.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–46. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 18.Abaza I, Gebauer F. Functional domains of Drosophila UNR in translational control. Rna. 2008 doi: 10.1261/rna.802908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–4. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 20.Shull NP, Spinelli SL, Phizicky EM. A highly specific phosphatase that acts on ADP-ribose 1″-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:650–60. doi: 10.1093/nar/gki211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–5. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lassmann T, Sonnhammer EL. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–9. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 26.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 27.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–11. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 29.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–85. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 30.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 32.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]