Abstract
Methanothermobacter thermautotrophicus uses lysine for both protein synthesis and cross-linking pseudomurein in its cell wall. A diaminopimelate aminotransferase enzyme from this methanogen (MTH0052) converts tetrahydrodipicolinate to L,L-diaminopimelate, a lysine precursor. This gene complemented an Escherichia coli diaminopimelate auxotrophy, and the purified protein catalyzed the transamination of diaminopimelate to tetrahydrodipicolinate. Phylogenetic analysis indicated this gene was recruited from anaerobic Gram-positive bacteria. These results expand the family of diaminopimelate aminotransferases to a diverse set of plant, bacterial and archaeal homologs. In contrast marine methanogens from the Methanococcales, which lack pseudomurein, appear to use a different diaminopimelate pathway for lysine biosynthesis.
1. Introduction
Bacteria and archaea use at least five different biosynthetic pathways to produce lysine [1]. Three of these pathways produce a diaminopimelate (DAP) intermediate (Figure 1), while two others produce an α-aminoadipate intermediate. Genome sequence analysis predicts that most Crenarchaea and the euryarchaeal Thermococcales produce lysine by a variation of the α-aminoadipate pathway that was first identified in fungi [2]. Alternatively, NMR studies using 13C-labeled glycerol suggested that the euryarchaeon Haloarcula hispanica uses a DAP dehydrogenase [3]. Similar studies in the methanogens Methanospirillum hungatei and Methanococcus voltae using 13C-labeled acetate indicated that these organisms also use a DAP pathway, but could not discriminate among the three DAP pathways [4,5]. Finally, enzyme assays of Methanothermobacter thermautotrophicus ΔH extracts detected dihydrodipicolinate synthase and DAP decarboxylase activities that are characteristic of all the DAP pathways [6].
Figure 1.

Alternative biosyntheses of meso-diaminopimelate from tetrahydrodipicolinate [8,25]. L-Tetrahydrodipicolinate exists in equilibrium with the enamine product and the acyclic L-2-oxo-6-aminopimelate product [24]. In the succinylate pathway (left), the acyclic form is protected by the tetrahydrodipicolinate N-succinyltransferase enzyme (DapD; EC 2.3.1.117). N-Succinyl-L,L-diaminopimelate aminotransferase (DapC; EC 2.6.1.17) transfers an amino group from L-glutamate, and then N-succinyl-L,L-diaminopimelate desuccinylase (DapE, EC 3.5.1.18) deprotects the product. The L,L-diaminopimelate epimerase enzyme (DapF; EC 5.1.1.7) produces meso-diaminopimelate for lysine and peptidoglycan biosynthesis. Some Gram-positive bacteria use an acetyl protecting group instead of the succinyl group. Alternatively, L,L-diaminopimelate aminotransferase (DapL; EC 2.6.1.83) directly transaminates tetrahydrodipicolinate to produce L,L-diaminopimelate. Finally, some bacteria use a diaminopimelate dehydrogenase enzyme (Ddh; EC 1.4.1.16) to reductively aminate tetrahydrodipicolinate.
Methanogen genome sequences lack some genes required for lysine biosynthesis. A metabolic reconstruction of Methanocaldococcus jannaschii predicted this organism uses an acylated DAP pathway, based on the identification of homologs of the Escherichia coli asd, dapA, dapB, dapF and lysA genes [7]. However, no dapD or dapC homologs were found in M. jannaschii, and the dapE homolog shares only 22% amino acid identity with the E. coli protein. Therefore the pathway from tetrahydrodipicolinate to L,L-diaminopimelate is unresolved; the other methanogen genomes are also missing these genes.
Plants and the bacterium Chlamydia trachomatis use an L,L-diaminopimelate aminotransferase enzyme (DapL) to bypass the need for an acyl protecting group in lysine biosynthesis (Figure 1) [8,9]. Sequence similarity searches suggested that M. thermautotrophicus and two other members of the Methanobacteriales order have homologs of the plant and chlamydial protein that share 39-43% amino acid sequence identity [10]. Unlike other archaea, these methanogens produce pseudomurein –a carbohydrate-based cell wall that is cross-linked through a pentapeptide containing L-alanine, L-lysine and L-glutamate [11]. While most bacteria use DAP in their peptidoglycan, lysine serves the same purpose in some Gram-positive bacterial cell walls and in pseudomurein [12,13]. It is possible that the Methanobacteriales ancestor acquired a dapL homolog by horizontal gene transfer, along with glycosyltransferase and peptide ligase genes required for pseudomurein formation.
A crystal structure model of the Arabidopsis thaliana DapL protein showed that it belongs to the aspartate aminotransferase family of pyridoxal 5′-phosphate dependent proteins [14]. Because members of this enzyme family catalyze a diverse set of transamination reactions [15], careful sequence analysis and genetic or biochemical characterization is often required to identify gene function [16]. The discovery of DapL homologs in cyanobacteria, diverse Gram-positive and Gram-negative bacteria, and the Methanobacteriales suggested these organisms also use this pathway for lysine biosynthesis. However, the bacterial and archaeal homologs form a separate, diverged cluster in the aminotransferase phylogeny. No representative gene has been characterized from this group, therefore it is possible that these homologs have a different function.
To test whether the M. thermautotrophicus homolog functions as a DAP aminotransferase, we cloned the MTH0052 homolog of dapL in E. coli. Expressed in an E. coli DAP auxotroph, this gene restored the cells’ ability to grow in the absence of DAP. Cell-free extracts and affinity purified MTH0052 protein catalyzed the transamination of DAP with 2-oxoglutarate to produce tetrahydrodipicolinate and glutamate. Combined with phylogenetic analysis, these results suggest that lysine biosynthesis in the Methanobacteriales proceeds through the DAP aminotransferase pathway using a gene that was acquired from a Gram-positive bacterium by horizontal gene transfer. The other orders of methanogens appear to produce lysine by a different mechanism.
2. Materials and Methods
2.1. Plasmids and Strains
Escherichia coli MG1655 (CGSC 7740) was obtained from the E. coli Genetic Stock Center (Yale). E. coli DH5α (Invitrogen) was used as a general cloning host, and E. coli LMG194 (Invitrogen) was used for protein expression. E. coli ATM769 (MG1655 ΔdapD∷Tet ΔdapE∷Kan), E. coli ATM782 (MG1655 ΔdapE∷Kan), ATM780 (Chlamydia trachomatis gene CT390 in pUC19), and pNEA15 (E. coli dapE in pBAD18) were gifts from Anthony Maurelli (Uniformed Services University of the Health Sciences) [9]. Plasmid pBAD/HisA was from Invitrogen. Methanothermobacter thermautotrophicus ΔH cells were a gift from Eric Johnson and Biswarup Mukhopadhyay (Virginia Polytechnic Institute and State University). Chromosomal DNA was purified from Methanococcus maripaludis S2 cells by standard methods and from M. thermautotrophicus as described by Mukhopadhyay et al. [17].
The gene encoding protein MTH0052 (RefSeq accession NP_275195.1) was amplified from M. thermautotrophicus DNA by PCR using oligonucleotide primers 5MTH0052X and 3MTH0052H (Operon). The PCR product was cloned into XhoI and HindIII sites of vector pBAD/HisA to produce vector pDG428. The gene encoding protein MMP1527 (RefSeq accession NP_988647.1) was amplified from M. maripaludis DNA using primers 5MMP1527XN and 3MMP1527H. The PCR product was cloned into XhoI and HindIII sites of pBAD/HisA to produce pDG422. The gene encoding protein MMP1398 (RefSeq accession NP_988518.1) was amplified from M. maripaludis DNA using primers 5MMP1398XN and 3MMP1398H. The PCR product was cloned into XhoI and HindIII sites of pBAD/HisA to produce pDG419. Oligonucleotide sequences are shown in Supplementary Table 2. Recombinant DNA was sequenced at the Institute for Cellular and Molecular Biology Core Labs DNA Sequencing facility (UT-Austin).
2.2. Complementation
Plasmids were transformed into E. coli by electroporation. Cells were grown aerobically in LB Miller broth containing ampicillin (100 μg ml-1) and DL-α,ε-diaminopimelate (100 μg ml-1) at 37°C. For complementation, this broth was supplemented with L-arabinose (0.2% w/v). Overnight cultures were streaked onto selective agar medium and incubated at 37°C for 22 h.
2.3 Protein expression and purification
E. coli LMG194 (pDG428) cells were grown with continuous shaking at 37°C (250 rpm) in LB broth with ampicillin. When cultures reached an optical density at 600 nm of 0.6 to 0.7, protein expression was induced by the addition of L-arabinose (0.2% w/v). After four hours of incubation with the inducer, cells were harvested by centrifugation and stored at -20°C. Cell lysis, Ni2+-affinity chromatography and sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) were performed using standard methods [16].
2.4 Aminotransferase activity analysis
The formation of tetrahydrodipicolinate was measured using o-aminobenzaldehyde to form a yellow-colored derivative [18]. Neither the structure nor molar absorbance coefficient of this derivative is known, therefore aminotransferase activity is expressed in absorbance units (AU). Standard reactions included 1 mM DL-α,ε-diaminopimelate (Sigma), 4 mM potassium 2-oxoglutarate, 10 mM o-aminobenzaldehyde (Sigma) dissolved in acetonitrile, 100 mM Hepes/KOH (pH 7.5) and enzyme. The absorbance of the product derivative at 440 nm was measured in a quartz cuvette using a DU-800 spectrophotometer (Beckman) equipped with a Peltier heated stage at 37°C. Reactions with alternative amine acceptors included oxaloacetate or 2-oxoadipate [19] instead of 2-oxoglutarate.
For product analysis, reactions containing 1 mM DAP and 4 mM 2-oxoglutarate were incubated with His6-MTH0052 protein for 3 h at 50°C. The reaction product (20 μl) was mixed with 40 μl of 5 mM 2,4-dinitrophenylhydrazine (DNPH) in 2 M HCl for 10 min at room temperature. The hydrazone derivatives were separated by reversed phase HPLC using an octadecylsilane column (Axxi-Chrom, 250 by 4.6 mm) with a guard column (Phenomenex, 4 by 3 mm). Chromatography was performed using an isocratic elution with buffer A for 3 min, followed by a gradient to 100% buffer B over 12 min. Buffer A contained 25 mM ammonium phosphate (pH 2.5) and 8% (v/v) acetonitrile. Buffer B contained 25 mM ammonium phosphate (pH 2.5) and 80% (v/v) acetonitrile. DNPH derivatives were detected by their absorbance at 360 nm.
For mass spectrometry, enzymatic reaction mixtures were evaporated to dryness by heating under a flow of nitrogen, and derivatized with trifluoroacetic anhydride (TFAA) for 30 min at 55°C. Excess TFAA was evaporated and the residue was dissolved in water. The acyl derivatives were analyzed by liquid chromatography-mass spectrometry (LC-MS) using a Thermo LTQ-XL instrument. The sample was applied to a reversed phase column (Thermo Hypersil Gold 3 μm; 50 × 2.1 mm) and eluted with a gradient from 5% to 80% acetonitrile with 0.1% formic acid in water at a flow rate of 0.5 ml min-1. Ultraviolet-visible light absorption was measured using a photodiode array detector, and electrospray ionization mass spectrometry was performed in the negative ion mode. Tandem mass spectra (MS/MS) were acquired using collision-induced dissociation of the [M-H]- ions.
2.5 Phylogenetic analysis
An alignment of 34 homologs of the diaminopimelate aminotransferase proteins was prepared using the T-Coffee program (ver. 4.96) [20]. From the full alignment 409 positions were chosen for phylogenetic analysis. The phylogeny was inferred using the proml program (with 100 bootstrap replicates) from the Phylip package (ver. 3.67) [21]. This program used the Jones-Taylor-Thornton model of amino acid changes and assumed a γ-distribution of rates (α=2.4) approximated by three states. Complete organism names and sequence accession numbers are listed in the Supplementary Material.
3. Results
3.1 MTH0052 gene complements dapDE mutations
The E. coli strain ATM769 (dapD dapE) is a diaminopimelate auxotroph and requires supplemental DAP for growth on LB medium [9]. When expressed from the arabinose-inducible PBAD promoter in plasmid pDG428, the MTH0052 gene complements this mutation (Figure 2). The ATM769 (pDG428) strain grows as well as wild-type E. coli MG1655 and the ATM769 (pATM780) strain that expresses the chlamydial homolog CT390. As expected, the ATM769 (pDG428) strain failed to grow without arabinose. With the arabinose inducer, the ATM769 (pDG428) strain had a specific growth rate of 0.95 ± 0.007 h-1, compared to a rate of 0.99 ± 0.03 h-1 for the same strain supplemented with DAP. Therefore expression of the MTH0052 protein substitutes for the succinyl-DAP enzymes of E. coli.
Figure 2.
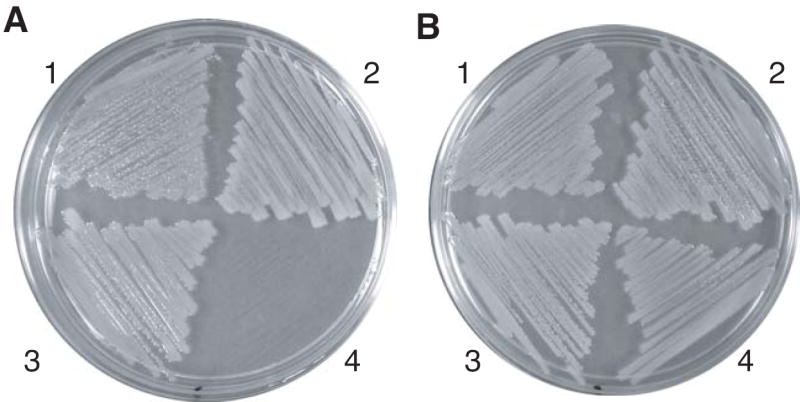
Expression of MTH0052 complemented diaminopimelate auxotrophy in E. coli ATM769. Part A, LB agar supplemented with ampicillin (100 μg ml-1) and arabinose (0.02% w/v) supported the growth of ATM769 (pDG428) in sector 1, ATM769 (pATM780) in sector 2, MG1655 (pBAD/HisA) in sector 3, but not ATM769 (pBAD/HisA) in sector 4. Part B, The same medium as in part A supplemented with DL-α,ε-diaminopimelate (100 μg ml-1) supported the growth of all the strains listed for part A.
3.2 MTH0052 protein catalyzes the diaminopimelate aminotransferase reaction
In reactions containing DAP, 2-oxoglutarate and o-aminobenzaldehyde, cell-free extracts of the E. coli ATM769 strain expressing the MTH0052 protein catalyzed the transamination reaction with an activity of 25 ± 9 milliAU min-1 mg-1 total protein (Figure 3). For comparison, cell-free extracts of the same strain with the empty pBAD/HisA vector had no significant activity (2 ± 5 milliAU min-1 mg-1 total protein) after an initial burst of color formation.
Figure 3.
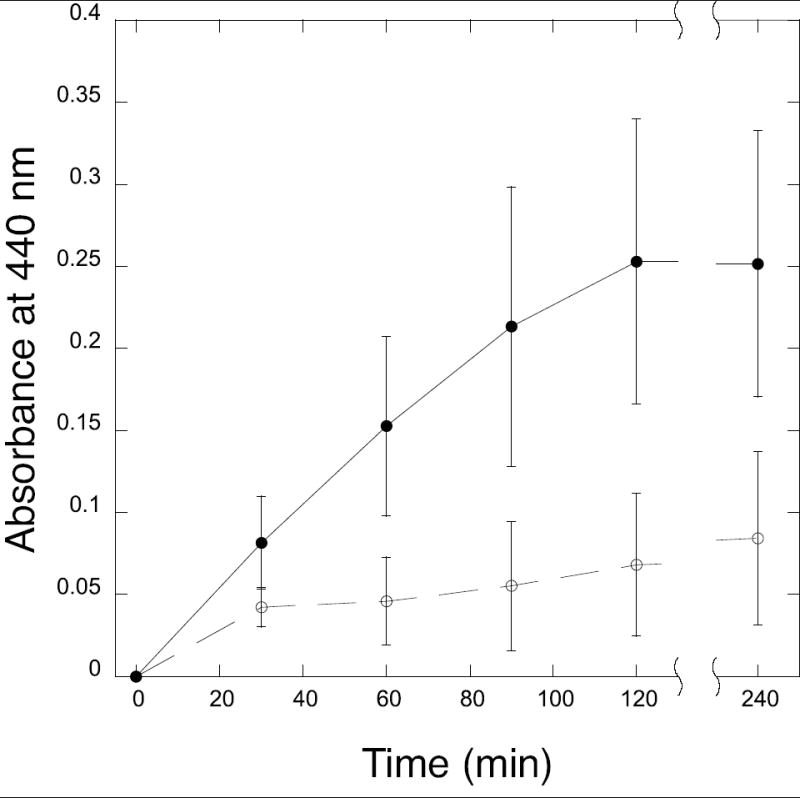
Reaction profile of diaminopimelate aminotransferase activity in E. coli ATM769 (pDG429) extracts expressing MTH0052 (filled circles) and E. coli ATM769 (pBAD/HisA) vector control extracts (open circles). Cell-free extract (100 μg) was incubated with 4 mM 2-oxoglutarate, 2 mM DL-α,ε-diaminopimelate (racemic), 50 μM pyridoxal 5′-phosphate, 10 mM o-aminobenzaldehyde and 100 mM Tris/HCl (pH 8) in a 300 μl reaction at 37°C. The absorbance of the yellow o-aminobenzaldehyde derivative was measured at 440 nm. Error bars show the standard deviation for three replicates of each sample.
To confirm that DAP-AT activity was catalyzed by the heterologously expressed MTH0052 protein, the His6-MTH0052 protein was partially purified by nickel affinity chromatography. The protein had an apparent mass of 47 kDa determined by SDS-PAGE, close to its expected mass of 50.6 kDa. It had an absorption maximum at 412 nm, characteristic of PLP bound to lysine as a Schiff base. This protein catalyzed the transamination of DAP with a specific activity of 0.35 ± 0.12 AU min-1 mg-1. It has maximal aminotransferase activity using 2-oxoglutarate as an amino group acceptor. No activity was observed when oxaloacetate replaced 2-oxoglutarate, although 2-oxoadipate could substitute with 21% relative activity.
The products of the MTH0052 DAP aminotransferase reaction were analyzed by liquid chromatography. Carbonyls derivatized with DNPH formed the hydrazones of the 2-oxoglutarate substrate and the 2-oxo-6-aminopimelate product, which were resolved by HPLC (Figure 4). Reaction mixtures treated with TFAA formed N-acyl derivatives that were analyzed by LC-MS and tandem MS/MS. Using this method, the derivatives had the following retention times, UV-visible absorption maxima and mass spectral data. Peaks corresponding to the molecular ions ([M-H]-) are shown first, followed by characteristic complexes (usually [2M-H]-), and ion fragments listed in decreasing order of intensity. The acyl derivative of glutamate eluted at 0.55 min (233 nm) producing peaks at 242, 485, 113 and 216 m/z; MS/MS of the ion at 242 m/z produced peaks at 199 and 225 m/z. The acyl derivative of tetrahydrodipicolinate eluted at 0.88 min (243 nm) producing peaks at 266, 533, 113, and 222 m/z; MS/MS of the ion at 266 m/z produced peaks at 223, 113 and 153 m/z. The diacyl derivative of DAP eluted at 1.4 min (229 nm) producing peaks at 381, 763, 113 and 240 m/z); MS/MS of the ion at 381 m/z produced peaks at 207, 268, 240, 338, 324, 137 and 113 m/z.
Figure 4.
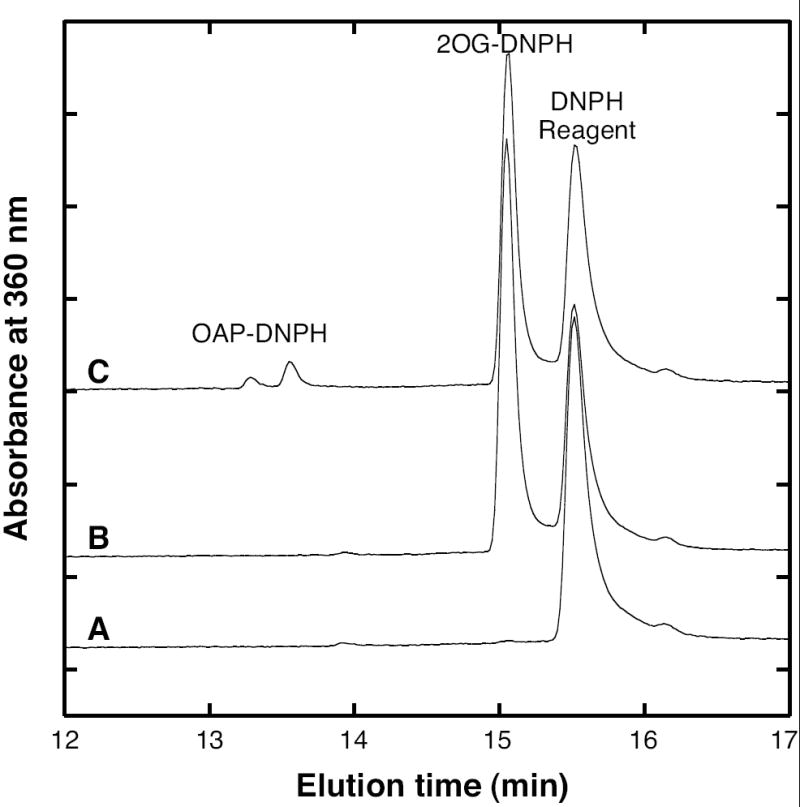
Reaction products of MTH0052 derivatized with 2,4-dinitrophenylhydrazine (DNPH) and separated by reversed-phase HPLC. Curve A shows only a DNPH reagent peak from a control reaction with His6-MTH0052 enzyme alone. Curve B shows the 2-oxoglutarate (2-OG) DNPH hydrazone from a control reaction with 2-OG and DAP substrates. Curve C shows that a reaction containing His6-MTH0052 protein, DAP and 2-OG produced tetrahydrodipicolinate in equilibrium with 2-oxo-6-aminopimelate (OAP), which forms hydrazones with DNPH reagent.
3.3 Diaminopimelate and lysine biosynthesis in the Methanococcales
Both M. jannaschii and Methanococcus maripaludis are marine methanogens that belong to the order Methanococcales. Their genomes encode homologs of the asd, dapA, dapB, dapF and lysA genes that compose a gapped diaminopimelate biosynthetic pathway, as in M. thermautotrophicus (see Supplementary Table 1). The closest M. maripaludis homolog of MTH0052 is MMP1527 (29% amino acid identity). But expression of the MMP1527 protein in E. coli ATM769 (pDG422) failed to rescue growth in the absence of diaminopimelate. Therefore the MMP1527 protein probably belongs to a different group of aminotransferases.
E. coli strain ATM782 lacks the dapE gene that encodes an N-succinyl-DAP desuccinylase. The M. maripaludis protein MMP1398 is 21% identical to E. coli DapE, and both are annotated as succinyl-DAP desuccinylase enzymes. Therefore it was surprising that expression of MMP1398 from plasmid pDG419 failed to complement the dapE mutation. Only expression of the native dapE in strain ATM782 (pNEA15) restored growth in the absence of DAP. Although the DAP aminotransferase circumvents the entire succinyl-DAP pathway in E. coli, expression of neither MTH0052 nor the chlamydial homolog complemented the dapE mutation.
While these results do not identify the functions of the MMP1527 or MMP1398 proteins, they suggest that the Methanococcales use a different pathway than M. thermautotrophicus to transaminate tetrahydrodipicolinate to L,L-diaminopimelate. No homolog of DAP dehydrogenase was identified in the M. maripaludis genome. Furthermore, no DAP dehydrogenase activity was detected in spectrophotometric assays containing 0.2 mg ml-1 M. maripaludis cell-free extract, 3 mM DAP, 1 mM NAD+ or NADP+, and 50 mM glycine/NaOH (pH 10).
3.4 Methanobacteriales acquired DAP aminotransferase by horizontal gene transfer
Phylogenetic analysis of the DAP aminotransferase family identified orthologs in all three genomes of the sequenced Methanobacteriales: M. thermautotrophicus, Methanobrevibacter smithii and Methanosphaera stadtmanae (Figure 5). These genes share 63-73% nucleotide sequence identity. The three proteins are more closely related to the bacterial and plant DAP aminotransferases than they are to aminotransferase homologs from other archaea. They also share up to 64% nucleotide sequence identity with homologs from the anaerobic δ-proteobacteria and Gram-positive bacteria. The sporadic phylogenetic distribution and the unusually high similarity between the archaeal and bacterial homologs indicate that these genes were horizontally transferred into numerous lineages. A phylogeny constructed using nucleotide sequences from a subset of genes represented in Figure 5 confirmed the specific relationship between the methanobacterial dapL and clostridial homologs (Supplementary Figure 1). The genomes that have DAP aminotransferase homologs lack dapD or dapE homologs that are required for the succinyl-DAP pathway.
Figure 5.
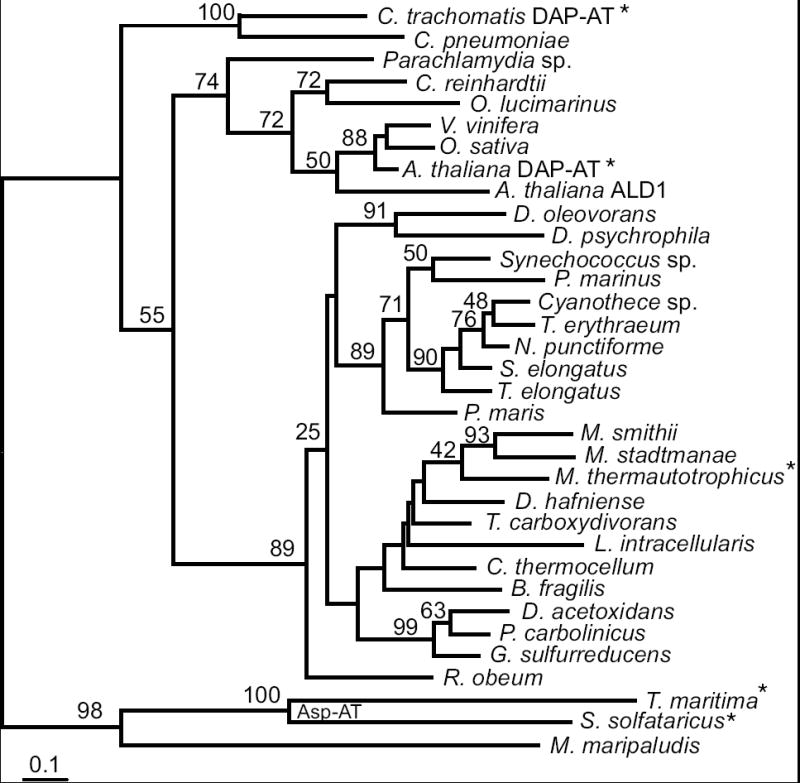
Phylogram of the DapL homologs inferred by the protein maximum likelihood method. Bootstrap values from are shown near branches supported by a plurality of 100 trees. This tree is rooted using known aspartate aminotransferases enzymes and the closest homolog from M. maripaludis (MMP1527). Enzymes whose functions have been experimentally confirmed are indicated by an asterisk. The scale bar indicates one amino acid substitution per 10 positions. Sequence details are described in the Supplementary materials.
4. Discussion
Convergent pathways for the production of lysine have evolved several times, although in most organisms there is little redundancy in lysine biosynthesis. Archaea also have two different classes of lysyl-tRNA synthetases [22]. Horizontal gene transfer has subsequently shuffled these modular pathways and displaced ancestral modes of lysine formation. Crenarchaea and the deeply-branching euryarchaeal Thermococcales use an α-aminoadipate pathway to produce lysine. Most methanogens have a DAP pathway, but they differ in their enzymology for transaminating tetrahydrodipicolinate to form DAP. We have shown here that Methanobacteriales have a DAP aminotransferase to catalyze this reaction; the analogous reactions in other methanogens have not yet been identified. Therefore the DAP aminotransferase was probably acquired by the Methanobacteriales ancestor through horizontal gene transfer from an anaerobic bacterium.
The previous identification and characterization of DAP aminotransferases in plants and Chlamydia led us to question how widespread is this pathway. Homologs of dapL were noted in all cyanobacterial genomes [8], yet phylogenetic analysis rejects the model that plants acquired dapL through chloroplast endosymbiosis (Figure 5). The M. thermautotrophicus protein is the first characterized member of this archaeal and bacterial clade that also includes homologs from anaerobic bacteria. The DapL phylogeny does not identify a single gene donor to the Methanobacteriales lineage, and there is only weak bootstrap support for grouping the three Methanobacteriales homologs. However, gene order is conserved: the orthologs in both M. thermautotrophicus and M. stadtmanae are adjacent to glutamyl-tRNA synthetase genes. Therefore most of the divergence in gene sequences is probably due to the high G+C base composition in MTH0052 (53%) versus M. stadtmanae (31%) and M. smithii (33%) orthologs. Supporting this artifact, a phylogeny derived from an alignment of DNA sequences groups the MTH0052 gene with homologs from the high-%G+C Gram-negative bacteria to the exclusion of the other methanogen homologs (Supplementary Figure 1). The most closely related bacterial homologs come from anaerobic Gram-positive or δ-proteobacterial lineages. These organisms share the same ecological niche, and could also be the donors of peptide ligase homologs (MurC, MurE and MurF) in the Methanobacteriales that are probably responsible for pseudomurein biosynthesis.
The set of conserved amino acid positions in an alignment of DapL homologs includes all of the active site residues identified in the A. thaliana crystal structure model [14]. This protein family evolved from an aspartate aminotransferase ancestor, and the two enzymes remain highly similar. Several amino acid substitutions may be responsible for the change in substrate specificity: A. thaliana DapL residues Glu97 and Asn309 are highly conserved in DapL sequences, but not in aspartate aminotransferases. Both residues form hydrogen bonds with the γ-carboxylate of glutamate bound in a co-crystal structure model. These residues could also recognize the amino group of L-2-oxo-6-aminopimelate and LL-DAP. By analogy with aspartate aminotransferase, the DapL enzyme probably transfers an amino group from pyridoxamine to L-2-oxo-6-aminopimelate. Chemically synthesized tetrahydrodipicolinate forms an equilibrium mixture of the enamine and acyclic forms shown in Figure 1 [23,24]. Future studies of inhibitors of this protein and co-crystal structure models may test whether DapL can catalyze the ring-opening reaction of tetrahydrodipicolinate, or whether its true substrate is the ketone form. Tetrahydrodipicolinate is unstable at neutral pH [23], so the evolution of the acylated pathway, using the DapD, DapC and DapE proteins to protect the ring-open form, suggests that substrate sequestration and isomerization could be rate-limiting.
Supplementary Material
Acknowledgments
This work was supported in part by grants from NIH (AI06444-01) and the Petroleum Research Foundation (44382-G4).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scapin G, Blanchard JS. Enzymology of bacterial lysine biosynthesis. Adv Enzymol Relat Areas Mol Biol. 1998;72:279–324. doi: 10.1002/9780470123188.ch8. [DOI] [PubMed] [Google Scholar]
- 2.Xu H, Andi B, Qian J, West AH, Cook PF. The α-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46:43–64. doi: 10.1385/CBB:46:1:43. [DOI] [PubMed] [Google Scholar]
- 3.Hochuli M, Patzelt H, Oesterhelt D, Wüthrich K, Szyperski T. Amino acid biosynthesis in the halophilic archaeon Haloarcula hispanica. J Bacteriol. 1999;181:3226–3237. doi: 10.1128/jb.181.10.3226-3237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekiel I, Smith IC, Sprott GD. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983;156:316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekiel I, Jarrell KF, Sprott GD. Amino acid biosynthesis and sodium-dependent transport in Methanococcus voltae, as revealed by 13C NMR. Eur J Biochem. 1985;149:437–444. doi: 10.1111/j.1432-1033.1985.tb08944.x. [DOI] [PubMed] [Google Scholar]
- 6.Bakhiet N, Forney FW, Stahly DP, Daniels L. Lysine biosynthesis in Methanobacterium thermoautotrophicum is by the diaminopimelic acid pathway. Curr Microbiol. 1984;10:195–198. [Google Scholar]
- 7.Selkov E, Maltsev N, Olsen GJ, Overbeek R, Whitman WB. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 8.Hudson AO, Singh BK, Leustek T, Gilvarg C. An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 2006;140:292–301. doi: 10.1104/pp.105.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT. L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci USA. 2006;103:17909–17914. doi: 10.1073/pnas.0608643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DR, et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.König H, Kandler O. The amino acid sequence of the peptide moiety of the pseudomurein from Methanobacterium thermoautotrophicum. Arch Microbiol. 1979;121:271–275. doi: 10.1007/BF00425067. [DOI] [PubMed] [Google Scholar]
- 12.Ghuysen J-M, Shockman GD. Biosynthesis of peptidoglycan. In: Leive L, editor. Bacterial Membranes and Walls. Marcel Dekker; New York: 1973. pp. 37–130. [Google Scholar]
- 13.Hartmann E, König H. Comparison of the biosynthesis of the methanobacterial pseudomurein and the eubacterial murein. Naturwissenschaften. 1990;77:472–475. doi: 10.1007/BF01135923. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe N, et al. Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia. J Mol Biol. 2007;371:685–702. doi: 10.1016/j.jmb.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 15.Mehta PK, Hale TI, Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur J Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- 16.Helgadóttir S, Rosas-Sandoval G, Söll D, Graham DE. Biosynthesis of phosphoserine in the Methanococcales. J Bacteriol. 2007;189:575–582. doi: 10.1128/JB.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay B, Purwantini E, Pihl TD, Reeve JN, Daniels L. Cloning, sequencing, and transcriptional analysis of the coenzyme F420-dependent methylene-5,6,7,8-tetrahydromethanopterin dehydrogenase gene from Methanobacterium thermoautotrophicum strain Marburg and functional expression in Escherichia coli. J Biol Chem. 1995;270:2827–2832. doi: 10.1074/jbc.270.6.2827. [DOI] [PubMed] [Google Scholar]
- 18.Yugari Y, Gilvarg C. The condensation step in diaminopimelate synthesis. J Biol Chem. 1965;240:4710–4716. [PubMed] [Google Scholar]
- 19.Nelson RB, Gribble GW. On the preparation of α-ketoadipic acid. Org Prep Proc Intl. 1973;5:55–58. [Google Scholar]
- 20.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP (phylogeny inference package) Department of Genetics, University of Washington; Seattle: 2005. [Google Scholar]
- 22.Ibba M, et al. A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- 23.Chrystal EJT, Couper L, Robins DJ. Synthesis of a key intermediate in the diaminopimelate pathway to L-lysine: 2,3,4,5-tetrahydrodipicolinic acid. Tetrahedron. 1995;51:10241–10252. [Google Scholar]
- 24.Caplan J, Sutherland A, Vederas JC. The first stereospecific synthesis of L-tetrahydrodipicolinic acid; a key intermediate of diaminopimelate metabolism. J Chem Soc Perkin Trans 1. 2001;1:2217–2220. [Google Scholar]
- 25.Born TL, Blanchard JS. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr Opin Chem Biol. 1999;3:607–613. doi: 10.1016/s1367-5931(99)00016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


