Abstract
Background
Progress in lung cancer is hampered by the lack of clinically useful diagnostic markers. The goal of this study was to provide a detailed evaluation of lung cancer tumor markers indicative of molecular abnormalities and to assess their diagnostic utility in non-small cell lung cancer (NSCLC) patients.
Methods
Quantitative real-time RT-PCR was used to determine LunX, CK19, CEA, VEGF-C and hnRNP A2/B1 mRNA levels in peripheral blood and pleural fluid from NSCLC patients, compared with those from patients with other epithelial cancer (esophagus cancer and breast cancer), benign lung disease (pneumonia and tuberculo pleurisy) and from healthy volunteers.
Results
In peripheral blood LunX mRNA was detectable in 75.0% (33/44) of patients with NSCLC, but not in patients with other epithelial cancer (0/28), benign lung disease (0/10) or in healthy volunteers (0/15). In contrast, all other genetic markers were detected in patients with either NSCLC, other epithelia cancer or benign lung disease, and in healthy volunteers. The expression level and positive rate of LunX mRNA in peripheral blood correlated with the pathologic stage of NSCLC (P < 0.001 and P = 0.010 respectively). Furthermore, LunX mRNA was detected in 92.9% (13/14) of malignant pleural fluid samples and was the only marker whose expression level was significantly different between malignant and benign pleural fluid (P < 0.001). Additionally, expression of LunX mRNA in the peripheral blood of NSCLC patients decreased shortly after clinical treatment (P = 0.005).
Conclusion
Of several commonly used genetic markers, LunX mRNA is the most specific gene marker for lung cancer and has potential diagnostic utility when measured in the peripheral blood and pleural fluid of NSCLC patients.
Background
Lung cancer is one of the leading causes of cancer death and has become an increasingly urgent worldwide health problem. Progress in lung cancer treatment is hampered by a lack of diagnostic markers useful in clinical practice. Tumor markers, including carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), squamous cell carcinoma (SCC) antigen, cytokeratin 19 (CK19), vascular endothelial growth factor-C (VEGF-C), heterogeneous ribonuclear proteins A2/B1 (hnRNP A2/B1), muc1, BJ-TSA-9, KS1/4 and lung-specific X protein (LunX), have been investigated for their putative diagnostic and prognostic value for lung cancer [1-7]. On the basis of RT-PCR analysis, CEA mRNA in blood cells or in lymph nodes and CK19 mRNA in mediastinal lymph nodes have been suggested as promising tools for the detection of micrometastatic cells in patients with lung cancer [8-11]. KS1/4 was shown to be the most sensitive marker for detecting metastatic NSCLC in mediastinal lymph nodes using real-time RT-PCR [6]. Determining CEA and NSE in pleural fluid could enhance the diagnostic yield for malignant effusion associated with lung cancer [12]. Additionally, accumulating evidence suggests that hnRNP B1 expression may be useful for the early diagnosis of lung cancer, provided that expression levels can be accurately quantified [5]. LunX, a novel human lung-specific gene, has been reported to be a superior diagnostic marker for the detection of micrometastases in lymph nodes and peripheral blood of NSCLC patients [1,6,13]. Although lots of studies have provided suggestive results, a definitive assessment of the relative value of these molecular markers for lung cancer is lacking.
A demonstration of the diagnostic utility of these various tumor markers requires a detailed, direct comparison using reliable, sensitive methodologies. Quantitative real-time RT-PCR is a development of the RT-PCR procedure, which is simple, rapid and automated. Most importantly, real-time RT-PCR analysis can yield accurate estimates of gene expression levels, differentiating between baseline levels of gene expression in normal tissue and increased levels in cancer cells [14,15]. Molecular diagnosis using RT-PCR technique can detect tumor marker-expressing cells undetectable by other means in patients with localized or metastatic cancer, and may offer the most effective solution for detecting micrometastases at the molecular level in various types of cancer patients [16].
The purpose of this study was to evaluate the known molecular markers,LunX, CK19, CEA, VEGF-C and hnRNP A2/B1, for their expression in lung cancer cells in peripheral blood and pleural fluid using real-time RT-PCR, with the ultimate goal of establishing a more reliable molecular diagnostic method as an adjunct to clinical decision-making.
Methods
Patients and clinical procedures
Patients with pathologically proven non-small cell lung cancer (NSCLC) identified by routine imaging and cytologic assessments were eligible for the study. The clinical characteristics of patients (peripheral blood and pleural fluid groups) were shown in Table 1. Patients with other epithelial cancer, including esophagus and breast cancer, were studied as a control. Additionally, 12 patients with NSCLC were investigated before and after treatment (Table 2). Patients with a history of malignancy were excluded from the study. Informed consent was obtained from each subject and the research was performed in compliance with the principles enunciated in the Helsinki Declaration with the approval of the Ethics Committee of the University of Science and Technology of China.
Table 1.
Clinical characteristics of patients in this study
| Peripheral blood | Pleural fluid | ||||||
| Lung cancer | Esophagus cancer | Breast cancer | Pneumonia | Healthy | Lung cancer | Tuberculo pleurisy | |
| Age mean (min-max) | 67(36–89) | 62(39–85) | 51(35–82) | 67(52–83) | 42(25–60) | 66(36–85) | 64(24–85) |
| Sex (M/F) | 31/13 | 5/3 | 0/20 | 8/2 | 8/7 | 9/5 | 12/2 |
| Pathologic stage | |||||||
| I | 8 | 1 | 3 | - | - | - | - |
| II | 8 | 3 | 8 | - | - | - | - |
| III | 14 | 2 | 5 | - | - | 5 | - |
| IV | 14 | 2 | 4 | - | - | 9 | - |
| Pathologic Typing | |||||||
| Squ 26 | Squ 6 | - | - | - | Squ 9 | - | |
| Ade 16 | Ade 2 | - | - | - | Ade 5 | - | |
| LCLC 2 | - | - | - | - | LCLC | - | |
| Total number | 44 | 8 | 20 | 10 | 15 | 14 | 14 |
Pathologic stage was determined as I, II, III or IV according to the American Joint Committee On Cancer (AJCC) TNM system. Squ = squamous epithelial carcinoma; Ade = adenocarcinoma; LCLC = large cell lung cancer.
Table 2.
Clinical characteristics and treatments of 12 NSCLC patients
| Patient | Sex | Age (y) | Pathologic Typing | Pathologic Stage | Means of treatment |
| 1 | F | 64 | LCLC | II | chemo-therapy for 7 days |
| 2 | M | 70 | Squ | III | chemo-therapy for 7 days |
| 3 | M | 70 | Squ | III | chemo-therapy for 7 days |
| 4 | F | 36 | Squ | III | chemo-therapy for 7 days |
| 5 | M | 75 | Ad | IV | chemo-therapy for 7 days |
| 6 | M | 62 | Squ | IV | interventional therapy |
| 7 | M | 65 | Ad | II | operation |
| 8 | F | 58 | Squ | IV | chemo-therapy for 7 days |
| 9 | M | 55 | Squ | III | interventional therapy |
| 10 | M | 58 | Squ | II | operation |
| 11 | M | 68 | Ad | I | operation |
| 12 | M | 61 | Ad | II | operation |
Peripheral blood samples from each patient were collected 1 day before and 7 days after treatment. Pathologic stage was determined as I, II, III or IV according to the American Joint Committee On Cancer (AJCC) TNM system. Squ = squamous epithelial carcinoma; Ade = adenocarcinoma; LCLC = large cell lung cancer.
Collection of specimens
3 ml peripheral blood (the first 2 ml peripheral blood had been discard for detection convenience) was collected and treated with RBC lysis buffer (RX-2-1-2, U-gene, China), and then nucleated cells were collected for the detection of biomarker mRNA. 10 ml pleural fluid was inspired from indicated patients, and centrifugated at 3500 rpm for 10 min to pellet cells.
RNA extraction and cDNA synthesis
All reagents were purchased from Invitrogen. Total cellular RNA was extracted using the TRIZOL reagent according to the protocol provided by the manufacturer. cDNA was generated form total RNA by reverse transcription (RT) in a reaction containing 4 μg total RNA, 5 μM oligo dT, 0.5 mM dNTP, 8 μl 5×Buffer, 10 mM DTT, 56 units RNase inhibitor, 400 units of M-MLV and distilled water (ultrapure, DNase and RNase free) in a total volume of 40 μl. The RT reaction was performed at 37°C for 50 minutes, followed by heating at 70°C for 15 minutes. All of the steps were performed using sterile technique in areas designated for RNA extraction and RT-PCR.
Real-time PCR
Quantitative real-time PCR was performed using real-time Taq-Man technology and an ABI PRISM 7000 sequence detector (Applied Biosystems, Foster City, CA). Gene-specific primers and Taq-Man probes were shown in Table 3. The standard reaction contained 25 μl 2×PCR buffer (ABsolute™ QPCR Mix, AB-1140/b), 0.5 U uracil N-glycosylase (UNG) Erase enzyme (Invitrogen, Cat NO.18054-015), 5 μl cDNA template, 0.4 μM forward and reverse primers, 0.25 μM hybridization probe in a total volume of 50 μl. The initial PCR step was at 37°C for 10 min to activate UNG erase, followed by a 15 min hold at 95°C. PCR reactions were performed using a total of 50 cycles consisting of a 15 s melt at 95°C, followed by a 1 min annealing/extension at 60°C for CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin or a 1 min annealing/extension at 56°C for LunX. During the DNA polymerization, the Taq-Man probe was hydrolyzed and fluorescence emitted. When the fluorescence signal reached 10 SDs of background, the threshold cycle (Ct) was noted. Each sample was analyzed in triplicate for each target gene, and mRNA was quantified by the standard curve method.
Table 3.
Primer pair and probe of each biomarker for real-time PCR
| Gene | Sequence of selected primer pair (5'-3') | Probe (5'-FAM---TAMRA-3') | Amplicon length (bp) | Ref |
| LunX | GCCTCATTGTCTTCTACGGGCTGTT; CTGAGGGCATTTGTCAAGCTTCCT |
CAGACCATGGCCCAGTTTGGAGGCCTG | 156 | This work |
| CK19 | ACTACAGCCACTACTACACGAC; CAGAGCCTGTTCCGTCTCAAAC |
TCTGGCTGCAGATGACTTCCGAACCA | 149 | [17] |
| CEA | AATAACGGGACCTATGCCTGTTT; CCCCAACCAGCACTCCAAT |
CATCTGGAACTTCTCCTGGTCTCTCAGCTG | 151 | This work |
| VEGF-C | TTCATTCCATTATTAGACGTTCCCT; GATTATTCCACATGTAATTGGTGGG |
CCAGCAACACTACCA CAGTGTCAGGCA | 94 | [18] |
| hnRNP A2/B1 | GACTGTGTGGTAATGAGGGATCCT; GCTCAACTACTCTCCCATCAATTGA |
CATGGCTGAGGTTGATGCTGCCAT | 133 | This work |
| β-actin | TTGCCGACAGGATGCAGAA; GCCGATCCACACGGAGTACTT |
TCATTGCTCCTCCTGAGC | 101 | This work |
Forward primer was shown as upper sequence and reverse primer was the lower sequence in the respective primer pair. The primers and probes for LunX (NM_130852), CEA (NM_004363), hnRNP A2/B1 (NM_031243) and β-actin (NM_001101.2) were designed using the Primer Express Software (Applied Biosystems). All primers were desalted when purified and the probes were HPLC purified.
Preparation of standard curves
LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin cDNA were generated from A549, SK-BR-3, SK-BR-3, K562, A549 and A549 cells (American Type Culture Collection, Rockville, MD), respectively, using the specific primers in Table 3. The PCR reaction consisted of an initial denaturation step at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C, annealing at 60°C for CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin or at 56°C for LunX, and extension at 72°C. After purification using a gel band purification kit (Invitrogen), the amplicon was ligated into a TA-cloning vector pCR 2.1 (Invitrogen, Groningen, The Netherlands) and then used to transform competent E. coli (DH5α). The sequence of the insert was verified using an ABI Prism BigDye terminator cycle sequencing ready reaction kit (PE Biosystems, Foster City, CA) and an ABI Prism 377 DNA sequencer. The number of plasmid copies was determined according to the formula: Copy number = . Serial dilutions from 1 × 102 to 1 × 108 copies of plasmids per microliter were made in TE buffer (10 mM Tris, 0.1 M EDTA, pH 8.0) in tubes lubricated with silicon and plasmids were detected by real-time PCR as described above. Standard curves were determined by plotting the Ct value against an initial copy number of standards (serially diluted plasmids) for LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin mRNA. Copy numbers of each gene marker of clinical samples were calculated by interpolating sample Ct value with standard curves of Ct values generated by serial dilution of the corresponding standard. The copy numbers of LunX, CK19, CEA, VEGF-C and hnRNP A2/B1were all further normalized to the copy number of β-actin in each sample.
Statistical analysis
SDS 1.0 software was used to analyze the results of real-time PCR. A regression analysis was applied to standard curves. K Independent Samples Test (Media Test) was used to compare the gene expression levels in peripheral blood among NSCLC patients at different pathologic stages. Mann-Whitney U Test was used to compare the gene expression levels in pleural fluid between NSCLC and tuberculo pleurisy patients. Wilcoxon Signed Ranks Test was used for the analysis of gene expression levels in peripheral blood of NSCLC patients before and after clinical treatment. In cases where the results of gene expression were negative, the data were treated as 0 for statistical convenience. χ2 test was used to analyze the positive detection rate. A value of P < 0.05 (2-tailed test) was considered significant.
Results
Establishment of real-time RT-PCR procedures for the gene markers
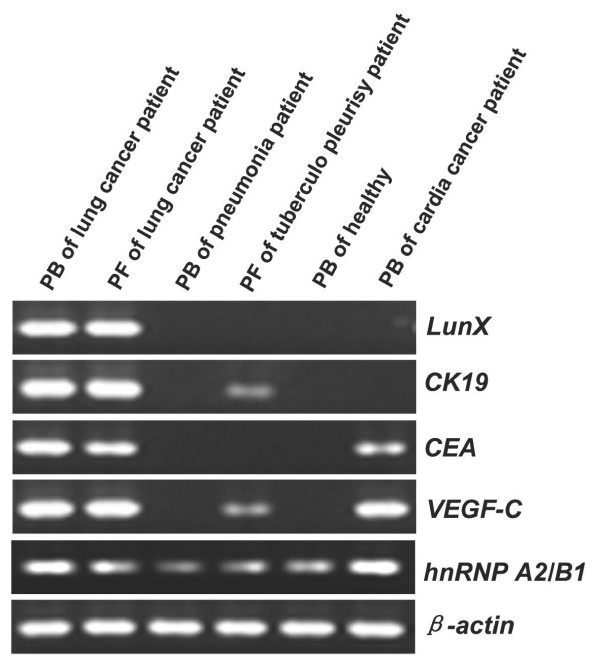
The standard curves for LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin mRNA detection were established using specific plasmids, as described in methods. The amplification efficiencies of the PCR reactions were 99.3% for hnRNPA2/B1, 100% for LunX, CEA and VEGF-C, 101% for β-actin and 103% for CK19, indicating a near-perfect doubling (100%) of product with each amplification cycle. Using established procedures, each biomarker was detected in specific specimens and the products were visualized on ethidium bromide-stained agarose gel (Figure 1). The expression patterns were consistent with the performance of each biomarker in these specimens, which further validated the RT-PCR procedures established for each biomarker in this study.
Figure 1.
Products of each gene marker by real-time RT-PCR. Each biomarker was detected by real-time RT-PCR in the indicated samples, as described in methods. Products were separated by electrophoresis and visualized in ethidium bromide-stained agarose gels. PB = Peripheral blood; PF = Pleural fluid.
LunX mRNA is the most specific gene marker for NSCLC cells in peripheral blood
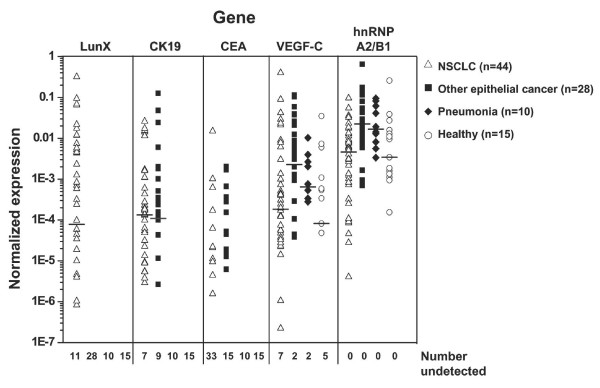
As shown in Figure 2, LunX mRNA was detectable in the peripheral blood from only NSCLC patients; LunX mRNA was not detectable in the peripheral blood from patients with other epithelial cancer, patients with pneumonia, or healthy volunteers. In contrast, CK19 mRNA could be detectable not only in peripheral blood from NSCLC patients but also in blood from other epithelial cancer patients, and there was no significant difference in the expression levels of CK19 mRNA between the two groups (P = 0.667). As with CK19, the difference in CEA mRNA between the two groups was not significant (P = 0.050). VEGF-C and hnRNP A2/B1mRNA were present at high levels in peripheral blood samples from NSCLC patients, other epithelial cancer patients, pneumonia patients and the healthy, obviously ruling out these genes as effective gene markers for lung cancer cells in peripheral blood.
Figure 2.
LunX mRNA is the most specific gene marker for lung cancer cells in peripheral blood. LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin mRNA from each peripheral blood sample were detected by real-time RT-PCR, and mRNA copy number was determined by reference to the standard curve, as described in methods. The copy number of each mRNA was further normalized as the ratio to the copy number of β-actin in all samples. For each gene marker, when the copy number was less than 100, it could not be detectable as a negative case. All the negative results of each indicated gene were shown as number undetected. The median is marked as "---" in each group. Where the frequency of negative cases is > 50%, the median cannot be shown.
Further, for NSCLC, the positive detection rate of LunX mRNA was 75.0% (33 of 44) in peripheral blood, which was similar to the positive detection rate of CK19 mRNA (84.1%, 37 of 44) (P = 0.290), but much higher than the positive detection rate of CEA mRNA (25.0%, 11 of 44) (P < 0.001) (Table 4). Although CK19 mRNA appeared to be a sensitive NSCLC detector in peripheral blood, there was no significant difference in the positive detection rate between NSCLC and other epithelial cancer groups (P = 0.106). The frequency of CEA mRNA detection in peripheral blood was likewise not significantly different between NSCLC and other epithelial cancer groups (P = 0.060), providing further confirmation that CK19 and CEA mRNA were not specific to lung cancer cells.
Table 4.
Number of positive cases by each biomarker in peripheral blood
| Group | Total number | Positive Detection Rate | ||||
| LunX | CK19 | CEA | VEGF-C | hnRNP A2/B1 | ||
| NSCLC | 44 | 33/44 (75.0%) | 37/44 (84.1%) | 11/44 (25.0%) | 37/44 (84.1%) | 44/44 (100%) |
| Other epithelial cancer | 28 | 0/28 (0%) | 19/28 (67.9%) | 13/28 (46.4%) | 26/28 (92.9%) | 28/28 (100%) |
| Pneumonia | 10 | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 8/10 (80.0%) | 10/10 (100%) |
| Healthy | 15 | 0/15 (0%) | 0/15 (0%) | 0/15 (0%) | 10/15 (66.7%) | 15/15 (100%) |
| P value | < 0.001* | < 0.001* | 0.002* | 0.174* | - | |
| < 0.001# | 0.106# | 0.060# | - | - | ||
| / | 0.290$ | < 0.001$ | - | - | ||
For each gene marker, when the copy number was less than 100, it could not be detectable as a negative case using our established real-time PCR assay. The positive detection rate was calculated by the number of positive cases/total number. * represented the analysis among NSCLC, other epithelial cancer, pneumonia and healthy groups for each biomarker. # represented the analysis between NSCLC and other epithelial cancer groups for each biomarker. $ represented the analysis between the biomarker and LunX for NSCLC. P values were calculated using χ2 test.
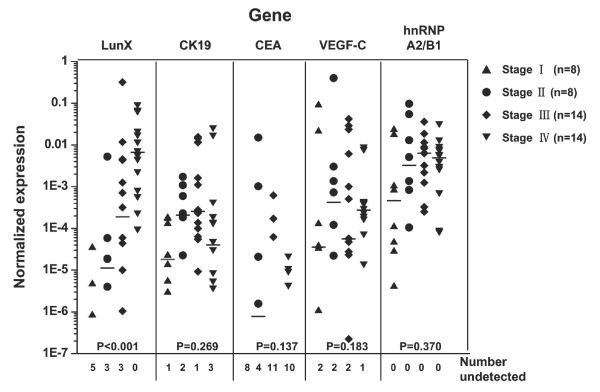
Expression of LunX mRNA in peripheral blood is correlated with the pathologic stage of NSCLC
The correlation of LunX mRNA expression in peripheral blood of NSCLC patients and clinical factors was further investigated, as shown in Table 5. Gender, age and pathologic type were not associated with the positive detection rate of LunX mRNA in peripheral blood (P = 0.461, 0.425 and 0.482 respectively). However, there was an association between different pathologic stages and the LunX mRNA positive detection rate in peripheral blood (P = 0.010), which increased with increasing clinical severity. A similar relationship between the levels of LunX mRNA expression in the peripheral blood of NSCLC patients and pathological stages was found (P < 0.001) (Figure 3). There was no significant difference in the mRNA expression levels of the other gene markers, CK19 (P = 0.269), CEA (P = 0.137), VEGF-C (P = 0.183) or hnRNP A2/B1 (P = 0.370), in peripheral blood among NSCLC patients at different pathologic stages (Figure 3).
Table 5.
Correlation between LunX mRNA in the peripheral blood and clinical factor in NSCLC patients
| Clinical factor | Positive cases/total cases |
| Sex | |
| P value | 0.461 |
| Male | 22/31 (71.0%) |
| Female | 11/13 (84.6%) |
| Age | |
| P value | 0.425 |
| ≤ 60 | 7/11 (63.6%) |
| > 60 | 26/33 (78.8%) |
| Pathologic Type | |
| P value | 0.482 |
| Squ | 18/26 (69.2%) |
| Ade | 13/16 (81.3%) |
| LCLC | 2/2 (100%) |
| Pathologic Stage | |
| P value | 0.010 |
| I | 3/8 (37.5%) |
| II | 5/8 (62.5%) |
| III | 11/14 (78.6%) |
| IV | 14/14 (100%) |
P values were calculated using χ2 test.
Figure 3.
Expression level of LunX mRNA in peripheral blood is correlated with the pathologic stage of NSCLC. LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin mRNA from each peripheral blood sample were detected by real-time RT-PCR, and mRNA copy number was determined by reference to the standard curve, as described in methods. The copy number of each mRNA was further normalized as the ratio to the copy number of β-actin. For each gene marker, when the copy number was less than 100, it could not be detectable as a negative case. All the negative results of each indicated gene were shown as number undetected. The median is marked as "---" in each group. Where the frequency of negative cases is > 50%, the median cannot be shown. The K Independent Samples Test (Media Test) was used to analyze gene expression levels among NSCLC patients at different pathologic stages. P < 0.05 (2-tailed test) was considered significant.
LunX mRNA is the most specific and sensitive gene marker for NSCLC cells in pleural fluid
Pleural fluid can be caused by several kinds of diseases, including malignant lung cancer and benign lung disease. Distinguishing between malignant pleural fluid and benign pleural fluid is an urgent clinical concern. As shown in Figure 4 and Table 6, the detection rates of LunX (92.9%), CK19 (100%), CEA (85.7%), VEGF-C (78.6%) and hnRNP A2/B1 (100%) in the malignant pleural fluid from NSCLC patients were all high. However, only LunX mRNA was appropriately infrequent in the benign pleural fluid from patients with tuberculo pleurisy (7.1%, 1 of 14); all remaining markers were present in the benign pleural fluid at high frequencies (100%, 42.9%, 64.3% and 100% for CK19, CEA, VEGF-C and hnRNP A2/B1 mRNA, respectively) (Figure 4, Table 6). There were also no significant differences in the expression levels of CK19 (P = 0.066), CEA (P = 0.074),VEGF-C (P = 0.054), or hnRNP A2/B1 (P = 0.613) mRNA between malignant and benign pleural fluid (Figure 4). Only the expression level of LunX mRNA was significantly different between malignant and benign pleural fluid (P < 0.001) (Figure 4). Thus, LunX mRNA was the most specific biomarker with high sensitivity (13/14, 92.9%) among these detected markers for the differential diagnosis of NSCLC from pleural fluid.
Figure 4.
LunX mRNA is the most specific gene marker with high sensitivity for NSCLC cells in pleural fluid. LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin mRNA from each pleural fluid sample were detected by real-time RT-PCR, and mRNA copy number was determined by reference to the standard curve, as described in methods. The copy number of each mRNA was further normalized as the ratio to the copy number of β-actin. For each gene marker, when the copy number was less than 100, it could not be detectable as a negative case. All the negative results of each indicated gene were shown as number undetected. The median is marked as "---" in each group. Where the frequency of negative cases is > 50%, the median cannot be shown. The Mann-Whitney U Test was used to compare the gene expression levels between NSCLC and tuberculo pleurisy groups. P < 0.05 (2-tailed test) was considered significant.
Table 6.
Number of positive cases by each biomarker in pleural fluid
| Group | Total number | Positive Detection Rate | ||||
| LunX | CK19 | CEA | VEGF-C | hnRNP A2/B1 | ||
| NSCLC | 14 | 13/14 (92.9%) | 14/14 (100%) | 12/14 (85.7%) | 11/14 (78.6%) | 14/14 (100%) |
| Tuberculo pleurisy | 14 | 1/14 (7.1%) | 14/14 (100%) | 6/14 (42.9%) | 9/14 (64.3%) | 14/14 (100%) |
| P value | < 0.001 | - | 0.018 | 0.678 | - | |
For each gene marker, when the copy number was less than 100, it could not be detectable as a negative case using our established real-time PCR assay. The positive detection rate was calculated by the number of positive cases/total number. P values were calculated using χ2 test.
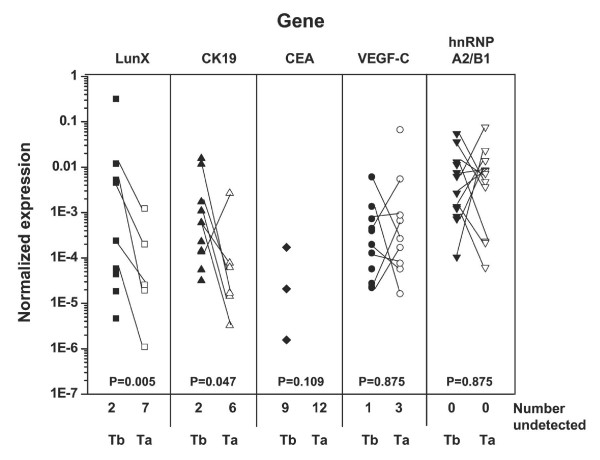
Expression of LunX mRNA in peripheral blood decreases shortly following treatment of NSCLC
An important attribute of a clinically useful diagnostic marker is the ability to promptly demonstrate the status of lung cancer after relevant treatments. In this study, LunX, CK19, CEA, VEGF-C and hnRNP A2/B1 mRNA in peripheral blood were tracked in 12 NSCLC patients before and after treatments (Table 2). As shown in Figure 5, the expression levels of LunX and CK19 mRNA in peripheral blood decreased significantly in NSCLC patients after treatment (P = 0.005 and P = 0.047, respectively). Treatment did not significantly affect the expression of VEGF-C or hnRNP A2/B1 mRNA (P = 0.875 and P = 0.875, respectively) (Figure 5), which, as demonstrated above, were detectable at high levels in peripheral blood from both patients and healthy (Figure 2). There was also no significant difference in the expression of CEA mRNA after treatment (P = 0.109) (Figure 5). These results, taken together with the demonstrated specificity of LunX mRNA for lung cancer cells and the correlation of LunX mRNA levels with NSCLC pathologic stages, indicated that LunX mRNA in peripheral blood might be a useful diagnostic marker for assessing the therapeutic effect on lung cancer.
Figure 5.
Expression of LunX mRNA in peripheral blood decreases shortly following the treatment of NSCLC. Peripheral blood samples from 12 NSCLC patients were collected 1 day before and 7 days after treatment as shown in Table 2. LunX, CK19, CEA, VEGF-C, hnRNP A2/B1 and β-actin mRNA were detected by real-time RT-PCR, and mRNA copy number was determined by reference to the standard curve, as described in methods. "Tb" represents 1 day before treatment and "Ta" represents 7 days after treatment. The copy number of each mRNA was further normalized as the ratio to the copy number of β-actin. For each gene marker, when the copy number was less than 100, it could not be detectable as a negative case. All the negative results of each indicated gene were shown as number undetected. The Wilcoxon Signed Ranks Test was used to analyze the gene expression levels before and after clinical treatment. P < 0.05 (2-tailed test) was considered significant.
Discussion
The current staging evaluation for lung cancer is based on the presence or absence of disease in lymph nodes (hilar and mediastinal) and distant organs (principally bone, brain, adrenal glands, and liver) [19]. Approximately 35% of patients are diagnosed at early stage and, as such, are candidates for curative lung resection. However, 50% of these patients will develop metastases and die from their disease. Furthermore, about one fourth of patients at the earliest stage of NSCLC (pathologically confirmed stage I) die of tumor recurrence after radical surgery, indicating that undetected metastases are present at the time of surgery and demonstrating that conventional staging techniques lack the sensitivity necessary to properly characterize patients [20,21]. Cancer cells can be released from a primary site and spread via the bloodstream to form a micrometastatic deposit in distant organs [1]. However, due to their extremely low concentration, these circulating tumor cells in peripheral blood are difficult to detect. Thus, developing sensitive and specific detection methods for cancer cells in peripheral blood may have important diagnostic, prognostic and therapeutic implications [22].
In this study, we have developed quantitative real-time RT-PCR procedures to detect potentially diagnostic tumor markers (Figure 1). The detection of metastatic cancer cells by RT-PCR is possible because cancer cells continue to express genetic markers specific to the tissue from which they originate, but which are not normally expressed in tissue compartments that frequently harbor metastatic foci [23,24]. Using this RT-PCR approach, which is ideal for the detection of genes expressed at low levels [13], we have assessed the expression of the known molecular markers,LunX, CK19, CEA, VEGF-C and hnRNP A2/B1, in lung cancer cells in peripheral blood and pleural fluid. Although LunX has been previously reported to be the most sensitive marker among the five genes LunX, muc1, KS1/4, CEA and CK19, for detecting circulating NSCLC cells by real-time RT-PCR in a study distinguishing patients with NSCLC from healthy volunteers, the specificity of LunX for lung cancer cells has not been tested [13]. Ours is the first study to directly compare the expression of LunX with other biomarkers in peripheral blood and pleural fluid, not only from NSCLC patients but also from patients with other epithelial cancer or benign lung disease and healthy volunteers. We have found that LunX mRNA is the most specific marker for lung cancer cells in peripheral blood (Figure 2, Table 4) and pleural fluid (Figure 4, Table 6). Compared with LunX mRNA, CK19 and CEA mRNA were over-expressed in other epithelial cancers, such as breast and esophagus cancer (Figure 2, Table 4), thus limiting their specificity for detection of lung cancer cells in peripheral blood. Because of lack of specificity, VEGF-C and hnRNP A2/B1mRNA were found to be similarly ineffective as genetic markers for lung cancer cells in our quantitative real-time RT-PCR assay (Figure 2, Table 4).
Further, it was demonstrated that when one lung cancer cell (A549 cell) was added into 3 ml peripheral blood, LunX mRNA could be detectable as a positive case in our established method (data not shown). For NSCLC patients, the positive detection rate of LunX mRNA in peripheral blood was high, almost as high as that of CK19 mRNA, and much higher than that of CEA mRNA (Table 4). Using RT-PCR, KS1/4 was previously reported to be the most sensitive marker among CEA, CK19, KS1/4, LunX, muc1 and PDEF for the detection of metastatic NSCLC in mediastinal lymph nodes, and LunX was with the second highest sensitivity distinguishing lung cancer from lung benign disease [6]. However,KS1/4 encodes a glycoprotein that is expressed on epithelial cells and is also present on epithelial cancers, thus, like CEA and CK19, KS1/4 is not specific to lung tissue. Another novel tumor-specific gene BJ-TSA-9 was reported to be a marker for circulating cancer cells in lung cancer patients, but BJ-TSA-9 alone was not sensitive enough to detect disseminated cancer cells in peripheral blood, and a combination of BJ-TSA-9 with LunX and SCC was required [7]. BJ-TSA-9 also suffers from the same tissue specificity problem that plagues CEA and CK19. In contrast, expression of the human LunX gene is lung-specific, and mRNA could be detected at a concentration of 10-4 μg cancer RNA in 1 μg normal lymph node RNA [1].
A malignant pleural effusion may be the initial presentation of cancer in 10–50% of patients [25]. Cytology is the standard method for the diagnosis of malignant effusion, but the sensitivity of cytology was not good enough [26]. Although an aggressive diagnostic technique thoracoscopy can be used to establish the diagnosis with a higher sensitivity (~90%), this procedure may not be available at all facilities and/or may be too invasive for many patients [27]. The evaluation of tumor markers in pleural fluid thus represents an alternative method for establishing the diagnosis of malignant pleural effusion. In this study, quantitative real-time RT-PCR was performed, for the first time, on pleural fluid. As was the case with peripheral blood, LunX mRNA was the most specific marker in malignant pleural fluid, showing a high positive detection rate in lung cancer (13 of 14, 92.9%), compared with the other gene markers CK19, CEA, VEGF-C and hnRNP A2/B1 mRNA (Figure 4, Table 6). Because determining the presence of circulating cancer cells in the peripheral blood of NSCLC patients is significant for early disease diagnosis and clinical therapy, a test for LunX mRNA that is able to reveal small amounts of lung cancer cells in peripheral blood with high specificity and sensitivity would be a great benefit in the clinical management of lung cancer. The potential value of differential LunX mRNA expression in pleural fluid for diagnosing malignant effusion reinforces this view.
Recent mass spectroscopy studies indicate that the human LunX gene product is also expressed in normal adult nasal lavage fluid. Further, this expression may be up-regulated in response to certain airway irritants, such as cigarette smoke and dimethylbenzylamine [28,29]. Nasal lavage fluid contains a large number of proteins that altogether comprise a potential source for detecting and characterizing biochemical alterations associated with airway diseases. In the present study, nasal lavage fluid samples were not investigated, so the element of smoking was not addressed in comparisons between NSCLC patients, other epithelial cancer patients, benign lung disease patients and healthy volunteers. Furthermore, as shown in Figure 2 and Table 4, LunX gene was not detectable in the peripheral blood of healthy volunteers, whether smokers or non-smokers. To date, the functional role of LunX protein remains unknown; however, LunX mRNA expression is known to be significantly enhanced in NSCLC tumors compared with corresponding cancer-free lung tissues [1]. In our study, we found that the expression level of LunX mRNA in peripheral blood correlated with the pathologic stage of NSCLC (Table 5, Figure 3). The more severe the disease was, the higher the expression level of LunX mRNA. Thus, the expression level of LunX mRNA in peripheral blood might be a valuable tool for staging NSCLC patients clinically. Furthermore, the expression of LunX mRNA in peripheral blood was sensitive to be influenced by the treatment of NSCLC patients (Figure 5), indicating that LunX mRNA expression might be associated with lung cancer progression.
Conclusion
In this study, we demonstrate that the detection of LunX mRNA in peripheral blood and pleural fluid by quantitative real-time RT-PCR provides a specific and sensitive indication of the presence of lung cancer cells. To our knowledge, LunX mRNA is the most specific NSCLC gene marker currently identified, and has tremendous clinical potential as an NSCLC diagnostic tool. Because LunX mRNA levels correlate with clinical severity and change in response to treatment protocols, this marker may prove valuable for NSCLC patients in the clinical decision-making process.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MC carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. YC performed the statistical analysis and drafted the manuscript. XY conceived of the study and participated in its coordination. ZT participated in the design of the study. HW participated in the design of the study and its coordination. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was supported by Natural Science Foundation of China (#30721002).
Contributor Information
Min Cheng, Email: chengmin@ustc.edu.cn.
Yongyan Chen, Email: ychen04@mail.ustc.edu.cn.
Xiaoqing Yu, Email: yu-xiaoqin@21cn.com.
Zhigang Tian, Email: tzg@ustc.edu.cn.
Haiming Wei, Email: ustcwhm@ustc.edu.cn.
References
- Iwao K, Watanabe T, Fujiwara Y, Takami K, Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M, Tanigami A. Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small-cell lung cancer. Int J Cancer. 2001;91:433–437. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1059>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Wieskopf B, Demangeat C, Purohit A, Stenger R, Gries P, Kreisman H, Quoix E. Cyfra 21-1 as a biologic marker of non-small cell lung cancer. Evaluation of sensitivity, specificity, and prognostic role. Chest. 1995;108:163–169. doi: 10.1378/chest.108.1.163. [DOI] [PubMed] [Google Scholar]
- Thomas D Chanin, Daniel T Merrick, Wilbur A Franklin, Fred R Hirsch. Recent developments in biomarkers for the early detection of lung cancer: perspectives based on publications 2003 to present. Current Opinion in Pulmonary Medicine. 2004;10:242–247. doi: 10.1097/01.mcp.0000130321.11513.13. [DOI] [PubMed] [Google Scholar]
- Lantuejoul S, Constantin B, Drabkin H, Brambilla C, Roche J, Brambilla E. Expression of VEGF, semaphorin SEMA3F, and their common receptors neuropilins NP1 and NP2 in preinvasive bronchial lesions, lung tumours, and cell lines. J Pathol. 2003;200:336–347. doi: 10.1002/path.1367. [DOI] [PubMed] [Google Scholar]
- Snead DR, Perunovic B, Cullen N, Needham M, Dhillon DP, Satoh H, Kamma H. hnRNP B1 expression in benign and malignant lung disease. J Pathol. 2003;200:88–94. doi: 10.1002/path.1292. [DOI] [PubMed] [Google Scholar]
- Wallace MB, Block MI, Gillanders W, Ravenel J, Hoffman BJ, Reed CE, Fraig M, Cole D, Mitas M. Accurate molecular detection of non-small cell lung cancer metastases in mediastinal lymph nodes sampled by endoscopic ultrasound-guided needle aspiration. Chest. 2005;127:430–437. doi: 10.1378/chest.127.2.430. [DOI] [PubMed] [Google Scholar]
- Li Y, Dong X, Yin Y, Su Y, Xu Q, Zhang Y, Pang X, Zhang Y, Chen W. BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia. 2005;7:1073–1080. doi: 10.1593/neo.05406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosotti M, Falleni M, Palleschi A, Pellegrini C, Alessi F, Bosari S, Santambrogio L. Quantitative Real-time Polymerase Chain Reaction Detection of Lymph Node Lung Cancer Micrometastasis Using Carcinoembryonic Antigen Marker. Chest. 2005;128:1539–1544. doi: 10.1378/chest.128.3.1539. [DOI] [PubMed] [Google Scholar]
- Castaldo G, Tomaiuolo R, Sanduzzi A, Bocchino ML, Ponticiello A, Barra E, Vitale D, Bariffi F, Sacchetti L, Salvatore F. Lung cancer metastatic cells detected in blood by reverse transcriptase-polymerase chain reaction and dot-blot analysis. J Clin Oncol. 1997;15:3388–3393. doi: 10.1200/JCO.1997.15.11.3388. [DOI] [PubMed] [Google Scholar]
- Le Pimpec-Barthes F, Danel C, Lacave R, Ricci S, Bry X, Lancelin F, Leber C, Milleron B, Fleury-Feith J, Riquet M, Bernaudin JF. Association of CK19 mRNA detection of occult cancer cells in mediastinal lymph nodes in non-small cell lung carcinoma and high risk of early recurrence. Eur J Cancer. 2005;41:306–312. doi: 10.1016/j.ejca.2004.09.021. [DOI] [PubMed] [Google Scholar]
- D'Cunha J, Corfits AL, Herndon JE, 2nd, Kern JA, Kohman LJ, Patterson GA, Kratzke RA, Maddaus MA. Molecular staging of lung cancer: real-time polymerase chain reaction estimation of lymph node micrometastatic tumor cell burden in stage I non-small sell lung cancer; preliminary results of Cancer and Leukemia Group B Trial 9761. J Thorac Cardiovasc Surg. 2002;123:484–491. doi: 10.1067/mtc.2002.119883. [DOI] [PubMed] [Google Scholar]
- Lee JH, Chang JH. Diagnostic utility of serum and pleural fluid carcinoembryonic antigen, neuron-specific enolase, and cytokeratin 19 fragments in patients with effusions from primary lung cancer. Chest. 2005;128:2298–2303. doi: 10.1378/chest.128.4.2298. [DOI] [PubMed] [Google Scholar]
- Mitas M, Hoover L, Silvestri G, Reed C, Green M, Turrisi AT, Sherman C, Mikhitarian K, Cole DJ, Block MI, Gillanders WE. Lunx Is a Superior Molecular Marker for Detection of Non-Small Lung Cell Cancer in Peripheral Blood. J Mol Diagn. 2003;5:237–242. doi: 10.1016/s1525-1578(10)60480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Olivi M, Champeme MH, Vidaud D, Lidereau R, Vidaud M. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int J Cancer. 1998;78:661–666. doi: 10.1002/(SICI)1097-0215(19981123)78:5<661::AID-IJC22>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assay. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Raj GV, Moreno JG, Gomella LG. Utilization of polymerase chain reaction technology in the detection of solid tumors. Cancer. 1998;82:1419–1442. doi: 10.1002/(SICI)1097-0142(19980415)82:8<1419::AID-CNCR1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Schröder CP, Ruiters MH, de Jong S, Tiebosch AT, Wesseling J, Veenstra R, de Vries J, Hoekstra HJ, de Leij LF, de Vries EG. Detection of micrometastatic breast cancer by means of real time quantitative RT-PCR and immunostaining in perioperative blood samples and sentinel nodes. Int J Cancer. 2003;106:611–618. doi: 10.1002/ijc.11295. [DOI] [PubMed] [Google Scholar]
- Hung CJ, Ginzinger DG, Zarnegar R, Kanauchi H, Wong MG, Kebebew E, Clark OH, Duh QY. Expression of vascular endothelial growth factor-C in benign and malignant thyroid tumors. J Clin Endocrinol Metab. 2003;88:3694–3699. doi: 10.1210/jc.2003-030080. [DOI] [PubMed] [Google Scholar]
- Mountain CF, Dresler CM. Regional lymph node classification for lung cancer staging. Chest. 1997;111:1718–1723. doi: 10.1378/chest.111.6.1718. [DOI] [PubMed] [Google Scholar]
- Martini N, Bains MS, Burt ME, Zakowski MF, McCormack P, Rusch VW, Ginsberg RJ. Incidence of local recurrence and second primary tumors in resected stage I lung cancer. J Thorac Cardiovasc Surg. 1995;109:120–129. doi: 10.1016/S0022-5223(95)70427-2. [DOI] [PubMed] [Google Scholar]
- Pantel K, Cote RJ, Fodstad O. Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst. 1999;91:1113–1124. doi: 10.1093/jnci/91.13.1113. [DOI] [PubMed] [Google Scholar]
- Matsunaga H, Hangai N, Aso Y, Okano K, Kawamura M, Kobayashi K, Kambara H, Hoger JH, Mitsuhashi M. Application of differential display to identify genes for lung cancer detection in peripheral blood. Int J Cancer. 2002;100:592–599. doi: 10.1002/ijc.10534. [DOI] [PubMed] [Google Scholar]
- Ghossein RA, Carusone L, Bhattacharya S. Molecular detection of micrometastases and circulating tumor cells in melanoma prostatic and breast carcinomas. In Vivo. 2000;14:237–250. [PubMed] [Google Scholar]
- Datta YH, Adams PT, Drobyski WR, Ethier SP, Terry VH, Roth MS. Sensitive detection of occult breast cancer by the reverse-transcriptase polymerase chain reaction. J Clin Oncol. 1994;12:475–482. doi: 10.1200/JCO.1994.12.3.475. [DOI] [PubMed] [Google Scholar]
- Fenton KN, Richardson JD. Diagnosis and management of malignant pleural effusions. Am J Surg. 1995;170:69–74. doi: 10.1016/S0002-9610(99)80257-8. [DOI] [PubMed] [Google Scholar]
- Nance KV, Shermer RW, Askin FB. Diagnostic efficacy of pleural biopsy as compared with that of pleural fluid examination. Mod Pathol. 1991;4:320–324. [PubMed] [Google Scholar]
- Light RW. Pleural diseases. 4. Philadelphia, PA: Lippincott, Williams & Wilkins; 2001. Pleural effusions related to metastatic malignancies; pp. 108–34. [Google Scholar]
- Lindahl M, Stahlbom B, Tagesson C. Identification of a new potential airway irritation marker, palate lung nasal epithelial clone protein, in human nasal lavage fluid with two-dimensional electrophoresis and matrix-assisted laser desorption/ionization time of flight. Electrophoresis. 2001;22:1795–1800. doi: 10.1002/1522-2683(200105)22:9<1795::AID-ELPS1795>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ghafouri B, Stahlbom B, Tagesson C, Lindahl M. Newly identified proteins in human nasal lavage fluid from nonsmokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics. 2002;2:112–120. doi: 10.1002/1615-9861(200201)2:1<112::AID-PROT112>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]