Abstract
In a previous study of sodium 4-phenylbutyrate (4-PBA)-responsive proteins in cystic fibrosis (CF) IB3-1 bronchial epithelial cells, we identified 85 differentially expressed high abundance proteins from whole cellular lysate (Singh, O. V., Vij, N., Mogayzel, P. J., Jr., Jozwik, C., Pollard, H. B., and Zeitlin, P. L. (2006) Pharmacoproteomics of 4-phenylbutyrate-treated IB3-1 cystic fibrosis bronchial epithelial cells. J. Proteome Res. 5, 562–571). In the present work we hypothesize that a subset of heat shock proteins that interact with cystic fibrosis transmembrane conductance regulator (CFTR) in common during chemical rescue and genetic repair will identify therapeutic networks for targeted intervention. Immunocomplexes were generated from total cellular lysates, and three subcellular fractions (endoplasmic reticulum (ER), cytosol, and plasma membrane) with anti-CFTR polyclonal antibody from CF (IB3-1), chemically rescued CF (4-PBA-treated IB3-1), and genetically repaired CF (IB3-1/S9 daughter cells repaired by gene transfer with adeno-associated virus-(wild type) CFTR). CFTR-interacting proteins were analyzed on two-dimensional gels and identified by mass spectrometry. A set of 16 proteins known to act in ER-associated degradation were regulated in common and functionally connected to the protein processing, protein folding, and inflammatory response. Some of these proteins were modulated exclusively in ER, cytosol, or plasma membrane. A subset of 4-PBA-modulated ER-associated degradation chaperones (GRP94, HSP84, GRP78, GRP75, and GRP58) was observed to associate with the immature B form of CFTR in ER. HSP70 and HSC70 interacted with the C band (mature form) of CFTR at the cell surface. We conclude that chemically rescued CFTR associates with a specific set of HSP70 family proteins that mark therapeutic interactions and can be useful to correct both ion transport and inflammatory phenotypes in CF subjects.
Cystic fibrosis (CF)1 is an autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (1). The most common mutation, ΔF508-CFTR, disrupts CFTR folding and trafficking (2) and impairs chloride conductance (3, 4). By an as yet unknown mechanism, the mutation also provokes a massive proinflammatory phenotype in the CF lung. The ΔF508-CFTR is rapidly ubiquitinated and either degraded by endoplasmic reticulum-associated degradation (ERAD) (5) or is short lived in the aggresome (6). We have shown previously that the chemical rescue agent sodium 4-phenylbutyrate (4-PBA) can induce correct trafficking of a portion of the mutant CFTR to the cell surface both in vitro (7) and in vivo (8). Chemical rescue with 4-PBA induces changes in a set of heat shock protein (HSP) 70 system proteins (9, 10). However, whether the chemical repair process that rescues mutant CFTR in any way resembles the trafficking process of wild type (wt) CFTR is not known.
Mapping the interactions of HSP70 system chaperones with CFTR during folding and transit from the ER is likely to shed light on the connection between mutant CFTR biogenesis and inflammatory signaling. It is well known that both CFTR (wt or mutants) and IKKβ (inhibitor of nuclear factor κB (NF-κB)) are processed through the 26 S proteasome (11, 12). The increased load of mutant CFTR on the 26 S proteasome likely interferes in proteasomal production of the inhibitor (13), thus unleashing NF-κB signaling. Because the complexity of both pathways is complicated by many interacting members (11, 14, 15), it is desirable to interrogate the networks of interacting proteins during perturbation with a potential therapeutic molecule.
Gene transfer with wtCFTR provides an alternative approach to repairing the mutant CF phenotype. However, whether the chemical repair process induced during the rescue of mutant CFTR by pharmacologic means resembles the trafficking pathway of wtCFTR introduced by genetic repair is not known. By controlling for diversity in the cell genomic background through the use of paired cell lines of the same origin, we can focus on the proteomics related to repair of the CFTR defect. We identified the proteins co-immunoprecipitated with CFTR in IB3-1 cells repaired chemically and compared them with the genetically repaired daughter (IB3-1/S9) cell line. We report here that chemically rescued ΔF508-CFTR is associated with changes in the network of a subset of HSP70 chaperones associated with ERAD (GRP94, HSP84, GRP78, GRP75, and GRP58). These relationships were also modulated by genetic repair and were specific to subcellular compartments. We suggest that a subset of HSP70 system proteins induced to interact with mutant CFTR by 4-PBA not only mark therapeutic interactions but may themselves be useful targets to both correct ion transport phenotypes and restrain the inflammatory phenotype.
EXPERIMENTAL PROCEDURES
Cell Culture—
IB3-1 cells (CF genotype ΔF508/W1282X, bronchial epithelial derivation) (16) and S9 (genetically repaired CF, IB3-1 corrected by adeno-associated virus-wtCFTR) (17) were grown in LHC-8 medium (BIOSOURCE, Rockville, MD) as described previously (Ref. 10; see supplemental Material SM.1). Where indicated, 5 mm sodium 4-phenylbutyrate (4-PBA; Medicis Pharmaceuticals) was added for 48 h (chemically rescued CF). Potential problems arising from clonal drift were avoided by using new batches of frozen cells after every 12 passages.
Formation of Immunocomplexes—
Whole cell lysates from CF, chemically rescued CF, and genetically repaired CF cells were generated in parallel as described previously (10). To accommodate the high concentration of urea, whole cell lysates were reconstituted in a modified 2-D immunoprecipitation (IP) dialysis buffer (0.1% Triton, 50 mm NaCl, 50 mm Tris, pH 8.0). The solution was dialyzed in benzoylated cellulose dialysis tubing (Sigma) with a molecular mass cutoff of 2,000 Da overnight with rotation in 2-D IP dialysis buffer at 4 °C. Total protein was quantified using the reducing agent compatible and detergent compatible protein assay kit (Bio-Rad) with BSA as the standard. The protein samples were stored in aliquots at −80 °C.
CFTR immunocomplexes were immunoprecipitated with polyclonal anti-R domain polyclonal antibody 169 by the addition of 10 μl of polyclonal antibody to 1 mg of whole cell lysate (18). After 1 h at 4 °C, immunocomplexes were collected on protein A-Sepharose 4B (Amersham Biosciences) that had been preabsorbed with BSA and incubated at 4 °C overnight with gentle end-over-end mixing. The beads were then centrifuged at 2,500 × g for 5 min and washed once with 2-D IP buffer (0.1% Triton, 50 mm NaCl, 50 mm Tris, pH 8.0) followed by three washes with PBS at 4 °C to remove nonspecifically absorbed protein. Bound antigen was eluted from the beads by brief incubation at room temperature in 2-D IP sample buffer (7 m urea, 2 m thiourea, 4% CHAPS) followed by centrifugation at 2,000 × g for 2 min at room temperature.
2-D Gel Electrophoresis and Mass Spectrometry—
CFTR immunocomplexes were evaluated by 2-D gel electrophoresis (2-DE) exactly as described previously (Ref. 10; see supplemental Material SM.2). Images of silver-stained 2-DE gels were obtained using Molecular Imager FX (Bio-Rad), and protein spot pattern analysis was performed as described in supplemental Material SM.3. Differentially expressed protein spots were identified as described earlier (10). A complete description of protein identification by peptide mass fingerprinting is given in supplemental Material SM.4.
Subcellular Fractionation—
ER fractions from CF (IB3-1) cells, chemically rescued CF (4-PBA-treated IB3-1 cells), and genetically repaired CF (wtCFTR-repaired IB3-1/S9 cells) were purified using the ER isolation kit (Sigma) to obtain Ca2+-precipitated rough ER enriched microsomes. The cytosolic fractions were purified in parallel using the Qproteome cell compartment kit (Qiagen). As described by the manufacturer, the extraction buffer CE1 selectively disrupts the plasma membrane without solubilizing it, resulting in the isolation of cytosolic proteins. PM and intact organelles are pelleted by centrifugation.
The PM fraction was obtained by cell surface biotinylation as described previously (19, 20) with modifications (see supplemental Materials SM.5 and SM.6). The PM samples were stored in aliquots at −80 °C.
Analysis of Whole Cell Lysates and Subcellular Fractions—
Immunocomplexes were generated with anti-CFTR polyclonal antibody as described above except that 2 mg of each subcellular fraction was subjected to immunoprecipitation. Bound antigen was eluted from the beads by incubation in SDS sample buffer for 15 min at 70 °C and resolved by 8% SDS-PAGE.
In parallel, 50 μg of protein from each isolated subcellular fraction was resolved by 8% SDS-PAGE. Electrophoretic transfer to nitrocellulose membrane and immunodetection were performed as described previously (10) using the highly sensitive ECL plus system (GE Healthcare). Nonspecific binding was eliminated by preblocking the nitrocellulose membrane with 5% nonfat dry milk in PBS, 0.05% Tween 20 for 1 h. Anti-CFTR polyclonal antibody 181 (1:1,000, directed against a peptide in NBD1) was used as described previously (18) to detect bands B and C. Other primary and secondary antibodies used in this study are described in supplemental Material SM.7.
Bioinformatics and Statistical Analysis of Protein Expression—
2-D gels undergoing direct comparison were produced and silver-stained in parallel. Images were acquired using Molecular Imager FX (Bio-Rad). Consistently replicative images were processed for spot detection, gel alignment, and spot quantification by match ratio using Nonlinear Dynamics Progenesis software (version 2005). Quantitative variations in proteins were expressed as volumes of spots with reference to a constant spot in each experiment. To compensate for variations in gel staining and differences in protein spot numbers for each gel, normalized spot volumes were used for comparison of protein expression levels. The normalized spot volume was automatically calculated by the software as the single spot volume divided by the total spot volume and then multiplied by the total spot area. Data are the means of nine gels, i.e. three sets of individual experiments, each with three replicate gels run in parallel. The detailed descriptions of statistical analysis of the variance associated with the measurements of selective proteins are given in supplemental Material SM.3.
The densitometry analysis on Western blots was performed as described earlier (10) by digitizing images using a Molecular Imager FX (Bio-Rad) scanner. The base-line density of the area surrounding the bands was determined by two-dimensional integration with Quantity One image analysis software (Bio-Rad), and local background was subtracted. Analysis was performed using the percent volume ratio of selected bands, i.e. total signal intensity inside a defined boundary drawn on an image. The data are presented here as the mean ± S.E., and p values were determined by one-way ANOVA followed by least significant differences (Tukey's honestly significant difference). A p value <0.05 was considered significant. CFTR immunocomplexes and total and subcellular fractions were analyzed by densitometry and then sorted for hierarchical clustering using Spotfire software (Spotfire, Inc., Somerville, MA). The average clustering was performed using the UPGMA method (unweighted pair group method with arithmetic mean) of the average values. Functional and physical connectivity among identified proteins was analyzed using the Pathway Studio version 4.0 software (Ariadne Genomics, Inc., Rockville, MD) and Pathway Architect software (Stratagene, La Jolla, CA). Additional information regarding this software is given in supplemental Material SM.8.
RESULTS
Chemical-modulated Subset of CFTR Interactome—
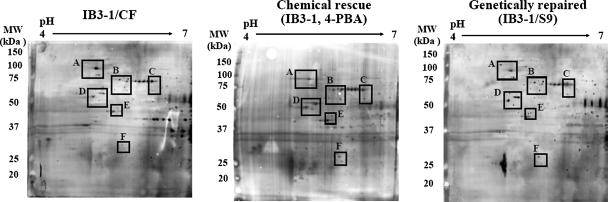
To test the hypothesis that proteins co-immunoprecipitated with chemically rescued mutant CF are similar to proteins co-immunoprecipitated with genetically repaired wtCFTR in IB3-1/S9 cells, we examined CFTR immunocomplexes at pH 4–7 in the first dimension followed by 10% SDS-PAGE in the second dimension. As depicted in Fig. 1 CFTR polyclonal antibody in chemically rescued and genetically repaired CF cells captured ∼821 protein spots with the best resolution occurring between pH 4.7 and 6.5 and 25 and 100 kDa. Our protocol was hindered by extremely basic (pH 6.8–7) proteins that were obscured by the heavy chains of the primary antibodies. Acidic proteins below 25 kDa were poorly visible (Fig. 1). Changes in expression of proteins over 2-fold in magnitude were chosen for further consideration. Six different regions common to CF, chemically rescued CF, and genetically repaired CF of CFTR immunocomplexes were investigated (Fig. 1). The protein pattern of our gels were highly reproducible (n = 9 independent gels from three independent experiments). The protein identities of spots from this study were compared with our existing high abundance CF proteome atlas (21).
Fig. 1.
2-D gel silver images of CFTR immunocomplexes captured by CFTR-169 polyclonal antibody. Immunocomplexes were generated with anti-CFTR antibodies from CF (IB3-1), chemically rescued CF (IB3-1, 5 mm 4-PBA for 48 h), and genetically repaired CF cells (S9, IB3-1 corrected by wtCFTR) and resolved on 10% SDS-PAGE 2-DE gels as described under “Experimental Procedures.” Six regions were selected (A–F) based on differential expression in common between rescue and non-CF conditions.
Overall the gel-estimated Mr and pI matched well with the corresponding theoretical values. The majority of the proteins were identified based upon five or more matching peptides. A sequence coverage exceeding 15% was obtained for over 95% of the spots (Table I). Based on a cutoff of at least a 2-fold change in magnitude during chemical rescue or genetically repaired CF conditions, we identified a set of 16 high abundance proteins common to both conditions. Of 16 informative proteins, 12 were up-regulated in chemically rescued CFTR-immunocaptured protein complex that correspond equally to those from the genetically repaired CF condition (Table I). Among these 16 proteins, the quantities of pulled down proteins often varied and were non-uniform with the polyclonal antibody as would be expected if CFTR-interacting molecular chaperones were participating in the proteolytic pathway for multiple proteins.
Table I.
Selective protein targets of chemically rescued CFTR protein interaction in IB3-1 cells and correlation with genetically repaired CF cells (IB3-1/S9)
| Accession no./Lucas ID(Swiss-Prot/TrEmBL) | Protein description | Estimated Z scorea/Mowse score | Molecular mass (kDa)/pI
|
Matches (n) | Unmatched (n) | Coverage(%) | Protein expressionb
|
Spot no. | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Theoretical | Experimental | Chemically rescued (4-PBA, IB3-1) | Genetically rescued (IB3-1/S9) | |||||||
| Protein processing-, protein trafficking-, ERAD-involved unique CFTR-associated HSP70 system molecular chaperones | ||||||||||
| P14625 | Glucose-regulated protein 94 kDa/TRA1 | 2.38 | 92.47/4.8 | 91.2/4.7 | 14 | 46 | 26 | U | U | IP-1 |
| P08238 | Heat shock protein 84 kDa/HSPCB | 2.43 | 83.59/5.0 | 85.6/4.8 | 16 | 32 | 34 | D | D | IP-2 |
| AAF13605 | Glucose-regulated protein 78 kDa (BiP)/HSPA5 | 2.43 | 71.03/5.2 | 77.5/5.1 | 27 | 41 | 49 | U | U | IP-3 |
| A29160 | dnaK-type molecular chaperone/HSPA1 | 2.43 | 70.14/5.4 | 71.6/5.5 | 14 | 30 | 26 | U | U | IP-4 |
| P54652 | Heat shock 70-kDa protein 2/HSPA2 | 2.43 | 73.95/5.9 | 70.6/6.3 | 24 | 28 | 51 | U | U | IP-5 |
| NP_002147 | Heat shock 60-kDa protein 1/HSPD1 | 2.43 | 61.21/5.7 | 62.4/5.2 | 20 | 52 | 38 | U | U | IP-6 |
| AAA67526 | Glucose-regulated protein 75 kDa/MTHSP75 | 2.29 | 73.95/5.9 | 72.6/5.7 | 25 | 11 | 48 | U | U | IP-7 |
| P34931 | Heat shock 70-kDa protein 1-HOM/HSP-HOM | 2.43 | 70.37/5.7 | 69.8/5.7 | 15 | 34 | 25 | D | D | IP-8 |
| NP_005304 | Glucose-regulated protein 58 kDa/PDIA3 | 1.84 | 56.78/6.0 | 58.5/5.7 | 19 | 46 | 41 | U | U | IP-9 |
| NP_006588 | Heat shock cognate protein 8 isoform 1, 71 kDa/HSPA8 | 2.43 | 71.11/5.4 | 71.8/5.9 | 25 | 21 | 58 | D | D | IP-10 |
| NP_004272 | BAG-3 | 1.86 | 61.62/6.7 | 62.5/6.1 | 7 | 18 | 13 | U | U | IP-11 |
| AAH12292 | Heat shock 27-kDa protein 1/HSPB1 | 2.43 | 22.81/6.0 | 27.8/5.8 | 15 | 9 | 52 | U | U | IP-16 |
| Protein aggregation-, protein trafficking-involved unique CFTR-associated cellular organization proteins | ||||||||||
| AAC31959 | α-Tubulin isoform 1/TUBA1B | 7.14e+0.4 | 50.82/4.9 | 54.5/4.9 | 17 | 45 | 54 | U | U | IP-12 |
| NP_821133 | β5-Tubulin/TUBB | 2.43 | 50.11/4.8 | 51.4/4.8 | 17 | 32 | 43 | U | U | IP-13 |
| NP_003371 | Vimentin/VIM | 2.43 | 53.65/5.1 | 51.6/4.7 | 26 | 57 | 68 | U | U | IP-14 |
| CAA31377 | Cytokeratin 18/KRT18 | 2.29 | 47.32/5.3 | 46.5/5.4 | 17 | 16 | 49 | D | D | IP-15 |
ProFound peptide mapping-based probability score.
U, up-regulated; D, down-regulated.
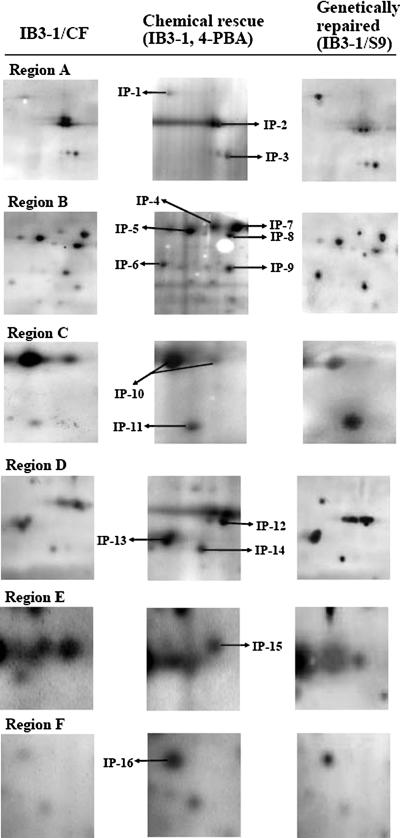
To assess the correlation between chemically rescued CF and genetically repaired CF, we focused on a subset of ER-resident molecular chaperones that were observed to be overexpressed, excluding HSP84 (a homologue of HSP90 1β) (Fig. 2, Region A, IP-2). A subset of major ER-resident proteins (GRP94, HSPA1, HSP60, GRP58, BAG-3, and HSP27) in CFTR immunocomplexes was 6-fold overexpressed in chemically rescued CF and similarly increased in genetically repaired CF in CFTR immunocomplexes (Fig. 2, Regions A, B, C, and F). HSP-HOM (Fig. 2, Region B, IP-8) was reduced among CFTR-interacting proteins. Protein spot HSP60 (Fig. 2, Region B, IP-6) that appeared in CF versus chemically rescued CF and genetically rescued CF and in CFTR immunocomplexes is known to induce the production of proinflammatory cytokines (22). Table I contains a subset of cytoskeletal proteins that was also immunoprecipitated with CFTR. The intensity of CFTR-interacting protein vimentin was over 4-fold highly expressed (Fig. 2, Region D). Another known CFTR cytoskeletal partner protein, KRT18, was diminished in chemically rescued CFTR immunocomplexes (Fig. 2, Region E, IP-15 and Table I). The biological differences in the proteins identified in chemically rescued and genetically repaired cells are shown in supplemental Material SM.3 and supplemental Fig. S.3.
Fig. 2.
Higher resolution map of six selected regions of Fig. 1. Left, center, and right panels of each region (A–F) contain proteins from CF (IB3-1), chemically rescued CF (5 mm 4-PBA-treated IB3-1 cells), and genetically repaired CF cells (IB3-1/S9 cells), respectively. Proteins labeled IP-1 through IP-16 were excised and identified as described under “Experimental Procedures.” The identified proteins are characterized in Table I.
Subcellular Fractions Respond to CFTR Rescue—
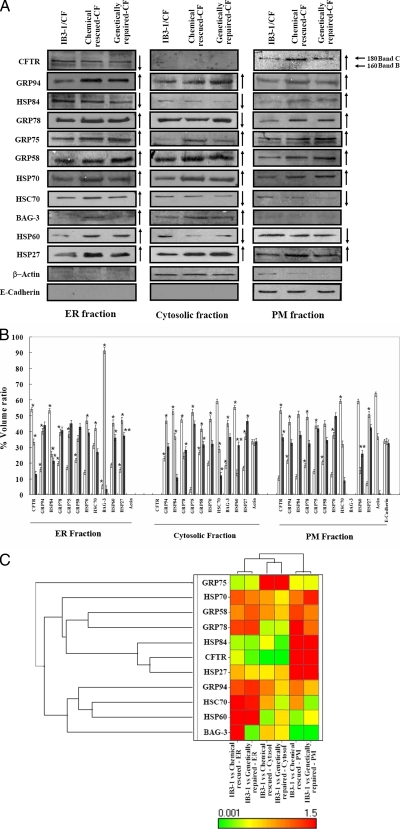
A subset of ER-resident (GRP94, -78, -75, -58, and HSP84) and cytosolic chaperones (HSP70, HSC70, and BAG-3) were modulated in chemically rescued CF in ER, cytosol, and PM, and the direction of the modulation mimicked genetically repaired CF interactions (Fig. 3). HSP84 (homologue of HSP90 1β) was the only ER-resident protein that was diminished by chemical treatment (Fig. 3A) in ER and cytosolic fractions. However, the level of HSP84 from rescued CF and genetically repaired CF was notably increased in the PM fraction. BAG-3 was abundant in cytosol fractions of rescued and genetically repaired CF, whereas a faint expression of BAG-3 was observed after rescue in ER that was undetectable in ER fractions of repaired CF (Fig. 3, A and B). Contrary to previous observations regarding rescue-mediated reduction in HSC70 from whole cells (10, 23), there was an increase in HSC70 with rescue in the ER fraction, suggesting that as overall levels of HSC70 decrease in cytosolic and PM fractions, the remaining HSC70 is recruited to the ER (Fig. 3A). Under the same conditions, HSP84 was recruited to the PM, and BAG-3 remained in cytosol. β-Actin was detected almost exclusively in cytosol, and E-cadherin was similarly confined to the PM (Fig. 3A), confirming a reasonable degree of separation of subcellular protein fractions. A comparative analysis of the differential targeting of 4-PBA treatment in IB3-1 cells is further illustrated by the hierarchical clustering in Fig. 3C where expression of the GRP94, HSP84, HSC70, HSP70, HSP27, and BAG-3 produces a greater class distinction between three subcellular protein fractions (ER, cytosol, and PM) (Fig. 3C).
Fig. 3.
Protein components in subcellular fractions from CF, rescued CF, and genetically repaired CF cells. A, total proteins from CF (IB3-1/CF), chemically rescued CF (IB3-1 treated with 4-PBA), and genetically repaired CF (IB3-1/S9 cells) were subfractionated into ER, cytosol, and PM and analyzed by immunoblot as described under “Experimental Procedures.” Representative blots are shown. B, densitometric analysis. The hollow bar represents CF, the light bar represents chemically rescued CF, and the solid bar represents the corresponding genetically repaired CF cellular lysates. Each data point is the mean ± S.E. of the density of gel bands relative to the average band density of control (IB3-1/CF) cells. The one-way ANOVA followed by least significant differences was calculated; a p value <0.05 was considered significant (*). C, hierarchical clustering of a chemical-modulated and genetically mimicked set of HSP70 system proteins in subcellular protein fractions.
Chemically Rescued Subcellular Fraction Modulates CFTR Interaction—
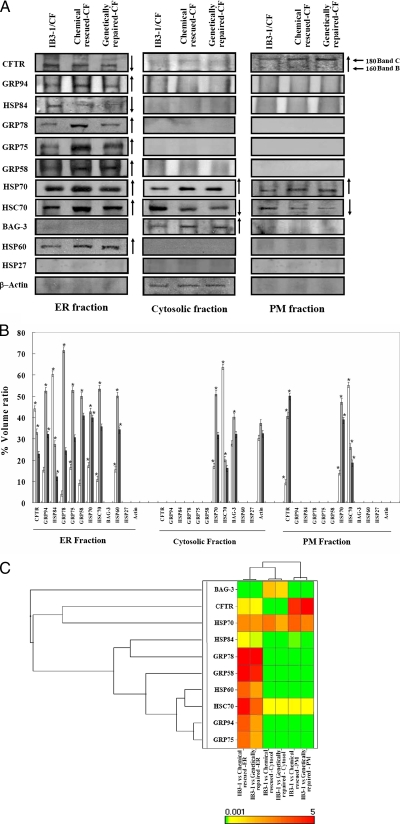
In an effort to compartmentalize specific CFTR-interacting proteins, we examined the CFTR immunocomplexes in ER, cytosol, and PM. CFTR was found exclusively as band B in the ER fraction and band C in the PM (Fig. 4A). BAG-3, HSP70, and HSC70 were co-immunoprecipitated with CFTR from the cytosol (Fig. 4A). Chaperone BAG-3 often associates with HSP70 (24), and the absence of BAG-3 from CFTR immunocomplexes in ER and PM (Fig. 4A) suggests that the cytosolic component of CFTR immunocomplexes is not membrane-bound, but without further investigation we cannot rule out aggresome contamination of cytosol.
Fig. 4.
Differential expression of multiple molecular chaperones interacting with ΔF and rescued and genetically repaired wtCFTR from subcellular fractions. A, total protein from three different cellular compartments, viz. ER, cytosol, and PM, was immunoprecipitated using CFTR-169 polyclonal antibody from CF (IB3-1), chemically rescued CF (4-PBA-treated IB3-1), and genetically rescued CF (wtCFTR, S9 cells). Immunoblotting was performed as described under “Experimental Procedures.” B, densitometric analysis was performed. The hollow bar represents CF, the light bar represents rescued CF, and the solid bar represents the corresponding non-CF. Each data point is the mean ± S.E. of the density of gel bands relative to the average band density of control (IB3-1/CF) cells. The one-way ANOVA followed by least significant differences was calculated; a p value <0.05 was considered significant (*). C, hierarchical clustering and dendrograms demonstrate functional drug relationships based on the indicated drug targets during ΔF508-CFTR trafficking.
To assess the wider reach of CFTR rescue on ERAD and trafficking, a subset of ER-resident proteins interacting with CFTR was examined. Fig. 4A shows that GRP94, -78, -75, -58, and HSP84 associate in abundance with the B form of CFTR (160 kDa) in ER and not with the folded C form (180 kDa) at the cell surface (Fig. 4A). Densitometry analysis revealed the intensity of protein abundance with CFTR in three subcellular fractions (Fig. 4B). This suggests that these chaperones remain within the ER regardless of whether mutant CFTR escapes or not.
The hierarchical clustering of ΔF508-CFTR- and chemically rescued ΔF508-CFTR-interacting proteins could be ordered along a continuum from strongly related to weakly related subgroups as shown in Fig. 4C. GRP94, -78, and -75 in ER, BAG-3 in cytosol, and CFTR are differentially expressed in subcellular compartments.
Functional Connectivity and Cytokine Expression of Rescued CFTR Interactome—
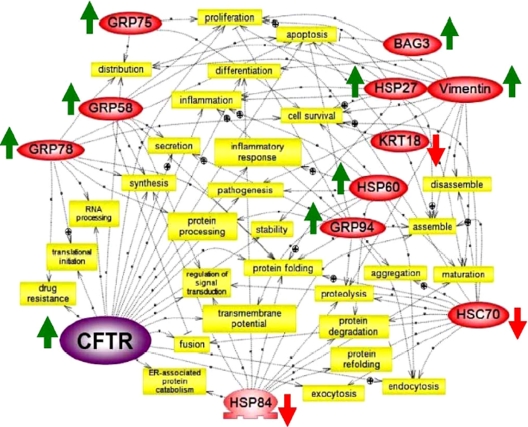
Using a set of 16 informative interactions, a scale-free network of biologically interacting pathways that are influenced by CFTR mutation state was built (Fig. 5). The literature-based connectivity map of our proteomics data reveals that 11 proteins are directly and/or indirectly connected to complex biological functions (Fig. 5). The connectivity map shows that the majority of ER stress chaperones are directly connected with protein folding, processing, degradation, inflammatory response, and inflammation. Most of the ER network proteins intersect with HSC70-regulated processes; however, some (HSPA1, HSP-HOM, α-tubulin, and β-tubulin) remain difficult to integrate. Perhaps the most interesting network is the set of inflammatory response and inflammation branches. While studying the HSP70-responsive cytokine expression we observed that the inflammatory response was highly sensitive to HSP60 and CFTR, and inflammation was greatly responsive to CFTR, GRP94, HSP60, and HSP27 (Fig. 6). This suggests that chemical regulation of GRP94, HSP60, and HSP27 as well as cytoskeletal proteins KRT18 and vimentin would be anti-inflammatory in CF.
Fig. 5.
Functional connectivity of chemically rescued CFTR-interacting proteins. Proteins highlighted in red were differentially expressed in chemically rescued IB3-1 cells and correlated with genetically repaired S9 cells. Pathways were created using the software Pathway Studio version 4.0. The biological function pathway shows common cellular targets (4-PBA-modulated proteins (red ovals) during cellular processing (yellow rectangles). ⊕, positive regulation; ▭, negative regulation; ○, regulation of protein expression.
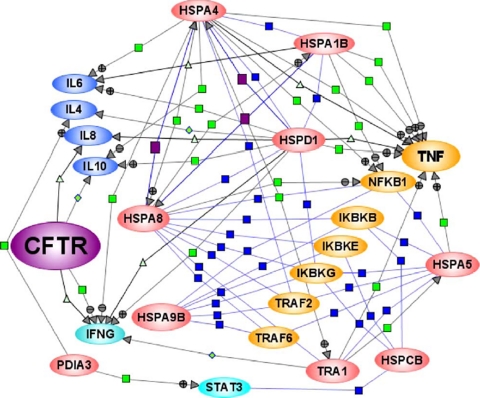
Fig. 6.
CFTR-interacting HSP70 chaperones regulate cytokine expression. Literature-based HSP70 system protein-regulated expression of cytokine connectivity map was created using Pathway Architect software by customizing software tools for regulatory pathway specific for cytokine regulation. Blue squares, protein-protein interactions; purple squares, protein expression; green squares, regulation with positive/negative signs; green triangles, transport mechanism of cytokines; red ovals, specific set of HSP70 system proteins modulated by 4-PBA; blue ovals, multiple cytokines; and yellow ovals, receptor-associated factors regulated by 4-PBA-modulated proteins. The entire map is documented by 88 literature citations. TRAF, TNF receptor-associated factor; IKB, inhibitor of κ light polypeptide gene enhancer in B cells; NFKB, nuclear factor of κ light polypeptide gene enhancer in B cells; IFNG, interferon γ; STAT3, signal transducer and activator of transcription 3).
DISCUSSION
In CF, CFTR protein interacts directly and/or indirectly with a variety of molecular chaperones and co-chaperones assisting in protein folding assembly, disassembly, and translocation (5, 23, 25–29). Some of these interactions could be targeted to redirect ΔF508-CFTR away from ERAD and toward the trafficking pathway. In our studies, of the 16 chemically rescued CFTR-interacting proteins, a subset of 12 also interacted with genetically repaired CF (wtCFTR).
CFTR Interactome Comparison in IB3-1 Cell Lines with Other CF Model—
Only a few attempts have been made to analyze the CFTR interactome signature of CF model systems (30, 31). Four chaperones, HSP70, HSC70, and HSP90 α and β, as well as velosin-containing protein/p97 were identified and characterized using specific CFTR polyclonal antibody to co-precipitate them from total cellular lysate from Calu-3 cells expressing endogenous wtCFTR (30). Here in chemically rescued ΔF508-CFTR we detected a set of four chaperones but not velosin-containing protein/p97.
In another recent study, a mass spectrometry approach using multidimensional protein identification technology (MudPIT) was utilized to define all CFTR protein interactions in baby hamster kidney cell lines expressing ΔF508-CFTR and in Calu-3, HT29, and T84 cell lines expressing wtCFTR (31). A large number of protein sets were identified, and the authors conclude that HSP90 co-chaperones (Aha1) modulate HSP90-dependent stability of CFTR protein folding in the ER (31). Alternatively we observed that a homologue of HSP90 1β, i.e. HSP84, was diminished during chemical rescue of ΔF508-CFTR in ER, correlating with the genetically repaired wtCFTR in IB3-1/S9 cells (Fig. 4). These reports suggest that members of a subset of HSP70 system proteins could serve as potential therapeutic targets.
Selective HSP(s) Intervenes in CFTR Processing and ERAD—
Several HSP70 system proteins have been observed to affect CFTR biogenesis, trafficking, and ubiquitin-mediated proteasomal degradation (5, 29, 31–33). It has been shown that HSC70 participates in the ubiquitination of misfolded CFTR protein (27–29, 33). Because of the prolonged association of ΔF508 with HSC70, a growing body of evidence suggests that the diminution in HSC70 alone may not be sufficient to interfere with ubiquitination of the immature form of CFTR (28). Instead induction of HSPBP1 type co-chaperones may be required to diminish ubiquitination and assist CFTR maturation (34). From our studies, the abundance of dnaK or HSPA1 and HSP-HOM with chemically rescued CFTR can modulate the CFTR-HSC70 complex. Evidently the co-chaperone dnaK transiently associates with HSP70, and a wide variety of newly synthesized polypeptides assist in folding and act in protein disaggregation to refold the native form (35–37). Therefore, a possible interaction of dnaK co-chaperone (ATPase chaperone) with CFTR and HSP70-HSC70 complex initiates refolding of CFTR.
The cytosolic BAG family proteins interact with HSP70 and facilitate the removal of injured proteins by ubiquitin-mediated proteasomal degradation (24). Appearance of BAG-3 in the cytosolic fraction (Fig. 3) and with CFTR immunoprecipitation (Fig. 4) supports the possibility of direct binding of BAG-3 to form an HSC70-HSP70-BAG-3 complex that assists in ERAD assembly. In a large data set, Wang et al. (31) reported that BAG-3 associates with ΔF508-CFTR. In IB3-1, we did not find immature CFTR band B associated with BAG-3 in ER protein fractions (Fig. 4); however, the cytosolic protein fraction reveals an abundance of BAG-3 in association with chemically rescued CFTR (Fig. 4, A–C). The BAG family proteins are HSP70 co-chaperones and are characterized by a common C-terminal BAG domain that interacts with the ATP-binding site of HSP70 (38). It is through this interaction and by competing for binding to the ATPase domain that the BAG family proteins, including HSP70/BAG-3, regulate the refolding chaperone properties of HSPs. Perhaps the appearance of BAG-3 in the cytosolic fraction with CFTR is a part of a direct association with HSP70 protein.
GRP78 (BiP) predominantly associates with the mutant form of CFTR in the rough endoplasmic reticulum (Fig. 4). A direct interaction of BiP with CFTR has not been demonstrated before (25); however, a major part of the ER machinery has been reported to be facilitated by GRP78 (25, 39). Among GRPs, GRP94 (gp96 homolog) (Table I) shows many similarities with BiP (40), but there are two exceptions. (i) GRP94 exists as both a transmembrane and luminal protein (41), and (ii) it forms a stable complex with the mutant polypeptides (42). Thereby GRP94 assists at more advanced steps in the folding and assembly process of misfolded proteins (42). We detected GRP94 and GRP78 in abundance with CFTR (Figs. 1 and 4) in both chemically rescued and genetically repaired CF cells in the ER. This supports a role for these chaperones in ERAD of mutant CFTR (Fig. 4).
HSP70 System Mediated CFTR Rescue and Inflammatory Response—
Trafficking of wt or ΔF508-CFTR is aided by transient interactions with molecular chaperones thought to interact with partially folded protein intermediates, which are continuously bound and released until the polypeptide has reached the completely folded or native state. Despite the complexity of the folding process, the intracellular trafficking of ΔF508-CFTR can be promoted by 4-PBA through (i) down-regulation of HSC70, an essential cofactor for ubiquitination and degradation of unfolded CFTR in association with other co-chaperones (26–28), or (ii) up-regulation of HSP70 (23, 29).
CFTR rescue modulation of ER-resident (GRP94, -78, -75, -58, and HSP84) and cytosolic (HSP70 and HSC70) chaperones was found to be interesting as they are mainly involved in ERAD, protein processing, and protein trafficking machinery (23, 26–29, 40, 43, 44), yet they tracked in relatively high abundance to the cell surface. These proteins have been detected in the cell surface proteome previously (45) where, under certain conditions, HSP(s) appears on the cell surface in a number of cell types (32, 45, 46). The presence of HSP(s) at the cell surface might be explained by heat shock protein-receptor interactions (47). The heat shock protein gp96-TLR2/4 interaction results in activation of NF-κB-driven reporter genes and mitogen- and stress-activated protein kinases (48). In addition, HSP70 activates the IL-1 receptor signaling pathway that is triggered by receptors at the cell surface (32). This suggests that 4-PBA may inadvertently increase HSP70 system proteins at the cell surface in CF, which might exacerbate the inflammatory milieu, and correspondingly, rescue of these proteins from this compartment might dampen inflammation.
Relationships between HSP70 System Chaperones and Cytokine Function—
HSPs play an active role in T cell regulatory function and in the induction of anti-inflammatory cytokines. HSPs can regulate via the CD14/Toll-like receptor (TLR2 and TLR4) complex signal transduction pathways leading to the activation of NF-κB and mitogen-activated protein kinases (MAPKs). The NF-κB connection therefore is extremely significant, and data implicate that HSP27 is an ATP-independent chaperone that functions as an inhibitor of the NF-κB pathway (49). Therefore, the reduced level of this negative regulator in CF cells can contribute to the increased activation of NF-κB and overproduction of IL-8 reported to occur in CF bronchial gland cells and CF lung (13, 50).
With specific respect to CFTR, our de novo biosynthetic profiling of the high abundance CF proteome showed that of 51 high abundance CFTR connective proteins 17 (33%) of them are linked to the inflammatory process either directly or through NF-κB (51). Here we show that a specific set of HSP70 system proteins directly associated with CFTR (HSPA5, TRA1, HSPA1B, and HSPD1) and positively regulated TNFα expression, whereas HSPA4 diminished serum levels of TNF and IL-10 (Fig. 6). Multiple factors of IκB and TNF receptor-associated factors were found connected with a specific set of HSP70 system proteins (Fig. 6). HSP60 and HSP70 contribute to the pathogenesis of autoimmune diseases and chronic inflammation. Both HSPs are highly immunogenic and capable of inducing antibody production and T cell activation. HSP60, an endogenous ligand for the TLR4 receptor, has been implicated in inflammatory conditions such as Type 1 diabetes in CF patients (52). Higher levels of serum IgG antibodies against HSP27, HSP60, and HSP90 (HSP84 homologue) have been detected in CF patients than in non-CF patients (53, 54). Other HSPs, viz. HSP70, HSP90, and GRP94 (gp96), have been shown to be potent activators of the innate immune system (40). Thus, the induction of proinflammatory cytokines by 4-PBA-modulated HSPs in this study may contribute to the pathogenesis due to chronic inflammation in CF.
Conclusions—
This 4-PBA-directed “pharmaco-interactomics” investigation highlights a specific subset of HSP70 system proteins that sense the rescue of ΔF508-CFTR from ERAD. These perturbations appear to correspond with patterns observed in wtCFTR both at the whole cell level and in subcellular compartments. 4-PBA-mediated rescue of ΔF508-CFTR is associated with changes in the network of ER-resident cytosolic chaperones and proinflammatory responses that mimic patterns of protein interaction observed with wtCFTR expression. We anticipate that this 2-DE-based virtual CFTR interactome for a specific subset of HSP70 system proteins will suggest potential therapeutic targets in CF subjects.
Supplementary Material
Acknowledgments
Technical assistance provided by Dmitry Minskovsky and Karen McKenzie is gratefully acknowledged. We thank Dr. Robert N. Cole, Director, The Johns Hopkins University Proteomics Core Facility, and bioinformatics assistance provided by The Johns Hopkins University Microarray Core Facility is gratefully acknowledged. A licensing agreement exists between The Johns Hopkins University, Ucyclyed Pharma, Inc., and Dr. Zeitlin. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies. IB3-1 cells are under a licensing agreement between Pfizer Inc., Japan Tobacco Inc., and The Johns Hopkins University. Dr. Zeitlin is entitled to a fee received by The University on sales of this cell line. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
Published, MCP Papers in Press, February 19, 2008, DOI 10.1074/mcp.M700303-MCP200
The abbreviations used are: CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; 4-PBA, sodium 4-phenylbutyrate; ER, endoplasmic reticulum; ERAD, endoplasmic reticulum-associated degradation; PM, plasma membrane; 2-D, two-dimensional; 2-DE, two-dimensional gel electrophoresis; HSP, heat shock protein; wt, wild type; IP, immunoprecipitation; ANOVA, analysis of variance; TNF, tumor necrosis factor; IL, interleukin; TLR, Toll-like receptor; BAG-3, BCL2-associated athanogene 3; Bip, immunoglobulin heavy chain-binding protein.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL 59410 and N01 BAA HL 02-04. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Tsui, L. C. ( 1992) Mutations and sequence variations detected in the cystic fibrosis transmembrane conductance regulator (CFTR) gene: a report from the Cystic Fibrosis Genetic Analysis Consortium. Hum. Mutat. 1, 197–203 [DOI] [PubMed] [Google Scholar]
- 2.Ward, C. L., Omura, S., and Kopito, R. R. ( 1995) Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 3.Gabriel, S. E., Clarke, L. L., Boucher, R. C., and Stutts, M. J. ( 1993) CFTR and outward rectifying chloride channels are distinct proteins with a regulatory relationship. Nature 363, 263–268 [DOI] [PubMed] [Google Scholar]
- 4.Stutts, M. J., Canessa, C. M., Olsen, J. C., Hamrick, M., Cohn, J. A., Rossier, B. C., and Boucher, R. C. ( 1995) CFTR as a cAMP-dependent regulator of sodium channels. Science 269, 847–850 [DOI] [PubMed] [Google Scholar]
- 5.Amaral, M. D. ( 2004) CFTR and chaperones: processing and degradation. J. Mol. Neurosci. 23, 41–48 [DOI] [PubMed] [Google Scholar]
- 6.Johnston, J. A., Ward, C. L., and Kopito, R. R. ( 1998) Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubenstein, R. C., Egan, M. E., and Zeitlin, P. L. ( 1997) In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing ΔF508-CFTR. J. Clin. Investig. 100, 2457–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubenstein, R. C., and Zeitlin, P. L. ( 1998) A pilot clinical trial of oral sodium 4-phenylbutyrate (Buphenyl) in ΔF508-homozygous cystic fibrosis patients: partial restoration of nasal epithelial CFTR function. Am. J. Respir. Crit. Care Med. 157, 484–490 [DOI] [PubMed] [Google Scholar]
- 9.Wright, J. M., Zeitlin, P. L., Cebotaru, L., Guggino, S. E., and Guggino, W. B. ( 2004) Gene expression profile analysis of 4-phenylbutyrate treatment of IB3-1 bronchial epithelial cell line demonstrates a major influence on heat-shock proteins. Physiol. Genomics 16, 204–211 [DOI] [PubMed] [Google Scholar]
- 10.Singh, O. V., Vij, N., Mogayzel, P. J., Jr., Jozwik, C., Pollard, H. B., and Zeitlin, P. L. ( 2006) Pharmacoproteomics of 4-phenylbutyrate-treated IB3-1 cystic fibrosis bronchial epithelial cells. J. Proteome Res. 5, 562–571 [DOI] [PubMed] [Google Scholar]
- 11.Jensen, T. J., Loo, M. A., Pind, S., Williams, D. B., Goldberg, A. L., and Riordan, J. R. ( 1995) Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129–135 [DOI] [PubMed] [Google Scholar]
- 12.Scherer, D. C., Brockman, J. A., Chen, Z., Maniatis, T., and Ballard, D. W. ( 1995) Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 92, 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knorre, A., Wagner, M., Schaefer, H. E., Colledge, W. H., and Pahl, H. L. ( 2002) ΔF508-CFTR causes constitutive NF-κB activation through an ER-overload response in cystic fibrosis lungs. Biol. Chem. 383, 271–282 [DOI] [PubMed] [Google Scholar]
- 14.Siebenlist, U., Franzoso, G., and Brown, K. ( 1994) Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 10, 405–455 [DOI] [PubMed] [Google Scholar]
- 15.Hehlgans, T., and Pfeffer, K. ( 2005) The intriguing biology of the tumor necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology 115, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeitlin, P. L., Lu, L., Rhim, J., Cutting, G., Stetten, G., Kieffer, K. A., Craig, R., and Guggino, W. B. ( 1991) A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection. Am. J. Respir. Cell Mol. Biol. 4, 313–319 [DOI] [PubMed] [Google Scholar]
- 17.Flotte, T. R., Afione, S. A., Solow, R., Drumm, M. L., Markakis, D., Guggino, W. B., Zeitlin, P. L., and Carter, B. J. ( 1993) Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 268, 3781–3790 [PubMed] [Google Scholar]
- 18.Crawford, I., Maloney, P. C., Zeitlin, P. L., Guggino, W. B., Hyde, S. C., Turley, H., Gatter, K. C., Harris, A., and Higgins, C. F. ( 1991) Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc. Natl. Acad. Sci. U. S. A. 88, 9262–9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao, Y., Zhang, W., Kho, Y., and Zhao, Y. ( 2004) Proteomic analysis of integral plasma membrane proteins. Anal. Chem. 76, 1817–1823 [DOI] [PubMed] [Google Scholar]
- 20.Lisanti, M. P., Le Bivic, A., Sargiacomo, M., and Rodriguez-Boulan, E. ( 1989) Steady-state distribution and biogenesis of endogenous Madin-Darby canine kidney glycoproteins: evidence for intracellular sorting and polarized cell surface delivery. J. Cell Biol. 109, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard, H. B., Ji, X. D., Jozwik, C., and Jacobowitz, D. M. ( 2005) High abundance protein profiling of cystic fibrosis lung epithelial cells. Proteomics 5, 2210–2226 [DOI] [PubMed] [Google Scholar]
- 22.Wallin, R. P., Lundqvist, A., More, S. H., von Bonin, A., Kiessling, R., and Ljunggren, H. G. ( 2002) Heat-shock proteins as activators of the innate immune system. Trends Immunol. 23, 130–135 [DOI] [PubMed] [Google Scholar]
- 23.Choo-Kang, L. R., and Zeitlin, P. L. ( 2001) Induction of Hsp70 promotes ΔF508 CFTR trafficking. Am. J. Physiol. 281, L58–L68 [DOI] [PubMed] [Google Scholar]
- 24.Doong, H., Rizzo, K., Fang, S., Kulpa, V., Weissman, A. M., and Kohn, E. C. ( 2003) CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation. Accumulation of poly-ubiquitinated Hsp90 client proteins. J. Biol. Chem. 278, 28490–28500 [DOI] [PubMed] [Google Scholar]
- 25.Yang, Y., Janich, S., Cohn, J. A., and Wilson, J. M. ( 1993) The common variant of cystic fibrosis transmembrane conductance regulator is recognized by Hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. U. S. A. 90, 9480–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farinha, C. M., Nogueira, P., Mendes, F., Penque, D., and Amaral, M. D. ( 2002) The human DnaJ homologue (Hdj)-1/heat-shock protein (Hsp) 40 co-chaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem. J. 366, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meacham, G. C., Lu, Z., King, S., Sorscher, E., Tousson, A., and Cyr, D. M. ( 1999) The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18, 1492–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meacham, G. C., Patterson, C., Zhang, W., Younger, J. M., and Cyr, D. M. ( 2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y., Nijbroek, G., Sullivan, M. L., McCracken, A. A., Watkins, S. C., Michaelis, S., and Brodsky, J. L. ( 2001) Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein, R. F., Niraj, A., Sanderson, T. P., Wilson, L. S., Rab, A., Kim, H., Bebok, Z., and Collawn, J. F. ( 2007) VCP/p97 AAA-ATPase does not interact with the endogenous wild-type cystic fibrosis transmembrane conductance regulator. Am. J. Respir. Cell Mol. Biol. 36, 706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X., Venable, J., LaPointe, P., Hutt, D. M., Koulov, A. V., Coppinger, J., Gurkan, C., Kellner, W., Matteson, J., Plutner, H., Riordan, J. R., Kelly, J. W., Yates, J. R., III, and Balch, W. E. ( 2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 32.Vabulas, R. M., Ahmad-Nejad, P., Ghose, S., Kirschning, C. J., Issels, R. D., and Wagner, H. ( 2002) Hsp70 as endogenous stimulus of the Toll/interleukin-1 receptor signal pathway. J. Biol. Chem. 277, 15107–15112 [DOI] [PubMed] [Google Scholar]
- 33.Farinha, C. M., and Amaral, M. D. ( 2005) Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol. Cell. Biol. 25, 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberti, S., Bohse, K., Arndt, V., Schmitz, A., and Hohfeld, J. ( 2004) The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 15, 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teter, S. A., Houry, W. A., Ang, D., Tradler, T., Rockabrand, D., Fischer, G., Blum, P., Georgopoulos, C., and Hartl, F. U. ( 1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97, 755–765 [DOI] [PubMed] [Google Scholar]
- 36.Ben-Zvi, A., De Los Rios, P., Dietler, G., and Goloubinoff, P. ( 2004) Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual Hsp70 chaperones. J. Biol. Chem. 279, 37298–37303 [DOI] [PubMed] [Google Scholar]
- 37.De Los Rios, P., Ben-Zvi, A., Slutsky, O., Azem, A., and Goloubinoff, P. ( 2006) Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl. Acad. Sci. U. S. A. 103, 6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gässler, C. S., Wiederkehr, T., Brehmer, D., Bukau, B., and Mayer, M. P. ( 2001) Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276, 32538–32544 [DOI] [PubMed] [Google Scholar]
- 39.Accili, D., Kadowaki, T., Kadowaki, H., Mosthaf, L., Ullrich, A., and Taylor, S. I. ( 1992) Immunoglobulin heavy chain-binding protein binds to misfolded mutant insulin receptors with mutations in the extracellular domain. J. Biol. Chem. 267, 586–590 [PubMed] [Google Scholar]
- 40.Lee, A. S., Bell, J., and Ting, J. ( 1984) Biochemical characterization of the 94- and 78-kilodalton glucose-regulated proteins in hamster fibroblasts. J. Biol. Chem. 259, 4616–4621 [PubMed] [Google Scholar]
- 41.Kang, H. S., and Welch, W. J. ( 1991) Characterization and purification of the 94-kDa glucose-regulated protein. J. Biol. Chem. 266, 5643–5649 [PubMed] [Google Scholar]
- 42.Melnick, J., Dul, J. L., and Argon, Y. ( 1994) Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature 370, 373–375 [DOI] [PubMed] [Google Scholar]
- 43.Liu, Y., Liu, W., Song, X. D., and Zuo, J. ( 2005) Effect of GRP75/ mtHsp70/ PBP74/ mortalin overexpression on intracellular ATP level, mitochondrial membrane potential and ROS accumulation following glucose deprivation in PC12 cells. Mol. Cell. Biochem. 268, 45–51 [DOI] [PubMed] [Google Scholar]
- 44.Ferrari, D. M., and Soling, H. D. ( 1999) The protein disulphide-isomerase family: unravelling a string of folds. Biochem. J. 339, 1–10 [PMC free article] [PubMed] [Google Scholar]
- 45.Shin, B. K., Wang, H., Yim, A. M., Le Naour, F., Brichory, F., Jang, J. H., Zhao, R., Puravs, E., Tra, J., Michael, C. W., Misek, D. E., and Hanash, S. M. ( 2003) Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem. 278, 7606–7616 [DOI] [PubMed] [Google Scholar]
- 46.Asea, A., Kraeft, S. K., Kurt-Jones, E. A., Stevenson, M. A., Chen, L. B., Finberg, R. W., Koo, G. C., and Calderwood, S. K. ( 2000) Hsp70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat. Med. 6, 435–442 [DOI] [PubMed] [Google Scholar]
- 47.Harada, M., Kimura, G., and Nomoto, K. ( 1998) Heat shock proteins and the antitumor T cell response. Biotherapy 10, 229–235 [DOI] [PubMed] [Google Scholar]
- 48.Vabulas, R. M., Braedel, S., Hilf, N., Singh-Jasuja, H., Herter, S., Ahmad-Nejad, P., Kirschning, C. J., Da Costa, C., Rammensee, H. G., Wagner, H., and Schild, H. ( 2002) The endoplasmic reticulum-resident heat shock protein Gp96 activates dendritic cells via the Toll-like receptor 2/4 pathway. J. Biol. Chem. 277, 20847–20853 [DOI] [PubMed] [Google Scholar]
- 49.Park, K. J., Gaynor, R. B., and Kwak, Y. T. ( 2003) Heat shock protein 27 association with the IκB kinase complex regulates tumor necrosis factor α-induced NF-κB activation. J. Biol. Chem. 278, 35272–35278 [DOI] [PubMed] [Google Scholar]
- 50.Tabary, O., Escotte, S., Couetil, J. P., Hubert, D., Dusser, D., Puchelle, E., and Jacquot, J. ( 2001) Relationship between IκBα deficiency, NF-κB activity and interleukin-8 production in CF human airway epithelial cells. Pfluegers Arch. Eur. J. Physiol. 443, S40–S44 [DOI] [PubMed] [Google Scholar]
- 51.Pollard, H. B., Eidelman, O., Jozwik, C., Huang, W., Srivastava, M., Ji, X. D., McGowan, B., Formas Norris, C., Todo, S., Darling, T., Mogayzel, P. J., Zeitlin, P. L., Wright, J., Guggino, W. B., Metcalf, E., Driscoll, W., Mueller, G., Paweletz, C., and Jacobowitz, D. M. ( 2006) De novo biosynthetic profiling of high abundance proteins in cystic fibrosis lung epithelial cells. Mol. Cell. Proteomics 5, 1628–1637 [DOI] [PubMed] [Google Scholar]
- 52.Jensen, P., Johansen, H. K., Carmi, P., Høiby, N., and Cohen, I. R. ( 2001) Autoantibodies to pancreatic hsp60 precede the development of glucose intolerance in patients with cystic fibrosis. J. Autoimmun. 17, 165–172 [DOI] [PubMed] [Google Scholar]
- 53.de Graeff-Meeder, E. R., Rijkers, G. T., Voorhorst-Ogink, M. M., Kuis, W., van der Zee, R., van Eden, W., and Zegers, B. J. ( 1993) Antibodies to human HSP60 in patients with juvenile chronic arthritis, diabetes mellitus, and cystic fibrosis. Pediatr. Res. 34, 424–428 [DOI] [PubMed] [Google Scholar]
- 54.al-Shamma, M. R., McSharry, C., McLeod, K., McCruden, E. A., and Stack, B. H. ( 1997) Role of heat shock proteins in the pathogenesis of cystic fibrosis arthritis. Thorax 52, 1056–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.