Abstract
Lentiviral vectors are attractive for hematopoietic stem cell (HSC) gene therapy because they do not require mitosis for nuclear entry, they efficiently transduce hematopoietic repopulating cells, and self-inactivating (SIN) designs can be produced at high titer. Experiments to evaluate HIV-derived lentiviral vectors in nonhuman primates prior to clinical trials have been hampered by low transduction frequencies due in part to host restriction by TRIM5α. We have established conditions for efficient transduction of pigtailed macaque (Macaca nemestrina) long-term repopulating cells using VSV-G–pseudotyped HIV-based lentiviral vectors. Stable, long-term, high-level gene marking was observed in 3 macaques using relatively low MOIs (5-10) in a 48-hour ex vivo transduction protocol. All animals studied had rapid neutrophil engraftment with a median of 10.3 days to a count greater than 0.5 × 109/L (500/μL). Expression was detected in all lineages, with long-term marking levels in granulocytes at approximately 20% to 30%, and in lymphocytes at approximately 12% to 23%. All animals had polyclonal engraftment as determined by analysis of vector integration sites. These data suggest that lentiviral vectors should be highly effective for HSC gene therapy, particularly for diseases in which maintaining the engraftment potential of stem cells using short-term ex vivo transduction protocols is critical.
Introduction
Nonhuman primate models have been invaluable for developing hematopoietic gene therapy strategies for humans because gene transfer and stem cell clonality closely simulate that seen in patients. Gene therapy has now cured several hematopoietic diseases, as evidenced by successes for immunodeficiencies including X-linked severe combined immunodeficiency (SCID-X1), adenosine deaminase (ADA) deficiency, and more recently chronic granulomatous disease (CGD). However, the risk of malignancy from vector integration is significant, as evidenced in the French SCID-X1 trial where 4 of 10 patients have developed leukemia that can be directly attributed to vector-mediated oncogene activation.1 These findings have highlighted the importance of developing safer vector systems and developing animal models to evaluate vector integration and genotoxicity
HIV-based lentiviral vectors are now being used in human trials because they offer several advantages for hematopoietic stem cell (HSC) gene therapy. Lentiviral vectors can enter the nucleus independent of mitosis, and are able to efficiently deliver complex transgenes such as globin genes that are not delivered efficiently by gammaretroviral vectors.2 Lentiviral self-inactivating (SIN) vectors have deletions of enhancer and promoter sequences in the long terminal repeats (LTRs) designed to reduce the potential for transactivation of host genes and can be produced at high titer. While murine leukemia virus (MLV)–based gammaretroviral vectors favor integration into regions near promoters,3 human immunodeficiency virus 1 (HIV1)–based lentiviral vectors integrate preferentially within genes.4 In addition, in canine and primate long-term repopulating cells, gammaretroviral vectors were found more frequently near (within 5000 bp) proto-oncogene start sites than HIV-based lentiviral vectors.5,6 A comparison of the genotoxicity of SIN lentiviral vectors to gammaretroviral vectors with LTR-driven transgenes similar to the ones used in the SCID-X1 trial suggests that SIN lentiviral vectors may be safer, at least in a murine model.7
We have previously shown that HIV-based lentiviral vectors can mediate gene transfer to long-term repopulating cells in the canine model,8 but gene transfer into primate models including the rhesus macaque9,10 and the baboon has been inefficient,11 likely due to host restriction factors including TRIM5α.12 The rhesus macaque TRIM5α potently restricts HIV-1 replication by accelerating capsid uncoating after cell entry.13 Restriction has also been seen with SIV-based vectors in human hematopoietic cells.14
We found that pigtailed macaque (Macaca nemestrina) CD34+ cells are highly permissive for transduction by HIV-1–derived vectors and describe efficient transduction of pigtailed macaque long-term repopulating cells using HIV-based lentiviral vectors at relatively low MOIs. This nonhuman primate model will be very useful for preclinical studies to evaluate the safety and efficacy of HIV-1–based vectors proposed for clinical studies.
Methods
Animal care
Healthy juvenile baboons (Papio cynocephalus anubis) and pigtailed macaques (Macaca nemestrina) were housed at the University of Washington National Primate Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. Study protocols were approved by the Institutional Review Board and the Institutional Animal Care and Use Committee. Prior to hematopoietic cell collection, animals were administered recombinant human granulocyte colony-stimulating factor (rhG-CSF, 100 μg/kg), and also given recombinant human stem cell factor (rhSCF, 50 μg/kg) daily as subcutaneous injections for 5 days. On day 5, bone marrow was harvested from the humeri and/or femora. In preparation for transplantation, all animals received myeloablative total-body irradiation. Twenty-four hours after transplantation, the animals were started on intravenous G-CSF at 100 μg/kg daily until the animals attained stable neutrophil engraftment with an absolute neutrophil count of greater than 0.5 × 109/L (500/μL). Standard supportive care, including blood product transfusions, fluid and electrolyte management, and antibiotics, was administered as needed. Hematopoietic recovery was monitored by daily complete blood counts.
Retroviral vectors
The HIV vectors were SIN pRRL15 or pWPT-GFP (provided by Didier Trono, Lausanne, Switzerland) vector backbones containing a central polypurine tract, and a woodchuck posttranscriptional regulatory element. The HIV-based vector internal promoters were either an internal elongation factor 1α promoter for animal J02043, or both a spleen-focus forming virus SFFV expressing a P140K methylguanine methyltransferase (MGMT) transgene and a phosphoglycerate kinase (PGK) promoter driving expression of enhanced green fluorescent protein (EGFP) for animals J02370 and T04228. For the in vitro studies, the lentiviral vectors expressed EGFP from the PGK promoter. HIV-based vectors were pseudotyped with VSV-G envelope and produced by transient transfection of 293T cells and concentrated 100-fold as previously described.8
Comparison of HIV vector transduction in human, macaque, and baboon cells in vitro
Human HT1080 fibrosarcoma cells were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum and with 100 U/mL penicillin and 100 μg/mL streptomycin. Macaque and baboon CD34+ cells were collected from rhG-CSF and stem cell factor (SCF)–primed bone marrow, and human CD34+ cells were collected from G-CSF–mobilized human volunteers. For CD34+ transductions, cells were cryopreserved and then thawed and immediately exposed to a lenti vector at an MOI of 5 or 50 in Iscove modified Dulbecco medium (IMDM) with 10% FBS with 100 U/mL penicillin and 100 μg/mL streptomycin, supplemented with 100 ng/mL recombinant human stem cell factor (rhSCF), 100 ng/mL recombinant human Flt-3 ligand (rhuFlt3-L), 100 ng/mL interleukin-3 (IL-3), 100 ng/mL IL-6, 100 ng/mL thrombopoietin (TPO), and 100 ng/mL G-CSF in CH-296–coated tissue culture plates. After 18 hours, the cells were replated in growth media plus cytokines, and percentage of cells expressing EGFP was determined by flow cytometry at day 3 or day 5 after transduction.
Primate CD34+ isolation, retrovirus transduction, and transplantation
CD34+ cell enrichment was performed using the 12.8 IgM anti-CD34+ antibody and MACS IgM microbeads (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions and as described previously.16,17 CD34-enriched primate cells were cultured in IMDM with 10% FBS with 100 U/mL penicillin and 100 μg/mL streptomycin, supplemented with 100 ng/mL rhSCF, 100 ng/mL rhuFlt3-L, 100 ng/mL interleukin-3 (IL-3), 100 ng/mL IL-6, 100 ng/mL TPO, and 100 ng/mL G-CSF for 15 to 18 hours before transduction. For transduction, cells were supplemented with the same cytokine combination in flasks previously coated with the CH-296 fragment of fibronectin (Takara, Shiga, Japan). The transduction medium was also supplemented with 4 μg/mL protamine sulfate. For animals J02043 and T04228, 1 μg/mL cyclosporine was also added. The cells were exposed to an initial dose of vector for 6.5 to 8 hours and then exposed to second dose of vector overnight for 17 to 18 hours. Following transduction, cells were infused into recipients after myeloablative total body irradiation (TBI) conditioning. Animal J02043 received a myeloablative dose of 1020 cGy TBI in 2 doses of 510 cGy, and animals J02370 and T04228 received 800 cGy in 4 doses of 200 cGy from a single source linear X-ray accelerator (Linac Systems, Lakewood, NJ) at 7 cGy/minute. After infusion of autologous gene-modified cells, the animals received recombinant G-CSF at 100 μg/kg daily until the animals maintained an absolute neutrophil count (ANC) of greater than 0.5 × 109/L (500/μL). The animals also received standard supportive care including intravenous hydration and broad spectrum antibiotics (ceftazadime, vancomycin, gentamicin), an antiviral agent (acyclovir), an antifungal agent (fluconozol), and transfusions with irradiated pigtailed macaque whole blood for treatment of posttransplantation thrombocytopenia.
CFU assay
Transduced and mock-transduced CD34-enriched cells were cultured in a double-layer agar culture system as previously described.6,18 Colonies were enumerated and evaluated for GFP expression on day 14 of culture using fluorescence microscopy.
Retrovirus-specific LAM-PCR
Retroviral integration site analysis by linear amplification–mediated polymerase chain reaction (LAM-PCR) was performed on primate DNA isolated from peripheral blood leukocytes and has been described in detail elsewhere.5,19,20 Briefly, initial linear amplification PCR was performed with biotin-tagged LTR-specific primers and after further processing, samples were digested to create compatible genomic restriction site overhangs with either Tsp509I or MspI restriction enzymes (New England Biolabs, Ipswich, MA). Internal vector amplicons were eliminated with retrovirus-specific restriction enzyme digestion, and an enzyme-specific linker cassette was ligated to the restriction enzyme site of the genomic DNA bound to beads creating binding sites for reverse primers. Nested PCR was carried out and the PCR reactions were separated on an acrylamide gel and stained with ethidium bromide to visualize PCR products.
Flow cytometric analysis of macaque hematopoietic cells
Leukocytes isolated by ammonium chloride red cell lysis from heparinized peripheral blood and bone marrow samples drawn at multiple time points after transplantation were analyzed for EGFP expression on a FACSVantage or FACSCalibur (Becton Dickinson, San Jose, CA). Transgene expression in granulocyte, monocyte, and lymphocyte populations was determined by gating based on either forward and right-angle light scatter characteristics or expression of lineage-specific CD markers. The antibodies used for lineage-specific markers included CD3 (clone SP34-2), CD13 (clone L138), CD20 (clone L27), and CD34 (clone 563). All antibodies were supplied by Becton Dickinson (Franklin Lakes, NJ) and conjugated to phycoerythrin. Red cells and platelets from whole blood diluted in phosphate-buffered saline were delineated by forward and right-angle light scatter properties and assessed for EGFP expression.
DNA analysis of gene marking
Relative marking levels were analyzed with the TaqMan 5′ nuclease quantitative real-time polymerase chain reaction (PCR) assay.21 We amplified 300 ng DNA in at least duplicate with an EGFP-specific primer/probe combination (5′-CTG CAC CAC CGG CAA-3′ and 5′-GTA GCG GCT GAA GCA CTG-3′; probe, 5′-FAM-CCA CCC TGA CCT ACG GCG TG-TAMRA-3′) (Synthegen, Houston, TX) or a primer/probe combination specific to the lentiviral cis-acting region (5′-TGA AAG CGA AAG GGA AAC CA-3′, 5′-CCG TGC GCG CTT CAG-3′; probe, 5′-FAM-AGC TCT CTC GAC GCA GGA CTC GGC-TAMRA-3′). Standards consisted of dilutions of DNA extracted from a cell line containing a single lentiviral vector provirus. Negative controls consisted of DNA extracted from peripheral blood mononuclear cells (PBMCs) obtained before transplantation from control animals or water. A monkey-specific betaglobin primer/probe combination was used to adjust for equal loading of DNA per reaction (5′-TGG CTA ATG CCC TGG CCC ACA AGT A-3′; primer sequences BG73F, 5′-CCT ATC AGA AAG TGG TGG CTG G-3′ and BG149R, 5′-TTG GAC AGC AAG AAA GTG AGC TT-3′). Reactions were run by the ABI master mix (Applied Biosystems, Branchburg, NJ) on the ABI Prism 7700 sequence detection system (Applied Biosystems) under the following thermal cycling conditions: 50°C for 2 minutes and 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
For Southern blot analysis,22 peripheral blood DNA was digested with the restriction enzymes AvaI and XmnI (New England Biolabs) that cleave within the lentiviral vector to produce an internal 1083-bp fragment from the cis-acting region. DNA from the single copy control cell line was mixed at different ratios with DNA from a cell line with no lentiviral vector provirus to generate copy number standards. Following digestion, approximately 10 μg DNA was electrophoresed, transferred to a Hybond-N+ membrane (GE Healthcare, Waukesha, WI), and hybridized using QuickHyb hybridization solution (Stratagene, La Jolla, CA) following manufacturers' recommendations. The 1083-bp lentiviral vector cis-acting region probe was generated by nested PCR amplification from 10 pg lentiviral vector plasmid DNA, then digested with XmnI and AvaI and gel-purified.
Detection of replication-competent virus
Serum of all 3 animals was tested for the presence of recombinant replication-competent lentiviral vectors by the p24 Antigen Assay (Beckman Coulter, Miami, FL) according to the manufacturer's recommendations. The p24 antigen was not detectable, indicating the absence of helper-virus production.
Results
Efficient transduction of pigtailed macaque cells in vitro
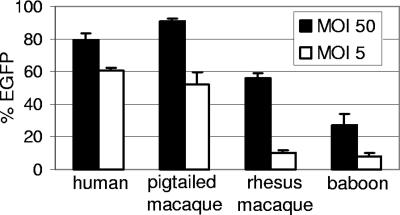
We had previously observed persistent but low-level transduction of baboon repopulating cells (Papio cynocephalus anubis) using HIV-1–derived lentiviral vectors,11 likely due to restriction mediated by TRIM5α.12 However, pigtailed macaques are permissive for HIV-1 replication23 and thus may not have as potent a TRIM5α-mediated restriction to HIV-1 infection as observed in other primate species. To determine the relative susceptibility of pigtailed macaque leukocytes to transduction by HIV-based vectors, we compared the efficiency of transduction of human, pigtailed macaque, rhesus macaque, and baboon CD34+ cells using HIV vectors pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G) that expressed enhanced green fluorescent protein (EGFP). Transduction of CD34+ cells was performed in vitro at 2 multiplicities of infection (Figure 1). As expected, compared with human cells, the baboon and rhesus cells were relatively refractory to transduction with HIV-based vectors. At a high MOI, the rhesus CD34+ cells were transduced efficiently; however, as would be expected at the low MOI, the rhesus cells were refractory to transduction with approximately 10% expressing EGFP. Conversely, at the low MOI both the human and pigtailed macaque CD34+ cells were efficiently transduced with greater than 50% of cells expressing EGFP. It has previously been reported that postentry restriction to HIV infection in monkey cells can be saturated by noninfectious capsids,24–26 which likely explains the ability of the HIV-1 vector to efficiently transduce both pigtailed and rhesus macaque cells at high MOI but not the rhesus macaque cells efficiently at a low MOI.
Figure 1.
Relative transduction efficiency of human, pigtailed macaque, rhesus, and baboon CD34+ cells with HIV-based vectors. Cryopreserved CD34+ cells were thawed and transduced with a lentiviral EGFP vector and the percentage of EGFP-expressing cells was determined 5 days after vector exposure. The mean and standard error from 3 experiments is shown.
Engraftment after transplantation of lenti vector transduced cells
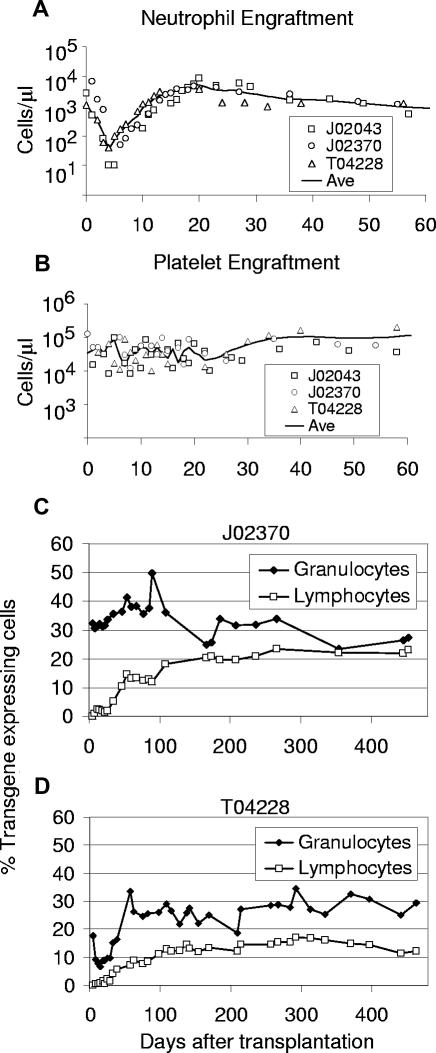
We performed a series of 3 transplantations in pigtailed macaques using HIV-based vectors. In the 3 animals, a stable absolute neutrophil count (ANC) greater than 0.5 × 109/L (500/μL) was reached by either day 10 or day 11 (Table 1). A platelet count greater than 20 × 109/L 20 000/μL was reached at an average of 29 days. Figure 2 displays the ANC and platelet counts after transplantation for all 3 animals in this study. Engraftment was slightly less rapid, but similar to engraftment of rhesus macaques that received CD34+ cells transduced with SIV-based vectors pseudotyped with an amphotropic envelope in which the mean time to ANC greater than 0.5 × 109/L (500/μL) was 7.3 days.14 These data show that monkey CD34+ cells exposed to lentiviral vectors pseudotyped with VSV-G at low MOIs5–10 retain robust capacity for engraftment.
Table 1.
Lentiviral transduction and engraftment of pigtailed macaque CD34+ cells
| Monkey* | No. of CD34-enriched cells/kg ×106 before culture | Purity of CD34-enriched cells, % | No. of infused cells/kg ×106 | MOI† | Preinfusion transduction efficiency,‡ | Days to ANC greater than 0.5 × 109/μL (500/μL) | Days to platelet count greater than 2 × 1010/L (20 000/μL) |
|---|---|---|---|---|---|---|---|
| J02043 (4.0 kg) | 20 | 82 | 4.6 | 10 | 4.3 (PCR) | 11 | 27 |
| J02370 (4.6 kg) | 30 | 97 | 11.7 | 10 | 30% (EGFP) | 10 | 35 |
| T04228 (3.6 kg) | 40 | 85 | 10.7 | 5 | 17.5% (EGFP) | 10 | 25 |
For all monkeys, the source of CD34+ cells was bone marrow.
Monkey and weight at time of transplantation in parentheses.
Multiplicity of infection based on titer determined by transduction of HT1080 cells.
Figure 2.
Engraftment and transgene expression levels in peripheral blood cells of monkeys that received lentivirally transduced stem cells from bone marrow using a 48-hour transduction protocol. Displayed for all 3 monkeys that underwent transplantation are the absolute neutrophil counts (A) and platelet counts (B) after transplantation. The solid line marks the interpolated time course of average cell numbers. The percentages of EGFP-expressing leukocytes detected by flow cytometry are shown for monkeys J02370 (C) and T04228 (D).
Gene transfer efficiency before transplantation
Transduction efficiency before transplantation was assessed by flow cytometric determination of EGFP-positive CD34-enriched cells for vectors that encoded a fluorescent marker (monkeys J02370 and T04228) or by performing real-time PCR for vector sequences (J02043). Gene transfer levels were high, especially given the relatively low MOI.5–10 Table 1 summarizes the results of the pretransplantation analysis of macaque CD34+ cells.
Efficient gene transfer into pigtailed macaque repopulating cells
Monkeys J02370 and T04228 received a transplant of cells exposed to an EGFP-expressing vector, so the transduction efficiency of hematopoietic repopulating cells after transplantation could be accurately measured by flow cytometric detection of EGFP in peripheral blood (PB) cells (Figure 2C,D). We observed stable long-term (> 450 and > 441 days) transgene expression with high marking levels in both granulocytes and lymphocytes, respectively. Long-term marking (more than 90 days after transplantation) rose to as high as 36% and 50% in T04228 and J02370, respectively, in short-lived granulocytes, and was typically between 20% and 35%, demonstrating efficient gene transfer to primitive repopulating cells. Marking in PB lymphocytes rose to as high as 23% and 17% in J02370 and T04228, respectively. The slow rise of transgene-expressing lymphocytes to a plateau in both animals suggests gene transfer to primitive repopulating cells with typical lymphocyte engraftment kinetics. The level of transgene-expressing cells was very stable over time, and marking was comparable between myeloid and lymphoid cells at later time points.
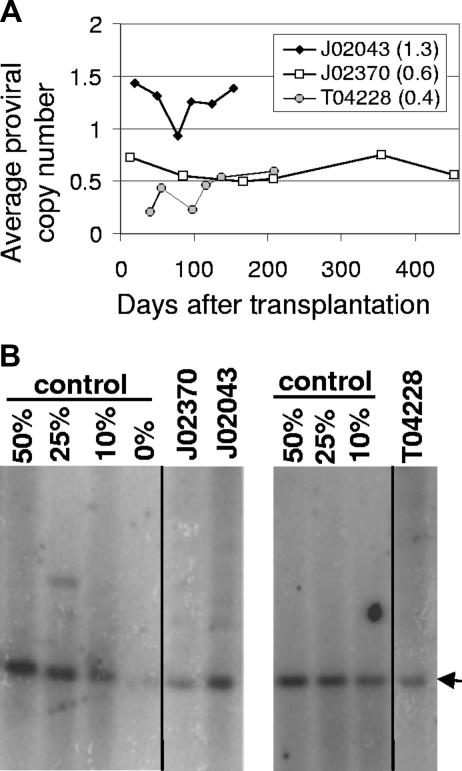
Gene transfer rates after transplantation were also determined at several time points by real-time PCR for all 3 monkeys (Figure 3A) using primers specific for the lentiviral cis-acting region. The average proviral copy number was determined using a control cell line that contains a single lentiviral vector provirus. The real-time PCR analysis showed that monkey J02043 that received cells transduced with a vector that did not encode a fluorescent marker also had very high marking with an average proviral copy number of 1.3 that varied between 0.9 and 1.4. The EGFP-expressing monkey J02370 had an average proviral copy number of 0.6 that varied between 0.5 and 0.75, and monkey T04428 had an average proviral copy number of 0.4 that varied between 0.2 and 0.6. In monkeys J02370 and T04228, these data confirm the high-level marking assessed using the EGFP transgene and correlate well with the relative marking observed using transgene expression. Given that approximately 25% of the PB leukocytes expressed transgene in J02370 and the average proviral copy number was approximately 0.6, the average copy number in repopulating cells that contain at least one vector provirus can be estimated to be approximately 2.4 if there was no silencing of transgene expression, or lower if there was some level of silencing. For monkey T04228, approximately 20% of PB leukocytes expressed the EGFP transgene, which would result in an upper limit of 2 for the average proviral copy number in repopulating cells that contain at least one vector provirus. LAM-PCR analysis of CFUs suggested that most colonies had more than one integrant (data not shown), indicating that there was very little silencing of transgene expression in this animal. High-level marking was confirmed by Southern blot analysis for all 3 animals, and the relative levels between monkeys correlated well with real-time PCR and EGFP expression data (Figure 3B). However, the marking levels were approximately 2-fold lower than observed by flow cytometry or real-time PCR. This difference might be explained in part by differences in loading of the DNA prior to Southern blot analysis (data not shown). It should be noted that all 3 monkeys contained a mutant MGMT gene for in vivo selection, but the marking data presented here were obtained without any in vivo selection.
Figure 3.
Gene marking levels in peripheral blood cells of monkeys that received HIV-based lentiviral transduced marrow stem cells. (A) The vector provirus genomes in PB from monkeys J02043, J02370, and T04228 were determined by Taqman PCR relative to a control cell line with a single integrated lentiviral vector to determine the average proviral vector copy number. The average copy number from all dates is indicated in parentheses. (B) Southern blot analysis confirms high-level marking in all 3 monkeys. Vertical lines indicate repositioned gel lanes.
Analysis of gene expression in hematopoietic lineages
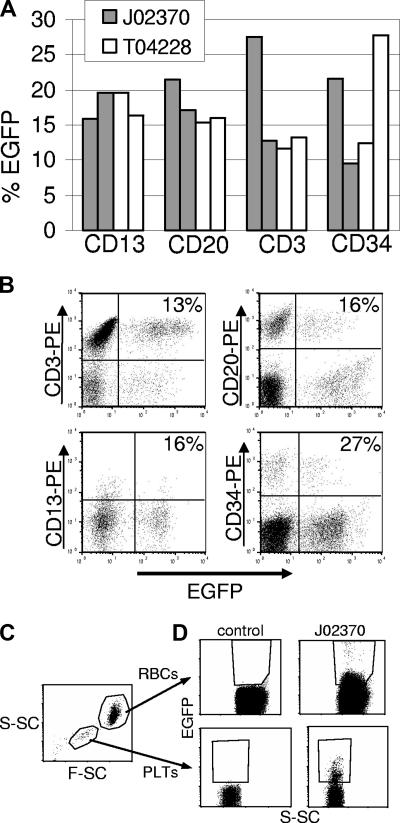
To assess gene expression in different hematopoietic lineages, PB cells were labeled with antibodies against granulocytes (CD13), T lymphocytes (CD3), and B lymphocytes (CD20), and analyzed by flow cytometry. We also determined the marking levels in BM-derived CD34+ cells. Figure 4A shows representative results obtained in the 2 monkeys that received the EGFP vectors at 67 and 174 days (J02370) and 116 and 441 days (T04228) after transplantation. Sustained EGFP expression could be detected in all subsets examined, and flow cytometric profiles are shown in Figure 4B. In both monkeys, we were also able to detect fluorescence-positive platelets and fluorescence-positive erythrocytes (Figure 4C,D). Since EGFP fluorescence intensity in these cell populations is significantly lower than in white blood cells, these data likely underestimate marking in erythropoietic progenitors. Both of these cell populations are enucleated, and are therefore transcriptionally inactive, so the EGFP protein half-life largely dictates the level of expression. We have monitored these animals by performing complete blood counts to monitor the potential development of myeloproliferation, lymphoproliferation, or leukemia. To date, there has been no evidence of malignancy and all 3 animals remain healthy.
Figure 4.
Flow cytometric analysis of transgene-expressing cells in peripheral blood subpopulations and BM CD34+ cells. (A) The percentage of transgene-positive cells in different leukocyte subpopulations in the peripheral blood of monkeys that received an EGFP-expressing vector at days 67 and 174 (J02370), or days 116 and 441 (T04228) after transplantation. In these monkeys, EGFP-expressing cells were found in all lineages examined. (B) Flow cytometry data for EGFP expression in different lineages from animal T04228 at day 441. Each lineage was identified with a phycoerythrin (PE)–labeled antibody. The percentages shown are for the total number of EGFP- and lineage-positive cells over the total number of lineage positive cells. (C) Gating on red blood cells (RBCs) and platelets (PLTs) was based on scatter characteristics (S-SC is side scatter and F-SC is forward scatter). (D) EGFP-expressing red blood cells (top panels) and platelets (bottom panels) are plotted with side scatter for a control animal and for animal J02370. Because of the overlapping positive and negative populations due to low fluorescence intensity, especially in red blood cells, the percentages of marked cells (5.6% in red blood cells, 6.7% in platelets) likely underestimate the actual percentage of EGFP-expressing cells.
Clonality of transduced macaque repopulating cells
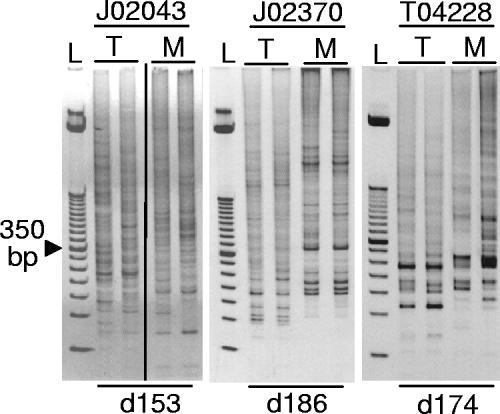
LAM-PCR was performed on the PB from all 3 animals to evaluate the clonality of the engrafted cells. All 3 monkeys engrafted with a polyclonal set of repopulating cells as evidenced by multiple LAM-PCR fragment sizes (Figure 5). We have identified more than 235 unique integration sites in these monkeys, and a detailed analysis of these integration sites has been published elsewhere.6 Animals J02370 and J02043 have since been moved to another study, but all monkeys are currently alive and well at 463 (T04228), 698 (J02370), and 981 days (J02043) after transplantation.
Figure 5.
Polyclonal repopulation with transduced hematopoietic cells. Peripheral blood samples were analyzed by LAM-PCR revealing polyclonal repopulation of all 3 monkeys. An ethidium bromide–stained acrylamide gel of LAM-PCR products is shown; L, 50 bp standard. In lanes labeled M, DNA samples were digested with the restriction enzyme MspI, and in lanes labeled T, DNA samples were digested with Tsp509I restriction enzymes that have compatible sticky ends. The day after transplantation on which the PB samples were collected is listed below (d153 is day 153 after transplantation). The vertical line indicates a repositioned gel lane.
Discussion
We evaluated the ability of HIV-derived viral vectors pseudotyped with VSV-G to transduce HSCs in a clinically relevant monkey model. A previous study had shown that pigtailed macaque cells are permissive for HIV infection,23 suggesting that they may have less potent restriction mechanisms for transduction by HIV vectors than other primate species. We show here for the first time that pigtailed macaque long-term hematopoietic repopulating cells can be efficiently transduced by HIV-1–derived lentiviral vectors. Transduction levels were considerably higher than seen with baboon or rhesus CD34+ cells. Pigtailed macaques and rhesus macaques have different amino acids in the TRIM5α SPRY domain27 that confer species-specific capsid restriction.28 Our findings are consistent with a recent report that identified truncated forms of TRIM5α in M nemestrina that do not restrict HIV-1 infection.29 These defective M nemestrina TRIM5α isoforms likely explain why we were able to efficiently transduce pigtailed macaque cells at low MOIs using the wild-type HIV-1 capsid, which is potently restricted by rhesus TRIM5α.12 The differences between rhesus and pigtailed macaque TRIM5α also likely explain the higher marking we observed here relative to previous studies using HIV-based vectors in the rhesus macaque model where long-term marking was typically lower than 3% with one animal having marking levels of approximately 10% and 2% in granulocytes and lymphocytes, respectively.9,10
In 3 monkeys that received autologous CD34+ cells transduced with VSV-G–pseudotyped HIV vectors, we observed stable, high-level gene transfer into long-term hematopoietic repopulating cells. Long-term marking was as high as 50% into short-lived PB granulocytes, demonstrating efficient transduction of primitive cells. Approximately 20% to 35% of granulocytes and 12% to 23% of PB lymphocytes expressed the EGFP transgene long term as determined by flow cytometry. This is a remarkable level considering the relatively short transduction period (48 hours) and the low MOIs5–10 used. Engraftment was relatively rapid with a mean of 10.3 days to an ANC greater than 0.5 × 109/L (500/μL) and was stable over time in both monkeys in which the EGFP fluorescent marker could be followed by flow cytometry of PB leukocytes. In all 3 monkeys, real-time PCR yielded consistently high level marking that could be readily detected by Southern blot analysis.
The most accepted assay for the transduction of stem cells is the ability to deliver genes to large animal repopulating cells that differentiate into all blood lineages and persist long term. We were able to track marking in the 3 monkeys for 5, 15, and 15 months, and for animals J02370 and T04228, EGFP-expressing cells were detected in all hematopoietic lineages including red blood cells and platelets. These data strongly suggest that long-term, multipotent repopulating cells were transduced with the VSV-G–pseudotyped lentiviral vectors. A previous study showed efficient gene transfer to rhesus macaque repopulating cells using simian immunodeficiency virus vectors pseudotyped with an amphotropic receptor14 at similar levels to what we observed here. However, in the rhesus study the SIV-derived vectors did not transduce human CD34+ cells as efficiently as rhesus macaque CD34+ cells. We have used HIV-1–based vectors that have a capsid that is not sensitive to human TRIM5α so the marking levels we have observed in the pigtailed macaque model using HIV-derived vectors should translate well to clinical studies.
Gene transfer levels detected in PB were similar to those determined in bone marrow leukocytes (Figure 4A). These data suggest that there is no block in differentiation or elimination of mature gene-modified cells. Comparison of the number of gene-expressing cells with the marking determined by real-time PCR showed that significant silencing did not occur over time. The clonality as assessed by the number of bands identified by LAM-PCR and the number of unique integrations sites that could be identified from sequencing these bands was higher than historic controls transduced with gammaretroviral vectors.6,18 Thus, transduction with HIV-based lentiviral vectors leads to engraftment of a large number of repopulating clones. These data, taken together with the rapid engraftment, show that repopulating cells retain robust engraftment capability after exposure to HIV vectors pseudotyped with VSV-G in a 48-hour ex vivo transduction protocol. In a previous study where VSV-G–pseudotyped gammaretroviral vectors were compared with gammaretroviral vectors pseudotyped with the amphotropic receptor, marking levels were less than 1% for both pseudotypes, but there was no evidence that the concentrated VSV-G–pseudotyped vectors were more toxic than unconcentrated vectors with the amphotropic receptor.30 Given that lentiviral vectors are able to transduce cells independently of mitosis and that either amphotropic14 or VSV-G–pseudotyped lentiviral vectors (our data) can lead to long-term, polyclonal repopulation in primates with marking levels in the range of 10% to 30%, it is unclear whether receptor levels, quiescence, or other factors limit even more efficient transduction. If the limiting factor is host receptor levels, a cocktail of HIV vectors with different envelope pseudotypes might further increase gene transfer to HSCs.
The risk of leukemia as a result of vector integration remains a significant concern for HSC gene therapy. Clinical studies have shown that activation of host genes by integrated gammaretroviral vector proviruses can lead to clonal expansion of repopulating clones31 or leukemia.32 In our study, we used SIN HIV vectors that expressed transgenes from internal promoters. A murine study has suggested that the genotoxicity associated with SIN HIV vectors is lower than for gammaretroviral vectors that express transgenes from the LTR.7 To date, we have not observed clonal expansion or malignant transformation in these monkeys, and all monkeys remain in good health. However, the leukemias observed in the French trial took approximately 3 years to develop so long-term follow-up for these monkeys will be important to further evaluate the safety of these SIN lentiviral vectors. The risk of malignancy as a result of insertional mutagenesis increases with the number of proviral copies, so limiting the number of integrations while still maintaining transgene expression is important for HSC gene therapy. We found no evidence of significant silencing, and comparing the real-time PCR data to the EGFP transgene expression data in PB suggests that the average proviral copy number in transduced long-term repopulating cells is approximately 2. Thus in the macaque model, SIN HIV vectors used at MOIs of 5 to 10 resulted in relatively low vector provirus copy numbers and provided robust transgene expression without significant silencing.
In conclusion, we report efficient and reproducible transduction of long-term, multipotent primate repopulating cells in a short overnight transduction protocol using HIV-based lentiviral vectors. These data show that using our described protocol, clinically relevant marking can be obtained in a primate model. Our data also show that pigtailed macaques are an excellent preclinical model for HSC gene therapy using HIV-based vectors. The relatively short transduction protocol described here should be particularly important for diseases such as Fanconi anemia where maintenance of stem cells during ex vivo transduction may be an obstacle to successful gene therapy. The rapid polyclonal engraftment observed in all 3 animals also suggests that VSV-G–pseudotyped HIV-based vectors may be effective for transplantations using less toxic nonmyeloablative preparative regimens, where maintenance of stem cell potential during ex vivo culture will be critical.
Acknowledgments
We thank John Ngo, David Dickerson, Garyfalia Karponi, Kate Beebe, and Julia Morris for technical assistance. We thank the staff of the Primate Center of the University of Washington and the technicians of the hematology and pathology laboratories of the University of Washington. We thank Amgen for providing growth factors. We thank Luigi Naldini and Didier Trono for supplying lentiviral vector constructs. We also acknowledge the assistance of Bonnie Larson and Helen Crawford in preparing the paper.
This work was supported in part by grants DK47754, HL53750, AI061839, AI063959, and DK56465 from the National Institutes of Health (Bethesda, MD). H.-P.K. is a Markey Molecular Medicine Investigator.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.D.T. designed research, performed research, analyzed and interpreted data, and drafted the paper; B.C.B. designed research, analyzed and interpreted data, and performed research; C.G. performed research and collected data; M.W., P.O., and J.F. performed research; P.M. designed research and analyzed and interpreted data; H.-P.K. designed research, analyzed and interpreted data, and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans-Peter Kiem, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D1-100, PO Box 19024, Seattle, WA, 98109-1024; e-mail: hkiem@fhcrc.org.
References
- 1.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 [Erratum in: Science. 2003;302:568]. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 2.May C, Rivella S, Callegari J, et al. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 4.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 5.Beard BC, Keyser KA, Trobridge GD, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, and foamy virus. Hum Gene Ther. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 6.Beard BC, Dickerson D, Beebe K, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 7.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 8.Horn PA, Keyser KA, Peterson LJ, et al. Efficient lentiviral gene transfer to canine repopulating cells using an overnight transduction protocol. Blood. 2004;103:3710–3716. doi: 10.1182/blood-2003-07-2414. [DOI] [PubMed] [Google Scholar]
- 9.An DS, Kung SK, Bonifacino A, et al. Lentivirus vector-mediated hematopoietic stem cell gene transfer of common gamma-chain cytokine receptor in rhesus macaques. J Virol. 2001;75:3547–3555. doi: 10.1128/JVI.75.8.3547-3555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An DS, Wersto RP, Agricola BA, et al. Marking and gene expression by a lentivirus vector in transplanted human and nonhuman primate CD34(+) cells. J Virol. 2000;74:1286–1295. doi: 10.1128/jvi.74.3.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn PA, Morris JC, Bukovsky AA, et al. Lentivirus-mediated gene transfer into hematopoietic repopulating cells in baboons. Gene Ther. 2002;9:1464–1471. doi: 10.1038/sj.gt.3301820. [DOI] [PubMed] [Google Scholar]
- 12.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 13.Stremlau M, Perron M, Lee M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanawa H, Hematti P, Keyvanfar K, et al. Efficient gene transfer into rhesus repopulating hematopoietic stem cells using a simian immunodeficiency virus-based lentiviral vector system. Blood. 2004;103:4062–4069. doi: 10.1182/blood-2004-01-0045. [DOI] [PubMed] [Google Scholar]
- 15.Dull T, Zufferey R, Kelly M, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn PA, Topp MS, Morris JC, Riddell SR, Kiem H-P. Highly efficient gene transfer into baboon marrow repopulating cells using GALV-pseudotype oncoretroviral vectors produced by human packaging cells. Blood. 2002;100:3960–3967. doi: 10.1182/blood-2002-05-1359. [DOI] [PubMed] [Google Scholar]
- 17.Kiem H-P, Andrews RG, Morris J, et al. Improved gene transfer into baboon marrow repopulating cells using recombinant human fibronectin fragment CH-296 in combination with interleukin-6, stem cell factor, FLT-3 ligand, and megakaryocyte growth and development factor. Blood. 1998;92:1878–1886. [PubMed] [Google Scholar]
- 18.Beard BC, Mezquita P, Morris JC, Kiem H-P. Efficient transduction and engraftment of G-CSF-mobilized peripheral blood CD34+ cells in nonhuman primates using GALV-pseudotyped gammaretroviral vectors. Mol Ther. 2006;14:212–217. doi: 10.1016/j.ymthe.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt M, Zickler P, Hoffmann G, et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002;100:2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- 20.Neff T, Horn PA, Peterson LJ, et al. Methylguanine methyltransferase-mediated in vivo selection and chemoprotection of allogeneic stem cells in a large-animal model. J Clin Invest. 2003;112:1581–1588. doi: 10.1172/JCI18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 22.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 23.Agy MB, Frumkin LR, Corey L, et al. Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science. 1992;257:103–106. doi: 10.1126/science.1621083. [DOI] [PubMed] [Google Scholar]
- 24.Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci U S A. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman RM, Hall L, Connole M, et al. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci U S A. 2006;103:19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan G, Kozyrev Y, Kodama T, Hu S-L. Novel TRIM5 isoforms expressed by Macaca nemestrina. J Virol. 2007;81:12210–12217. doi: 10.1128/JVI.02499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi PA, De Angioletti M, Donahue RE, Notaro R, Luzzatto L, Dunbar CE. In vivo gene marking of rhesus macaque long-term repopulating hematopoietic cells using a VSV-G pseudotyped versus amphotropic oncoretroviral vector. J Gene Med. 2004;6:367–373. doi: 10.1002/jgm.514. [DOI] [PubMed] [Google Scholar]
- 31.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 32.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]