SUMMARY
Epidermal lineage commitment occurs when multipotent stem cells are specified to three lineages: the epidermis, the hair follicle, and the sebaceous gland (SG). How and when a lineage becomes specified remains unknown. Here, we report the existence of a population of unipotent progenitor cells that reside in the SG and express the transcriptional repressor Blimp1. Using cell-culture studies and genetic lineage tracing, we demonstrate that Blimp1-expressing cells are upstream from other cells of the SG lineage. Blimp1 appears to govern cellular input into the gland since its loss leads to elevated c-myc expression, augmented cell proliferation, and SG hyperplasia. Finally, BrdU labeling experiments demonstrate that the SG defects associated with loss of Blimp1 lead to enhanced bulge stem cell activity, suggesting that when normal SG homeostasis is perturbed, multipotent stem cells in the bulge can be mobilized to correct this imbalance.
INTRODUCTION
Epidermal appendage formation involves an ordered set of developmental processes beginning with lineage commitment of undifferentiated stem cells, formation of a population of highly proliferative cells, and their subsequent terminal differentiation (Alonso and Rosenfield, 2003; Fuchs et al., 2001; Olivera-Martinez et al., 2004). These stages are well delineated in the developing hair follicle, where the highly proliferative cells, called matrix cells, are located at the base of the follicle in the hair bulb. The matrix cells surround specialized mesenchymal cells (dermal papillae, DP), which are thought to provide a growth stimulus. As matrix cells exit the cell cycle and move upward and away from the DP, they differentiate to specify a central hair shaft surrounded by its channel, or inner root sheath (IRS) and companion layer, that guide the shaft to its orifice at the skin surface. Exterior to these cell layers is the outer root sheath (ORS), which is contiguous with the interfollicular epidermis and contains a reservoir of quiescent stem cells, referred to as the bulge. It has been posited that when quiescent bulge stem cells become activated at the start of each new hair cycle, they exit the bulge niche and proliferate, moving downward to produce the matrix cells (Oshima et al., 2001).
In contrast to the hair follicle, much less is known about the formation of the sebaceous gland (SG), which buds from the upper ORS as a terminally differentiating structure that resides above the bulge. Differentiated sebocytes produce and secrete lipid-rich sebum into the hair canal that empties out to the skin surface (Alonso and Rosenfield, 2003; Stewart, 1992). Currently, two models, not necessarily exclusive, have been proposed to suggest how SG cells might arise. The most widely held view is that the bulge stem cells serve as the residence of the multipotent progenitors which then migrate upward and differentiate to generate the SG. This hypothesis is supported by transplantation experiments where isolated bulge stem cell populations are grafted to analyze cell fate plasticity and are able to form epidermis, hair follicles, and SGs (Blanpain et al., 2004; Morris et al., 2004; Oshima et al., 2001; Taylor et al., 2000). Studies analyzing the rudimentary follicle structures of hairless mutant mice further support a role for bulge stem cells in sebocyte formation (Panteleyev et al., 2000). It is possible that a population of self-renewing progenitor cells reside within the SG itself, and that these progenitors maintain and generate the sebocytes that typify this gland. Fate mapping via retroviral labeling of epithelial cells in vivo has provided some evidence in support of this notion (Ghazizadeh and Taichman, 2001).
Elucidating the mechanisms that drive sebocyte development is of fundamental importance not only to our understanding of stem cell biology and multipotency in the skin but also of two major and as yet unsolved health concerns, namely acne and sebaceous cancers. The end point in sebocyte differentiation and lipid production is likely to be controlled at least in part by the adipogenic transcription factor PPARγ, which is expressed in differentiating sebocytes (Rosenfield et al., 1998). Although PPARγ null mice are not viable, PPARγ null ES cells have been found to contribute poorly to the SGs of chimeric mice, lending some supporting functional evidence to this notion (Rosen et al., 1999). Most of the knowledge regarding earlier steps in the process stem from overexpression or dominant-negative studies in mice. Overexpression of c-Myc in the hair follicle induces SG hyperplasia at the expense of hair follicle differentiation (Arnold and Watt, 2001; Bull et al., 2005; Waikel et al., 2001), and alterations in hedgehog and Notch signaling proteins and their effectors result in perturbations in SG development (Allen et al., 2003; Niemann et al., 2003; Pan et al., 2004). How these factors contribute to the SG lineage is not yet clear.
In a screen to uncover new transcription factors in developing skin epithelium (to be published elsewhere), we identified B lymphocyte-induced maturation protein 1 (Blimp1) as a potential regulator of epidermal appendage development. Blimp1 is expressed in differentiating plasma cells, where it acts to repress genes involved in proliferation and promote differentiation (Lee et al., 2003; Lin et al., 1997, 2000; Tunyaplin et al., 2004). In primordial germ cells, Blimp1 regulates cell-lineage specification (Ohinata et al., 2005; Vincent et al., 2005). Blimp1 performs these functions by binding to DNA through its proline-rich zinc finger domain and recruiting transcriptional cofactors such as hGroucho, histone deacetylases, and histone methyltransferases (Gyory et al., 2004; Lee et al., 2003; Ren et al., 1999; Yu et al., 2000).
Given these interesting developmental roles of Blimp1, we wondered whether Blimp1 might regulate the proliferation/differentiation of skin progenitor cells. In the course of testing this hypothesis, we unexpectedly discovered a unique population of Blimp1-expressing cells that reside within the SG. Using conditional gene targeting, we show that loss of Blimp1 results in larger SGs, with enhanced pools of both slow-cycling progenitors and proliferative cells, accompanied by increased c-Myc expression. Finally, although bulge cells do not express Blimp1, they display signs of enhanced activity, which parallels the increased flux of BrdU-labeled cells through the Blimp1-deficient SGs. Taken together, our data suggest that the population within the SG that are marked by Blimp1 are unipotent progenitors, which control gland homeostasis and govern the activity of bulge stem cells.
RESULTS
Blimp1 Expression in a Novel SG Cell Population
In conducting microarray analyses, we identified Blimp1 (also known as PRDM1; Huang, 1994; Keller and Maniatis, 1991) as a transcription factor whose increased expression correlated with late stages of embryonic skin morphogenesis (data not shown). This finding was intriguing given a prior developmental study on Blimp1, in which immunohistochemical staining was noted in a wide variety of embryonic tissues, including E18 surface ectoderm, mesenchymal precursors of the dermal papillae (DP), and inner root sheath (IRS) of developing whisker follicles (Chang and Calame, 2002).
To explore Blimp1 expression in greater detail, we first generated Abs against the 2nd Blimp1 exon. We also employed two BAC transgenic mouse lines that express EGFP under the control of Blimp1’s regulatory elements (Blimp1GFP) (Ohinata et al., 2005). The patterns of expression of Blimp1GFP and Blimp1 Ab staining corresponded well, validating the efficacy of the anti-Blimp1 staining patterns as faithful reflections of where the Blimp1 gene was expressed. We subsequently used these tools interchangeably.
We first confirmed the embryonic expression of Blimp1 in nuclei of DP, suprabasal epidermis, and hair follicle IRS (Figures 1A–1C). As hair follicle morphogenesis proceeded, Blimp1 extended to all three mature IRS layers, and to cortical and medullar cells of the hair shaft, which trails IRS differentiation (Figure S1). While Blimp1 correlated with terminally differentiating cells of the epidermis and hair follicles, it was conspicuously absent in proliferative compartments (Figures 1, S1, and S4). Blimp1 was not detected in E15.5 skin or earlier, i.e., at times when embryonic epidermis exists as a single layer of multipotent progenitor cells (Figure 1A).
Figure 1. Blimp1 Expression in the Sebaceous Gland.

Immunofluorescence and/or epifluorescence microscopy was performed on skin sections of wild-type or Blimp1GFP transgenic mice at ages indicated. Color-coding is according to secondary Abs or GFP and is marked on each frame. (H), (K), and (M) are confocal images. Schematic in (N) summarizes the localization of Blimp1-expressing cells in the SG. Abbreviations: β4, β4 integrin; SG, sebaceous gland; ep, epidermis; DP, dermal papilla; IRS, inner root sheath; HS, hair shaft, positive for Blimp1GFP (*); NB, newborn; Ad, adult; Bu, bulge (bracketed). Arrows denoted Blimp1+ or Blimp1GFP+ cells in or near the orifice to the sebaceous gland. Note that patterns of anti-Blimp1 Ab and Blimp1GFP expression were comparable. Blimp1 was first observed at E17.5, in the suprabasal epidermis and dermal papillae (DP). By birth, Blimp1 expression expanded to the IRS and to a site in upper outer root sheath (ORS) where the sebaceous gland (SG) buds. Blimp1-expressing SG cells were K14/K5+, Ki67−, ORO−, PPARγ −, and CD34−.
Beginning late in embryonic development and extending into postnatal life, Blimp1+ nuclei appeared in the upper portion of maturing hair follicles (Figures 1C and 1D). Blimp1GFP and anti-K14 IgG colabeled these interesting cells (Figure 1E), and anti-Blimp1+ nuclei were contained within K14+ cytoplasms of cells (Figure 1F). These findings identified these cells as keratinocytes and were distinct from previous reports of Blimp1 localization in the hair follicle.
The Blimp1+ cells in the upper hair follicle appeared to be within or adjacent to SGs, which express the early differentiation marker PPARγ, known to govern a battery of genes involved in lipid synthesis (Figures 1F–1H). The Blimp1+ cells were also not Oil Red O+, which stains later in the maturation process as sebocytes become densely filled with lipids (Figure 1I) (Rosenfield et al., 2002). Surprisingly, the Blimp1+ keratinocytes were also negative for Ki67, a nuclear marker of actively cycling keratinocytes, and β4 integrin, which marks the basal row of ORS, bulge, and SG keratinocytes adhering to the surrounding basement membrane (Figures 1J–1L). Blimp1 and the bulge marker CD34 did not colocalize (Figure 1M). These cells were also distinct from and resided externally to the K6-positive companion layer, trichohyalin-positive IRS (AE15+) and hair keratin-positive hair shaft (AE13+) (Figure S1).
Summarized in Figure 1N, these data reveal that Blimp1 is expressed by a group of keratinocytes that reside at or near the bud site of the SG and possess molecular properties that distinguish them from the ORS, the bulge, and the differentiated cells of the hair follicle. These cells are also distinct from the proliferative and terminally differentiating sebocytes that were previously thought to be the sole constituents of the SG.
Blimp1 Regulates the Size and Activity of Glands Composed of Sebocytes
Mice null for the Blimp1 gene die as early embryos (Ohinata et al., 2005; Vincent et al., 2005; and references therein). To determine Blimp1’s role in skin, we bred Blimp1fl/fl mice (Ohinata et al., 2005) to transgenic mice expressing the Cre recombinase under the control of the K14 promoter, which is active by E15.5 in embryonic skin keratinocytes, including multipotent skin progenitors (Vasioukhin et al., 1999).
Neonatal mice genotypic for K14-Cre and Blimp1fl/fl alleles were born in the expected Mendelian numbers and grew to normal size as adults (Figures 2A–2C). Real-time PCR of epidermal mRNAs and anti-Blimp1 immunohistochemistry verified that expression was conditionally ablated as a consequence of the targeting event (data not shown and Figure 2D). Blimp1 conditional knockout (cKO) mice appeared indistinguishable from their wild-type (wt) counterparts for their first 4 weeks of postnatal life. Histological and biochemical studies revealed no overt differences in the epidermis or hair follicles. The hairs were comparable in types and lengths, and the hair cycle appeared to be morphologically comparable over 2 months of age (Figure S2F and data not shown). As cKO mice aged, however, their hair coat developed an oily appearance (Figure 2B). Hematoxylin and Oil Red O staining revealed an enlargement of the SGs, and quantification of stained intact glands indicated that they were nearly 4× larger than normal (Figures 2E–2G).
Figure 2. Blimp1 Regulates Sebocyte-Containing Gland Size.

K14-Cre:Blimp1fl/fl conditional knockout (cKO) mice and organs are compared to wild-type (WT) littermate controls.
(A and B) Images of hair coats of 2-month-old mice.
(C) PCR genotype analyses.
(D) Anti-Blimp1 immunostaining of skin sections from 6 day WT and Blimp1 cKO mice. Sections were counterstained with hematoxylin.
(E–L) Data from 2-month-old WT and Blimp1 cKO mice. All SG analyses were performed in telogen phase of the hair cycle.
(E and F) Brightfield images of Oil Red O and hematoxylin-stained skin sections (E) and whole mounts of Oil Red O-stained tail skins (F).
(G) Sebaceous gland measurements. The average area of 50 individual WT and cKO sebaceous glands was determined. n = 3 mice for each genotype.
(H) Thin-layer chromatography of skin lipids. CE, cholesterol esters; TG, triglycerides; C, cholesterol.
(I) Real-time PCR of skin mRNAs tested for sebocyte markers (Mc5R, PPARγ, and SCD3). n = 4 for each genotype.
(J) Brightfield images of hematoxylin and eosin staining of eyelid skin showing meibomian glands.
(K) Brightfield images of preputial glands from male mice. Abbreviations: Meibomian glands (meib), epidermis (ep), and muscle (m).
(L) Measurements of meibomian and preputial gland area. n = 4–5 for each genotype. Asterisks indicate significance, p < 0.05.
The increased gland size was also reflected at the biochemical level. Thin layer chromatography analyses of sebum lipids displayed an overall increase in cholesterol esters, triglycerides, and cholesterol per unit area of cKO skin (Figure 2H). Furthermore, real-time PCR indicated a 5–10× increase in levels of mRNAs encoding the sebocyte markers Mc5R, SCD3, and PPARγ but not the epidermal marker PPARα (Michalik et al., 2001) (Figure 2I). Following the paradigm set for Blimp1-deficient SGs, the meibomian glands of the eyelid and the preputial glands of the male reproductive system were also significantly enlarged in Blimp1 cKO animals (Figures 2J–2L). Taken together, these data provide compelling evidence for an essential role for Blimp1 in controlling the sizes and proportionate activity of different glands within the body that contain sebocytes.
Finally, since skin lipids can contribute to epidermal barrier function and differentiation (Dai and Segre, 2004) we analyzed barrier formation and epidermal differentiation (Figure S2). As judged by toluidine blue dye penetration assays, both wt and cKO embryos acquired full barrier function between E16.5 and E17.5, and similarly wt and Blimp1 cKO skins displayed indistinguishable patterns of differentiation markers both in embryos (shown) and adult mice (not shown). These data further substantiated that the epidermis is not markedly affected by Blimp1 ablation, and that the defects in lipids result from enlarged SG and increased sebum production. The differences in PPARγ expression but not PPARα were also consistent with this notion.
Blimp1 Regulates Cell Numbers within SGs
To determine if alterations in cell size or cell number might underlie the increased size of sebocyte-containing glands in Blimp1 cKO mice, we first counted nuclei within the SGs of stained skin tissue sections. Based upon these data, the average cell number per gland in cKO sections was more than 50% larger (Figures 3A and 3B). Similar increases were observed when PPARγ was used to distinguish the differentiating gland cells (Figures 3C and 3D). This was true for both adult and juvenile cKO glands, indicating that the enhanced numbers of sebocytes preceded the overt appearance of the excessively oily coat of the cKO mice. Finally, when Ki67+ cells in the glands were quantified, an increase of ~5× was detected in cKO skin (Figures 3E and 3F). Based upon these data, the increase in sebocytes in Blimp1-deficient glands was attributable to enhanced proliferation and expansion of the proliferative sebocyte pool.
Figure 3. Increased Cell number and Proliferation in Blimp1 Null Sebaceous Glands.
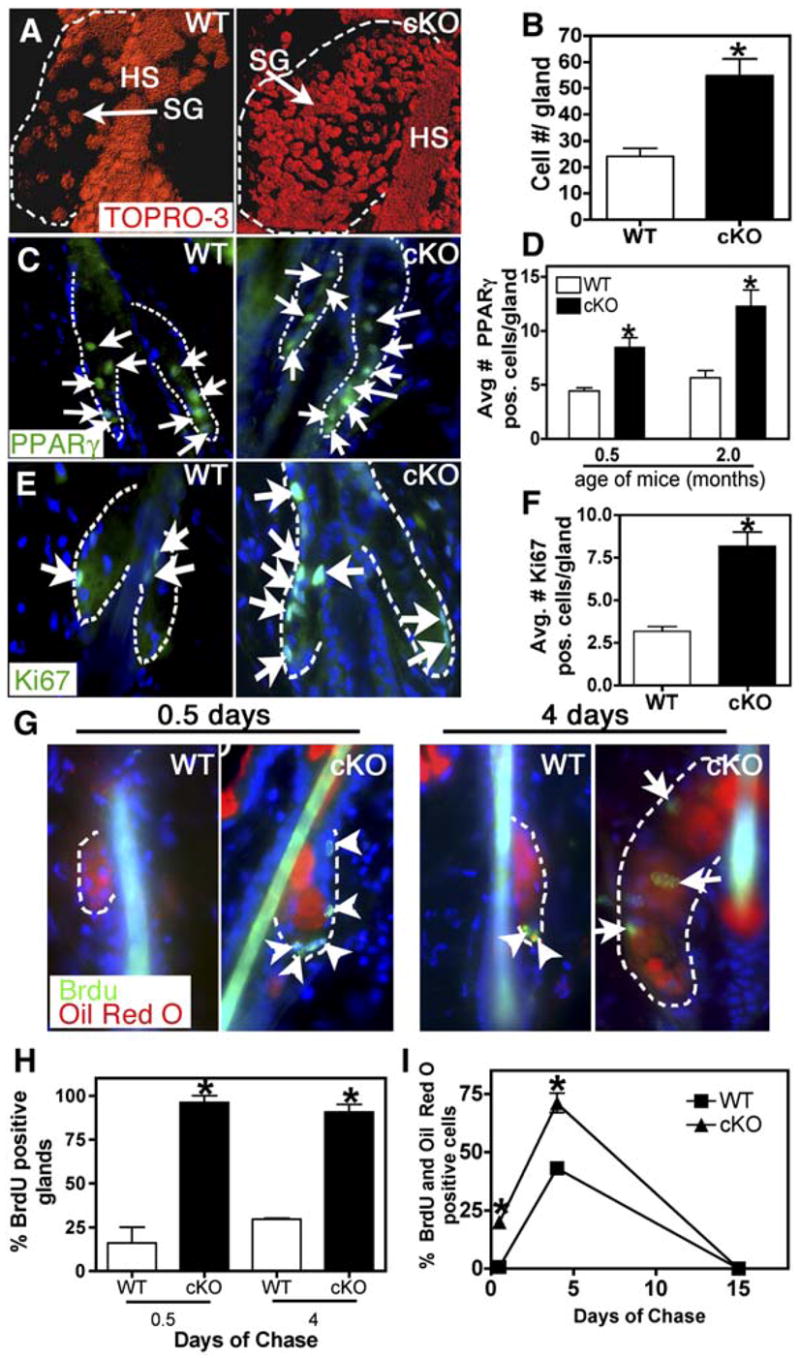
All analyses of SG cell number and proliferation were performed on telogen phase hair cycles in 2-month-old mice.
(A) Confocal images of tail whole-mount skin sections stained with the nuclear dye TOPRO-3. Images were edited with Imaris software.
(B) Quantification of cell number in SGs. n = 3 for each genotype.
(C and D) Immunostaining with anti-PPARγ Abs of skin sections from 2-month-old mice. Nuclei are counterstained with DAPI. Graph shows quantification of PPARγ-positive nuclei. n = 3 for each genotype.
(E and F) Immunostaining with anti-Ki67 Abs of skin sections from 2-month-old mice. Graph shows quantification of Ki67-positive nuclei in individual SGs. n = 3 for each genotype.
(G) Epifluorescence of Oil Red O and anti-BrdU immunofluorescence of 2-month-old mice pulsed with BrdU and chased for either 0.5 or 4 days. Arrows indicate nuclei within the Oil Red O+ cells, while arrowheads indicate nuclei outside of these cells.
(H) Quantification of SGs containing BrdU+ nuclei at indicated time points.
(I) Quantification of the percentage of BrdU+/Oil Red O+ cells after 0.5, 4, and 15 days of chase. Asterisks indicate signifcance, p < 0.05.
To monitor the proliferative residents and their fate within the SG, we performed a BrdU pulse-chase experiment on 2-month-old mice when their hair follicles were in telogen (Figure S2F). Following a BrdU pulse, skin sections were analyzed for anti-BrdU labeling (green) and Oil Red O counterstaining after 0.5, 4, or 15 days of chase (Figures 3G–3I). These data indicated that significantly more Blimp1-deficient SG cells were in the S phase of the cell cycle than their wt counterparts. Correspondingly, following the 4 day chase period, many of these BrdU-labeled cells were now also Oil Red O+, indicating that the BrdU-labeled cells that were proliferating at 0.5 days had now entered a terminal differentiation program to produce lipids. Consistent with the flux of proliferative cells through and out of the gland, BrdU labeling was undetectable in the skins of either genotype after 15 days of chase. Taken together, these findings establish that the defects in Blimp1 cKO sebaceous glands arise from an initial increase in Ki67+ proliferating SG cells. This results in elevated numbers of PPARγ+/Oil Red O+ differentiating sebocytes, which are eventually sloughed into the sebum canal that exits at the skin surface.
Loss of Blimp1 Results in Misregulation of c-myc in the Sebaceous Gland
Blimp1 cKO mice bore some phenotypic resemblance to K14-c-myc-overexpressing transgenic mice, which also displayed enlarged SGs (Arnold and Watt, 2001; Bull et al., 2005). Given that Blimp1 represses c-myc promoter activity in plasma cells (Lin et al., 1997; Yu et al., 2000), we tested whether the phenotype in Blimp1 cKO SGs is directly attributable to c-myc misregulation.
To identify which if any of the cells within the SG naturally express c-Myc, we first colabeled Blimp1GFP P6 skin with anti-c-Myc Abs. Anti-c-Myc labeled nuclei of cells within the basal and first suprabasal layers of epidermis and upper ORS, as well as a subset of Blimp1− cells within the SG (Figures 4A and 4B). c-Myc+ SG cells were uniformly K5+ and Oil Red O−, but only a subset of them labeled with anti-Ki67 (Figures 4B–4D). This pattern was maintained as animals aged, showing c-Myc expression in both proliferating and differentiating sebocytes.
Figure 4. c-Myc Expression Is Elevated in Blimp1 cKO Skin.
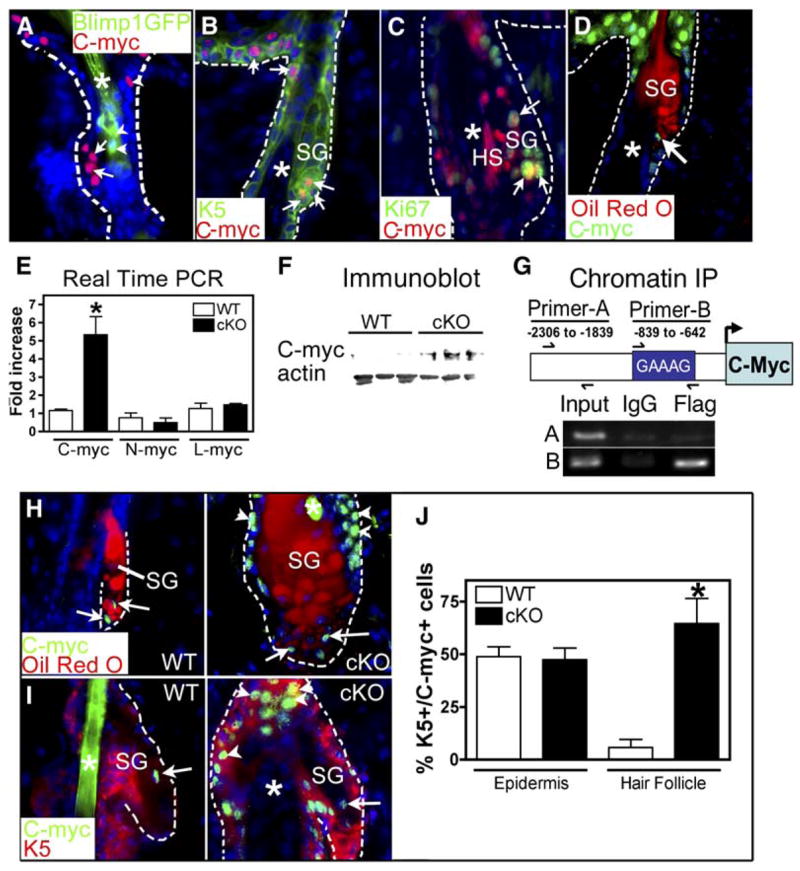
(A–D) Localization of c-Myc within SGs of P6 Blimp1GFP (A) or WT (B–D) mice. Skin sections were labeled with Abs against c-Myc, K5, and/or Ki67 or costained with Oil Red O as indicated. Nuclei were counterstained with DAPI. Arrowheads, Blimp1GFP-expressing cells; arrows, c-Myc+ nuclei; asterisks, hair shaft (HS).
(E) Real-time PCR analysis of c-myc, L-myc, and N-myc mRNAs in backskins of 2-month-old mice. Asterisks indicate significance, p < 0.05.
(F) Anti-c-Myc immunoblot analysis of 2-month-old backskin proteins.
(G) Chromatin immunoprecipitation (IP) assays were performed on keratinocytes expressing a flag epitope-tagged, recombinant Blimp1 protein with anti-FLAG or control IgG antisera. PCR analyses on the input or immunoprecipitated fragments were performed using primers encompassing a Blimp1 binding site previously shown to bind Blimp1 (Primer-B) or primers spanning an upstream region of the c-myc promoter lacking Blimp1 binding sites (Primer-A).
(H and I) Immunostaining with anti-c-Myc Abs and Oil Red O counterstain (H) or anti-K5 antibodies (I). Arrows indicate c-Myc+ nuclei within the SG. Arrowheads indicate c-Myc-positive nuclei outside the gland. Asterisks denote hair shaft.
(J) Quantification of the number of K5+ c-Myc+ cells in epidermis and hair follicles of 2-month-old mice. Asterisks indicate significance, p < 0.05.
If Blimp1 acts upstream of c-Myc to transcriptionally repress c-myc in undifferentiating cells of the SG, as it does in plasma cells, then Blimp1 ablation should raise c-myc expression levels. Real-time PCR revealed that mRNAs encoding c-myc, but not N-myc or L-myc, were markedly elevated by >5-fold in skin lacking Blimp1 (Figure 4E). Immunoblot analyses further confirmed that although c-Myc was barely visible in protein extracts from wt skin, it was readily detected in extracts from cKO skin (Figure 4F). Finally, if Blimp1 directly regulates c-myc gene expression, it should bind to the c-myc promoter. When chromatin-protein complexes from flag epitope-tagged, Blimp1-expressing keratinocytes were immunoprecipitated with anti-flag Abs, an ~200 bp DNA fragment was pulled down that encompassed a Blimp1 binding site in the c-myc promoter that had previously been shown by electrophoretic mobility assays to bind to Blimp1 protein in B plasma cells (Yu et al., 2000) (Figure 4G). By contrast, PCR control primers corresponding to an upstream c-myc promoter segment failed to produce product in samples immunoprecipitated with either control IgG or Flag Abs.
Finally, the number of anti-c-Myc-labeled cells was markedly elevated in Blimp1-deficient SGs (Figures 4H and 4I). As in wt glands, the majority of c-Myc+ cells in cKO glands were Oil Red O− and K5+. Further analyses of K5+c-Myc+ cells in the skin revealed that while numbers were unchanged in the epidermis, they were ~6× higher in the upper portion of cKO hair follicles compared to wt (Figure 4J). When taken together with the phenotype of K14-c-myc overexpression studies (Arnold and Watt, 2001; Bull et al., 2005; Waikel et al., 2001), our findings suggest that Blimp1 governs the size of SGs by repressing c-myc gene expression.
The Blimp1+ Cells Represent a Resident Population of Sebocyte Progenitor Cells
To test whether the Blimp1-expressing cells in the SGs might be resident progenitor cells of the sebaceous gland, we began by culturing primary mouse keratinocytes in vitro from Blimp1GFP+ animals (Figure 5A). Blimp1GFP+ and Oil Red O+ cells adhered to a fibroblast feeder layer and were independently dispersed throughout the dish (Figure 5A). After 5 days in culture, the numbers of Oil Red O+ cells had increased markedly. However, they now appeared as colonies of sebocytes and ~95% of the Oil Red O+ colonies that developed were associated with Blimp1GFP+ cells (Figures 5A and 5B). In addition, sebocyte colony numbers increased upon subsequent passage (data not shown), as did the numbers of GFP+ cells (Figure 5A), suggesting that the Blimp1+ cells are able to proliferate and give rise to sebocytes in vitro.
Figure 5. Blimp1+ SG Cells Are a Resident Population of Progenitor Cells with Properties of Unipotent Progenitor Cells.
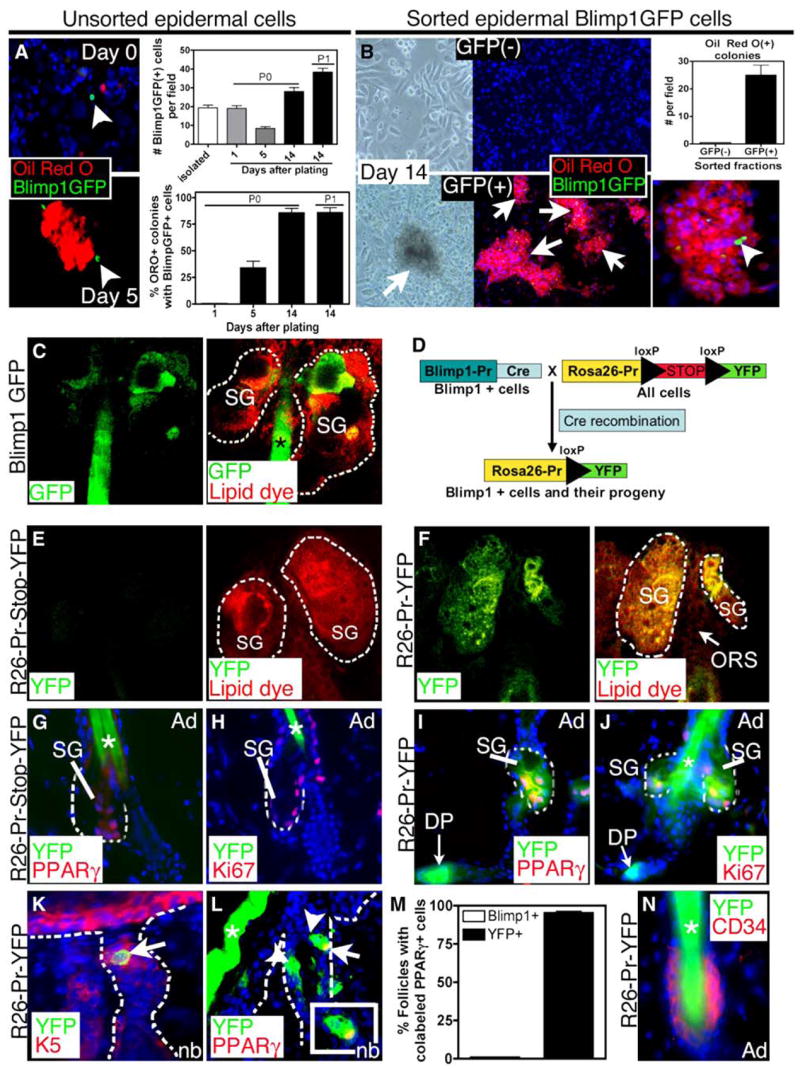
(A and B) Cell culture. Isolated Blimp1GFP keratinocytes are initially distinct from Oil Red O+ cells, but after day 5, sebocyte colonies composed of GFP+ and Oil Red O+ cells were detected. Numbers of Blimp1GFP+ cells increased with passage. (B) FACS isolated Blimp1GFP+ and (−) cells were cultured for 14 days and stained for Oil Red O. Oil Red O colonies were only found in cultures of GFP+ cells.
(C) Confocal projection of GFP and lipid dye (FM 4-64) epifluorescence from a whole mount of Blimp1GFP Bac transgenic tail skin.
(D) Schematic illustrating the genetic strategy to lineage-trace Blimp1+ SG cells in the skin.
(E and F) YFP epifluorence of tail skin whole mounts (left) stained a membrane dye (merge, right). Note YFP expression in the entire sebaceous gland but exclusion of signal in the ORS and bulge.
(G–L) Backskin sections of R26-Pr-Stop-YFP or R26-Pr-YFP mice were immunostained with Abs against GFP and either PPARγ (G, H, and L), Ki67 (H and J), or K5 (K).
(M) Quantification of the percentage of hair follicles containing either Blimp1+ (Blimp1GFP skin sections) or YFP+ (R26-Pr-YFP skin sections) cells that colocalize with PPARγ.
(N) Backskin sections of R26-Pr-YFP mice were immunostained with Abs against GFP and CD34. Note the colocalization of YFP with PPARγ- and Ki67-positive sebocytes and in the DP in R26-Pr-YFP mice. SG, Sebaceous gland; HS, hair shaft; DP, dermal papillae.
To more rigorously analyze the ability of Blimp1GFP+ cells to act as sebocyte progenitors, we used fluorescence-activated cell sorting (FACS) to sort Blimp1GFP+ and GFP− cells prior to culturing. As shown in Figure 5B, while the GFP− population gave rise to epidermal keratinocytes, the GFP+ cells gave rise to colonies with distinct morphology and with lipid-filled, Oil Red O+ cells. Similar to the experiments using unsorted cells, the Oil Red O+ colonies contained GFP+ cells, The increased number of Blimp1GFP+ cells in culture and with passage revealed a capacity to self-renew, and their ability to form sebocyte colonies underscored their action as progenitors for the terminally differentiated cells of the SG.
To document the behavior of Blimp1+ cells as progenitors in vivo, we next performed a genetic lineage tracing experiment. For this purpose, we crossed Blimp1Cre-BAC transgenic mice with mice expressing a transgene containing a floxed-Stop-floxed sequence situated between a ubiquitously expressed Rosa26 (R26) promoter and the yellow fluorescent protein (YFP) coding sequence (Ohinata et al., 2005; Srinivas et al., 2001). By expressing Cre recombinase under the control of the Blimp1 promoter, the floxed stop codon at the Rosa26 locus should be excised to activate YFP expression exclusively in Blimp1+ cells and their descendents, irrespective of Blimp1 promoter status (Figure 5D). If Blimp1+ cells are a part of the sebocyte lineage, then their position along the lineage should be readily assessed by analyzing the expression of Rosa26-YFP within the gland.
To evaluate Rosa26-YFP expression in SGs, initially we investigated whole, unfixed SGs of tail skins of Rosa26 promoter-Stop-YFP single transgenic adult mice (R26-Pr-Stop-YFP), and Blimp1-Cre/Rosa26-floxed-stop-floxed-YFP double transgenic mice (R26-Pr-YFP). After treating skins with dispase, the isolated skin epithelium was then counterstained with a lipid dye (Red), which intercalates into cell membranes. We then visualized the whole mounts by double fluorescence confocal microscopy.
As expected, YFP was not detected in single transgenic tail skins expressing either R26-Pr-Stop-YFP alone (Figure 5E) or Blimp1-Cre alone (not shown). By contrast, YFP was detected broadly in SG cells of R26-Pr-YFP tail skin (Figure 5F). The expression of R26-Pr-YFP in glands of these mice provided compelling evidence that most if not all of the cells of the SG arise from the resident population of Blimp1-expressing keratinocytes. To further verify that Blimp1-expressing SG cells contribute to both the differentiating and proliferating sebocytes, we immuno-stained R26-Pr-YFP skin sections with anti-PPARγ and anti-Ki67 antibodies. Although Blimp1 was not detected in either of these compartments (see Figure 1), YFP was detected in both differentiated PPARγ+ sebocytes (Figures 5G and 5I) and proliferative Ki67+ cells (Figures 5H and 5J).
To further analyze the contribution of Blimp1-expressing cells to the SG lineage, we examined newborn mice in which the initial genesis of SGs occurs, as indicated by PPARγ in 50% of developing hair follicles. In newborn hair follicles from R26-Pr-YFP skin sections, YFP+K5+ cells were seen in the upper hair follicle (Figure 5K), similar to the expression of Blimp1 in newborn hair follicles (Figure 1C). Additionally, in ~98% of follicles displaying PPARγ+ cells, YFP exhibited coexpression (Figures 5L and 5M). By contrast, Blimp1+ cells did not coexpress PPARγ (Figures 1G and 5M).
Based upon these collective analyses, we conclude that the Blimp1+ population constitutes a pool of resident SG progenitor cells. The expression of YFP in SG cells that do not express the Blimp1 gene was in marked contrast to YFP expression patterns in the epidermis, hair follicle, and DP (Figures S3 and S4). These data confirmed that the Blimp1+ cells in the epidermis and hair follicle reside downstream from the proliferative compartment, which remained negative for R26-Pr-YFP expression (Figures S3 and S4). Finally, it was notable that R26-Pr-YFP expression was not detected in the CD34+ follicle bulge (Figure 5N).
Increased Activity within the Bulge Stem Cell Compartment of Blimp1 cKO Follicles
Although bulge stem cells were not Blimp1+ themselves (see Figure 1E), they can reconstitute SGs in skin grafts (Blanpain et al., 2004), leading us to wonder whether they might be the precursors of the Blimp1+ SG progenitor cells. Moreover, if the Blimp1−, label-retaining cells (LRCs) of the bulge have the capacity to generate the Blimp1+ progenitors of the SG, then they might be expected to be utilized more frequently when Blimp1 is conditionally ablated. To test whether the activity of bulge cells is enhanced, we first examined proliferation in the bulge and in the ORS bridge between the bulge and the SG, in 2-month-old animals whose hair coats were in the telogen phase of the hair cycle (Figure S2). As shown in the representative images in Figure 6A, cKO follicles exhibited more Ki67+ cells in the bulge and in the ORS segment just above the bulge than their wt counterparts. By contrast, the numbers of cycling cells within the cKO and wt epidermis were comparable, and no differences in epidermal differentiation were observed (Figure S2).
Figure 6. Enhanced Activity of Bulge Stem Cells in Blimp1 cKO Skin.

(A) Increased Anti-Ki67 along anti-β4 ORS above the bulge (Bu).
(B) After a 12 hr BrdU pulse, 2-month-old back-skin samples were analyzed for anti-BrdU (arrows in the SG, arrowhead nucleus in ORS above the bulge) and anti-K5 immunofluorescence. Note: The anti-K5 chicken Ab used displayed atypically reduced staining in the bulge. Asterisks denote hair shaft.
(C and D) After a 36 hr pulse, 2-month-old backskin samples were analyzed for BrdU and CD34. For quantification, n = 3 animals each.
(E–I) At P6, matched WT and Blimp1 cKO litter-mates were given a 72 hr pulse of BrdU and were analyzed after a 28 day chase.
(E) FACS analysis of backskin cells shows that the proportions of α6+/CD34+ bulge cells are similar.
(F and G) Representative histogram (F) and quantification (G) of the number of α6+/CD34+ cells with BrdU incorporation after BrdU pulse-chase. n = 3 for each genotype. Asterisks indicate significance, p < 0.05.
(H and I) Immunofluorescence was used to analyze the numbers of BrdU (green, arrowheads) label-retaining bulge cells (CD34+, red). For quantification, n = 3 mice each.
(J and J′) GFP+ epifluorescence and anti-Blimp1 immunofluorescence colocalize at the SG base of sections of skin grafts derived from a single K14-actinGFP bulge stem cell, after clonal expansion in vitro. (J′) shows DIC phase-contrast.
(K–N) Samples from (H) revealed BrdU+CD34− LRCs above the CD34+ (red) cKO bulge (K). This population at the SG base is positive for K5 (L and N) and shows partial colocalization with anti-c-Myc Abs (M). (N) shows quantification of K5- and BrdU-positive cells at the SG base in WT and cKO mice. n = 3 for each genotype. Asterisks indicate significance, p < 0.05.
In response to short pulses of BrdU (5–12 hr), occasional cells within the SG and ORS intersegment were labeled, but the bulge compartments were not (Figure 6B). With longer BrdU pulses (36 hr), however, marked differences were observed in the CD34+ bulge cells of telogen phase follicles of Blimp1 cKO mice (Figures 6C and 6D). Most notable was an ~8× increase in the number of cKO follicles displaying BrdU-labeled cells within the bulge compartment. Enhanced labeling was also seen in the cKO sebaceous gland and ORS bridge, as we had already noted (Figures 4 and 6A).
To probe more deeply into the proliferative activity of bulge cells in Blimp1 cKO skin, we administered extended (3 day) BrdU pulses at either P6 or P28, then followed each with 28 day chases and assayed for label retention by FACS and immunofluorescence on the basis of their surface expression of CD34 and α6 (Blanpain et al., 2004; Trempus et al., 2003). As shown in Figure 6E, the patterns of CD34 and α6 expression were similar in wt and cKO skin cells, and no differences in bulge cell number or size were detected. However, the % of BrdU+ bulge cells was significantly reduced in cKO skin, and this was confirmed by immunofluorescence microscopy (Figures 6F–6I). The inability of cKO bulge cells to retain label was observed irrespective of whether the label retention was analyzed in anagen (Figures 6E–6G) or telogen (Figures 6H and 6I). At both of these times, hyperproliferation was prevalent in the cKO sebaceous glands, but not the epidermis (Figures 3 and S2). The dramatic reduction in LRCs within the bulge correlated well with the observed increase in proliferating ORS cells between the bulge and SG.
Based upon these data, the enhanced proliferation in the cKO bulge and upper ORS segment seemed likely to be a consequence of the defect in SG homeostasis caused by Blimp1 deficiency. To directly address whether bulge cells are capable of producing unipotent Blimp1+ sebocyte progenitors, we first addressed whether skin grafts derived from the individual K14-GFP+ bulge stem cells harbor GFP+ sebaceous gland progeny that are Blimp1 positive (for method, see Blanpain et al., 2004). Immunofluorescence and phase-contrast microscopy revealed Blimp1 expression in all of the expected epithelial locations of the graft. Most notably, Blimp1+ K14-GFP+ cells were detected in the SG, and by phase-contrast, it was clear that these cells resided at the mouth of the gland (arrowheads, Figures 6J and 6J′). These data support the hypothesis that the Blimp+ unipotent progenitor cells within the SG are descendants of the multipotent stem cells in the bulge.
Finally, although Blimp1 ablation resulted in increased proliferative activity and loss of LRCs within the bulge, the total number of LRCs was similar to that of wt mice (Figure 6G). Closer inspection revealed the existence of a new population of K5+ LRCs near or at the base of the SG (arrowheads in Figure 6K). In contrast to bulge LRCs, these LRCs lacked CD34 expression and instead often showed labeling with anti-c-Myc Abs (Figures 6K–6M). Quantification revealed that the loss in LRCs in the bulge (Figures 6H and 6I) was accompanied by an equivalent ~10× increase in follicles containing LRCs in this new residence (Figure 6N).
DISCUSSION
Our data suggest that late in embryonic development, near the birth of the animal, a population of Blimp1-expressing sebocyte progenitor cells is established to form and maintain the SG. Once the gland is formed, these progenitors reside near the entrance of the gland to control its homeostasis. Our genetic lineage-tracing experiments and cell-culture studies demonstrate that these cells are the progenitors of the cells within the gland, including the proliferative, i.e., transiently amplifying, sebocytes that subsequently differentiate to form the sebum-secreting cells. By contrast, the Blimp1+ SG cells are negative for both proliferation and differentiation markers. That said, Blimp1+ SG cells can multiply in culture, suggesting that these cells not only have the capacity to generate the SG but they also possess the capacity to self-renew.
The Blimp1+ stem cells are biochemically distinct from the multipotent stem cells of the bulge, which are negative for Blimp1 (this paper) and positive for CD34, Tcf3, and other markers (Blanpain et al., 2004; Morris et al., 2004; Tumbar et al., 2004). Moreover, our lineage-tracing experiments demonstrate that even though Blimp1+ cells are able to generate the entire SG, they do not migrate out of the gland to generate progeny within the epidermis or hair follicle as R26-Pr-YFP-expressing cells were not detected in proliferative compartments in these lineages. Our results are intriguing in light of lineage-tracing experiments in mice that were transduced with lacZ-marked retroviral vectors (Ghazizadeh and Taichman, 2001). In those studies, the authors noted chimeric lacZ expression in clusters of epithelial cells within some pilosebaceous units following five rounds of induced hair growth by depilation. In particular, it was notable that an appreciable number of retroviral transduced pilosebaceous units in these experiments displayed a lacZ-positive SG without labeling other cellular compartments. These results can best be explained by the existence of long-lived progenitor populations in the gland. Our studies provide strong support of this hypothesis and provide the first insights into the molecular characteristics of these resident progenitor cells.
The increase in the numbers of highly proliferative cells in Blimp1 null SGs supports a model whereby Blimp1 controls the production of transiently proliferative cells of the SG, which in turn differentiate to enhance the numbers of sebum-producing sebocytes (Figure 7). Since Blimp1 is a transcriptional repressor expressed in the SG exclusively by the sebocyte progenitors, we surmise that Blimp1 governs some aspect of gene expression in these cells that impacts directly on the flux of proliferating and differentiating cells within the gland. There are several ways in which Blimp1 might control this balance. Upstream, Blimp1 could regulate the contribution of bulge stem cells to the SG by controlling a pathway involved in communication between the bulge stem cell niche and the SG. Downstream, Blimp1 could control the transition from dormant progenitor cells into rapidly proliferating sebocytes by either repressing a key cell-cycle regulatory gene(s) that might be essential for converting the dormant progenitor cells into rapidly proliferating sebocytes or by repressing the expression of a secreted factor that regulates sebocyte proliferation through a nonautonomous mechanism.
Figure 7. Model for the Role of Blimp1 in Sebocyte Development.

Blimp1 is expressed in a population of cells that act as unipotent precursors for the proliferative sebocytes, which in turn differentiate into lipid-containing sebocytes. Blimp1 represses c-Myc expression in these sebocyte progenitors and regulates the size and number of cells within the gland. Upstream, the multipotent bulge stem cells are sensitive to the Blimp1-positive progenitors at the base of the gland and can replenish them as necessary. Blimp1 null glands may resemble a wound situation, as without Blimp1, the resident progenitor cells are pushed down a distinct differentiation pathway.
Our grafting studies with clonogenic bulge cells demonstrate clearly that bulge stem cells can give rise to Blimp1+ SG progenitor cells. In addition, our pulse-chase experiments demonstrate that bulge stem cells respond to the absence of Blimp1 by enhancing their proliferation rate. Since bulge cells do not express Blimp1, their activation must be a reflection of enhanced stem cell usage by one or more lineages. Of the three skin epithelial lineages, the only one with obvious hyperplasia and enhanced flux of proliferating and differentiating cells in Blimp1 cKO skin was the SG lineage. Together, these experiments provide compelling evidence that defects in the unipotent progenitors of the SG can elicit the activity of the multipotent stem cells of the bulge. This enhanced activity of the stem cells in Blimp1 cKO hair follicles may be similar to the contribution of bulge stem cells to the epidermis that only occurs during wounding (Ito et al., 2005; Levy et al., 2005). The fact that grafted bulge cells can produce Blimp1-positive cells supports the notion that in nonhomeostatic situations, bulge cells can form the sebaceous progenitor cells. This finding may be relevant for human disorders of the gland, like acne and cancers where the stem cells may be contributing to disease pathogenesis when sebaceous development is aberrant.
Our data also support a downstream role for Blimp1 in balancing SG homeostasis through the regulation of c-myc transcription. Prior studies had shown that overexpression of c-myc in the epidermis results in sebaceous hyperplasia as well as activation of the bulge stem cell compartment (Arnold and Watt, 2001; Bull et al., 2005; Waikel et al., 2001), and conversely, conditional deletion of c-myc in these cells results in SG hypoplasia (Zanet et al., 2005). Our studies document that c-myc gene expression is dramatically enhanced in the skin of Blimp1 cKO mice in K5+ cells surrounding Blimp1-deficient SGs and suggest that Blimp1 acts upstream of c-Myc in the SG. Thus, the regulation of c-Myc in progenitor cells may specify the number of sebocytes within the gland. Although c-Myc is known to promote proliferation by regulating cell-cycle genes (http://www.myc-cancer-gene.org/site/mycTargetDB.asp), enhanced c-Myc expression in Blimp1 cKO SGs is also associated with an accumulation of slowly cycling cells localized at the base of the SG. Since many of these c-Myc-positive cells were not actively cycling, we surmise that c-Myc may play additional roles besides regulating proliferation in the SG.
In closing, our studies illuminate the existence of a resident population of unipotent progenitor cells that exist within the sebaceous glands of the skin. A transcription factor, Blimp1, marks this population of cells and controls sebaceous gland homeostasis. Upstream from these Blimp1+ cells are mechanisms that control the activity of multipotent stem cells. Our findings provide a significant advance in our understanding of skin stem cells and lineage determination, and they have important clinical implications for therapeutic advances in the treatment of glandular disorders, ranging from acne to cancers.
EXPERIMENTAL PROCEDURES
Generation of Mice, Grafts, and BrdU Incorporation
The generation of K14Cre, Blimp1-floxed, Blimp1Cre, and Blimp1-eGFP mice were described previously (Ohinata et al., 2005; Vasioukhin et al., 1999). Bulge stem cell grafts were performed as in Blanpain et al. (2004). For 5-Bromo-2′-deoxyuridine (BrdU) (Sigma-Aldrich) pulse-chase experiments (Blanpain et al., 2004), mice were injected intraperitoneally with 50 μg/g BrdU (Sigma-Aldrich) and animals were then sacrificed thereafter either directly or following a chase period.
FACS Isolation and Cell Culture
Bulge cell isolation and FACS analysis for CD34+ and a6+ cells was performed as described (Blanpain et al., 2004). Isolation of epidermal cells from Blimp1GFP mice was performed by incubating P4 back-skins in trypsin overnight and scraping the epidermis from the underlying dermis. After mincing and triturating the epidermis, neutralized cell suspensions were strained with 70 μm pore cell filters. Single-cell suspensions were subject to FACS isolation, and equal numbers of cells were plated and cultured (Blanpain et al., 2004).
Histology and Immunofluorescence
Skins were embedded in OCT, frozen, sectioned, and fixed in 4% formaldehyde. For paraffin sections, skins were incubated in 4% formaldehyde at 4°C overnight, dehydrated with a series of increasing concentrations of ethanol and xylene, and embedded in paraffin. Paraffin sections were rehydrated in decreasing concentrations of ethanol and subjected to antigen unmasking in 10 mM Citrate, pH 6.0. Sections were subjected to immunofluorescence microscopy as described (Kaufman et al., 2003). For immunohistochemistry, HRP conjugate secondary antibodies (Abs) were used followed by the HRP substrate, diaminobenzidine. When applicable, the MOM Basic Kit (Vector Laboratories) was used to prevent nonspecific binding of mouse monoclonal Abs. Antibodies and dilutions are included in Supplemental Data. Oil red O (ORO) staining was performed by incubating skin samples in 0.18% ORO for 10 min, washing with PBS, and where indicated counterstaining with hematoxylin.
Tail samples of hair follicles were treated with dispase overnight at 4°C to separate skin epithelium from dermis. Hair follicle samples were mounted in lipid dye FM 4-64 (1:1000, Molecular Probes), nuclear dye, TOPRO-3 (1:1000, Molecular Probes) or stained with ORO.
For each type of analysis performed in this study, we assayed for hair cycle by both histology and proliferation status as indicated by BrdU incorporation during short pulses to confirm that the analyses were performed in the same stage of the hair cycle.
Real-Time PCR
For real-time PCR (Rendl et al., 2005), total RNAs were isolated (Absolutely RNA, Stratagene) from total skin, and equal amounts (1 μg) were added to reverse transcriptase reaction mix (Stratagene) with oligo-dT(12) as primer. RT-PCRs of RNA were used as negative controls, and glyceraldehyde phosphate dehydrogenase (GAPDH) was used to control for equal cDNA inputs. Real-time PCR was conducted with a LightCycler system (Roche Diagnostics, Basel, Switzerland) using the LightCycle DNA master SYBR Green kit for 45 cycles. Light-Cycle analysis software was used for quantifications. The number of cycles needed to reach the crossing point for each sample was used to calculate the amount of each product using the 2−ΔΔCP method. Levels of PCR product were expressed as a function of GAPDH. Primer sequences are included in the Supplemental Experimental Procedures.
Gland Area Measurements
Individual gland size was quantified using morphometric measurements with Image J software. To analyze sebaceous glands, tail skin was treated as above to remove dermal tissue, stained with ORO, mounted, and analyzed. Preputial glands were dissected and meibomian glands were analyzed from hematoxylin- and eosin-stained sections of eyelid skin.
Statistics
All quantification data are mean ± SEM. To determine significance between groups, comparisons were made using Student’s t tests. Analyses of multiple groups were performed using One-Way ANOVA with Bonferroni’s posttest with GraphPad Prism version for Macintosh (GraphPad Software). For all statistical tests, the 0.05 level of confidence was accepted for statistical significance.
Supplementary Material
Supplemental Data include Supplemental Experimental Procedures and four figures and can be found with this article online at http://www.cell.com/cgi/content/full/126/3/597/DC1/.
Acknowledgments
We thank the Rockefeller University core facility staff (Svetlana Mazel and Tamara Shengelia, FCRC Facility; Alison North, Bioimaging Facility; Fred Quimby, LARC); Fuchs’ lab staff for technical assistance (Maria Nikolova, Lisa Polak, and Nicole Stokes); Aaron Nagiel, for his assistance during his laboratory rotation; Andres Gottfried, Dr. Matt Rodeheffer, and all of the Fuchs Lab members for valuable discussions and advice. R.T. is supported by a MRC Clinician Scientist Fellowship. D.O. acknowledges the support of the Irvington Institute for Immunological Research and is their National Genetics Foundation fellow. V.H. is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-1802-04). E.F. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by grant R01-AR050452 from the NIH (E.F.).
References
- Allen M, Grachtchouk M, Sheng H, Grachtchouk V, Wang A, Wei L, Liu J, Ramirez A, Metzger D, Chambon P, et al. Hedgehog signaling regulates sebaceous gland development. Am J Pathol. 2003;163:2173–2178. doi: 10.1016/S0002-9440(10)63574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso LC, Rosenfield RL. Molecular genetic and endocrine mechanisms of hair growth. Horm Res. 2003;60:1–13. doi: 10.1159/000070821. [DOI] [PubMed] [Google Scholar]
- Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Pelengaris S, Hendrix S, Chronnell CM, Khan M, Philpott MP. Ectopic expression of c-Myc in the skin affects the hair growth cycle and causes an enlargement of the sebaceous gland. Br J Dermatol. 2005;152:1125–1133. doi: 10.1111/j.1365-2133.2005.06458.x. [DOI] [PubMed] [Google Scholar]
- Chang DH, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev. 2002;117:305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Merrill BJ, Jamora C, DasGupta R. At the roots of a never-ending cycle. Dev Cell. 2001;1:13–25. doi: 10.1016/s1534-5807(01)00022-3. [DOI] [PubMed] [Google Scholar]
- Ghazizadeh S, Taichman LB. Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. EMBO J. 2001;20:1215–1222. doi: 10.1093/emboj/20.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- Huang S. Blimp-1 is the murine homolog of the human transcriptional repressor PRDI-BF1. Cell. 1994;78:9. doi: 10.1016/0092-8674(94)90565-7. [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, Dai X, Alegre ML, Fuchs E. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Lee SC, Bottaro A, Insel RA. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol Immunol. 2003;39:923–932. doi: 10.1016/s0161-5890(03)00029-4. [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell. 2005;9:855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Lin KI, Lin Y, Calame K. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol Cell Biol. 2000;20:8684–8695. doi: 10.1128/mcb.20.23.8684-8695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Tan NS, Basu-Modak S, Escher P, Rieusset J, Peters JM, Kaya G, Gonzalez FJ, Zakany J, et al. Impaired skin wound healing in peroxisome proliferator-activated receptor (PPAR){alpha} and PPAR{beta} mutant mice. J Cell Biol. 2001;154:799–814. doi: 10.1083/jcb.200011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Niemann C, Unden AB, Lyle S, Zouboulis Ch C, Toftgard R, Watt FM. Indian hedgehog and beta-catenin signaling: role in the sebaceous lineage of normal and neoplastic mammalian epidermis. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11873–11880. doi: 10.1073/pnas.1834202100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Olivera-Martinez I, Thelu J, Dhouailly D. Molecular mechanisms controlling dorsal dermis generation from the somitic dermomyotome. Int J Dev Biol. 2004;48:93–101. doi: 10.1387/ijdb.15272374. [DOI] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, Kopan R. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Panteleyev AA, Rosenbach T, Paus R, Christiano AM. The bulge is the source of cellular renewal in the sebaceous gland of mouse skin. Arch Dermatol Res. 2000;292:573–576. doi: 10.1007/s004030000182. [DOI] [PubMed] [Google Scholar]
- Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Rosenfield RL, Deplewski D, Kentsis A, Ciletti N. Mechanisms of androgen induction of sebocyte differentiation. Dermatology. 1998;196:43–46. doi: 10.1159/000017864. [DOI] [PubMed] [Google Scholar]
- Rosenfield RL, Wu PP, Ciletti N. Sebaceous epithelial cell differentiation requires cyclic adenosine monophosphate generation. In Vitro Cell Dev Biol Anim. 2002;38:54–57. doi: 10.1290/1071-2690(2002)038<0054:SECDRC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart ME. Sebaceous gland lipids. Semin Dermatol. 1992;11:100–105. [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28:165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Pibre S, Jacquet C, Ramirez A, de Alboran IM, Gandarillas A. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J Cell Sci. 2005;118:1693–1704. doi: 10.1242/jcs.02298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Supplemental Experimental Procedures and four figures and can be found with this article online at http://www.cell.com/cgi/content/full/126/3/597/DC1/.


