Abstract
The outer segment is a specialized compartment of vertebrate rod and cone photoreceptor cells where phototransduction takes place. In rod cells it consists of an organized stack of disks enclosed by a separate plasma membrane. Although most proteins involved in phototransduction have been identified and characterized, little is known about the proteins that are responsible for outer segment structure and renewal. In this study we used a tandem mass spectrometry-based proteomics approach to identify proteins in rod outer segment preparations as an initial step in defining their roles in photoreceptor structure, function, renewal, and degeneration. Five hundred and sixteen proteins were identified including 41 proteins that function in rod and cone phototransduction and the visual cycle and most proteins previously shown to be involved in outer segment structure and metabolic pathways. In addition, numerous proteins were detected that have not been previously reported to be present in outer segments including a subset of Rab and SNARE proteins implicated in vesicle trafficking and membrane fusion. Western blotting and immunofluorescence microscopy confirmed the presence of Rab 11b, Rab 18, Rab 1b, and Rab GDP dissociation inhibitor in outer segments. The SNARE proteins, VAMP2/3, syntaxin 3, N-ethylmaleimide-sensitive factor, and Munc 18 detected in outer segment preparations by mass spectrometry and Western blotting were also observed in outer segments by immunofluorescence microscopy. Syntaxin 3 and N-ethylmaleimide- sensitive factor had a restricted localization at the base of the outer segments, whereas VAMP2/3 and Munc 18 were distributed throughout the outer segments. These results suggest that Rab and SNARE proteins play a role in vesicle trafficking and membrane fusion as part of the outer segment renewal process. The data set generated in this study is a valuable resource for further analysis of photoreceptor outer segment structure and function.
Vertebrate photoreceptor cells are highly specialized, photosensitive neurons that function in the transduction of light into an electrical signal and the transmission of this signal to other neurons in the retina as the initial steps in vision. Rod and cone photoreceptor cells consist of five principle regions: the outer segment where the process of phototransduction takes place, a thin connecting cilium that joins the outer segment to the inner segment and allows for the passage of proteins and other molecules between the inner and outer segments, the inner segment that contains the biosynthetic and metabolic machinery of the cell, the cell body harboring the nucleus, and the synaptic region containing the synaptic vesicles and the ribbon synapse for transmission of electrical signals to secondary neurons of the retina.
The rod outer segment (ROS)1 consists of an ordered stack of over 1000 closed disks surrounded by a separate plasma membrane. Cone outer segments (COSs) have a similar stacked membrane organization, although the disk membranes are continuous with the plasma membrane. Outer segments undergo a continual renewal process in which new disk membrane is added at the base of the outer segment while packets of aged disks are shed from the distal end and removed by a phagocytic process mediated by adjacent retinal pigment epithelial (RPE) cells (1, 2). This enables the outer segment to be completely renewed over a period of 10 days.
Studies over the past several decades have led to a comprehensive understanding of phototransduction (3–5). In rod cells photoexcitation of rhodopsin in disk membranes activates the G-protein (transducin)-mediated visual cascade resulting in the stimulation of phosphodiesterase, hydrolysis of cGMP, closure of cGMP-gated channels in the plasma membrane, and hyperpolarization of the cell. Following photoexcitation, the rod cell returns to its dark state through a series of reactions involving inactivation of rhodopsin and other protein components of the visual cascade, resynthesis of cGMP, and regeneration of rhodopsin from 11-cis-retinal and opsin. Similar photoexcitation and recovery mechanisms take place in COSs although in many cases the proteins involved are encoded by different although related genes.
Photoreceptor outer segments also contain proteins that function in other essential cellular processes. Retinol dehydrogenase (RDH8) and the photoreceptor-specific ABC transporter ABCA4 (also known as the rim protein or ABCR) play important roles in the removal of all-trans-retinal from disk membranes following the photobleaching of rhodopsin as part of the visual cycle (6–9). The GLUT-1 glucose transporter and enzymes of the glycolytic and creatine phosphate shuttle pathways function in the production of energy in the form of ATP for phototransduction and other energy-dependent processes, whereas hexose monophosphate shunt and nucleotide-processing enzymes play essential roles in the generation of NADPH and interconversion of adenosine and guanine nucleotides (10–12). Finally a number of membrane and soluble proteins including peripherin-2 (peripherin/rds), rom-1, prominin-1, glutamic acid rich proteins, and RP1 have been implicated in outer segment structure and morphogenesis, but their exact roles remain to be determined (13–17).
To date, over 135 genes have been linked to various retinal diseases including retinitis pigmentosa, macular degeneration, congenital stationary night blindness, and related disorders (RetNet™ retinal information network). A significant number of these genes encode proteins that play key roles in phototransduction, the visual cycle, and outer segment structure and morphogenesis (18). However, recent genetics analysis of autosomal dominant retinitis pigmentosa suggests that less than 50% of the cases can be traced to mutations in genes known to be associated with this disease (19). This suggests that mutations in genes encoding other unidentified ROS proteins may be responsible for at least some retinal degenerative diseases for which the defective gene is unknown.
To determine the molecular and cellular mechanisms underlying outer segment structure, function, and renewal and to extend our knowledge about the genetics and molecular basis underlying retinal degenerative diseases, it is essential to identify and characterize all proteins in photoreceptor outer segments. Mass spectrometry has proven to be a powerful tool for determining the protein composition of cells and subcellular organelles (20, 21). Here we used a tandem mass spectrometry-based proteomics approach to identify proteins present in highly pure ROS preparations. Our studies indicate that, in addition to previously documented proteins, outer segments contain numerous proteins not previously known to reside in this photoreceptor organelle. Using biochemical and immunocytochemical techniques, we showed that photoreceptor outer segments contain a subset of Rab and SNARE proteins that may play a role in photoreceptor outer segment membrane trafficking and fusion as part of the outer segment renewal process.
EXPERIMENTAL PROCEDURES
Materials—
Monoclonal antibodies (mAbs) to ABCA4 (Rim3F4), the cGMP-gated channel (PMc 2G11), peripherin-2 (Per3B6), rom-1 (Rom1D5), glyceraldehyde-3-phosphate dehydrogenase (Gph 3E12), and the rod Na/Ca-K-exchanger (PMe2D9) have been described previously (6, 11, 13, 22–24). Polyclonal antibody (pAb) to syntaxin 3 was a generous gift from Dr. Vesa Olkkonen (National Public Health Institute, Helsinki, Finland), and Rab 11a and Rab 11b antibodies were generously provided by Dr. Lynne Lapierre (Vanderbilt University). Other antibodies were obtained from the following sources: synaptophysin mAb, Santa Cruz Biotechnology; Munc 18 (syntaxin-binding protein 1) pAb and Rab GDP dissociation inhibitor (RabGDI) (α) mAb, Synaptic Systems; vesicle-associated membrane protein 2 (VAMP2) pAb that cross-reacts with VAMP3 and calnexin pAb, Stressgen; N-ethylmaleimide-sensitive factor (NSF) pAb and mAb and prohibitin pAb, Abcam; and Na/K-ATPase α3 mAb, Affinity BioReagents. All antibodies labeled a single band (or a doublet in the case of syntaxin 3) of the predicted protein molecular weight in retinal extracts as analyzed by Western blotting.
Retinal Membranes and ROS Preparations—
Crude retina membranes were prepared by placing a bovine retina in 0.5 ml of homogenizing buffer (20 mm Tris-HCl, pH 7.4, 0.15 m NaCl, 1 mm MgCl2, 1 mm CaCl2, Complete protease inhibitor). The retina was first disrupted by repeatedly pipetting the suspension through a 1-ml Gilson pipette on ice for 20 min and subsequently further homogenized by passing the suspension through a 22-gauge needle 12 times. The homogenate was applied on top of a 50% (w/w) sucrose/TBS cushion and centrifuged in a TLS55 rotor (Beckman) at 25,000 rpm for 30 min. Retinal membranes were collected at the top of the 50% sucrose and washed one time in TBS by centrifugation. Protein concentration was determined using the BCA assay (Pierce).
ROSs were isolated on a continuous sucrose gradient from fresh or previously frozen, dark-adapted bovine retinas and fractionated into ROS membrane and soluble (cytoplasmic) fractions by hypotonic lysis of ROSs followed by centrifugation (25). The soluble fraction was concentrated on a Nanosep 10,000 spin column (Pall Corp., East Hills, NY) to 1 mg/ml protein. The ROS membrane pellet was washed twice in Tris-EDTA buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA) and stored in the same buffer. ROS membranes were further separated into a highly pure disk fraction and enriched plasma membrane fraction by a modification of the immunogold density perturbation method (25). Briefly the extracellular surface of ROSs was labeled for 2 h at 4 °C with the PMe 2D9 mAb conjugated to 10-nm gold-dextran particles (22). The labeled ROSs were washed three times in Tris-EDTA buffer by centrifugation at 12,000 rpm for 10 min and resuspended in 2.5 mm Tris, pH 7.4, containing 1 mm EDTA and 1 mm DTT. Dissociation of disk membranes from the plasma membrane was allowed to occur overnight at 4 °C. The sample was subsequently vortexed, pelleted, and resuspended in 7% sucrose, 20 mm Tris, pH 7.4. The sample was centrifuged on a 20–40% sucrose continuous gradient at 22,500 rpm for 2 h. Unlabeled disk membranes were collected as a band near the top of the gradient, whereas the enriched labeled plasma membrane was collected near the bottom of the tube.
SDS-PAGE and Western Blot Analysis—
For Western blotting, samples were denatured in 10 mm Tris-HCl, pH 6.8, 4% SDS, 20% sucrose, 4% β-mercaptoethanol and separated on 8 or 11.5% SDS-polyacrylamide gels. Proteins were transferred onto Immobilon-FL membranes (Millipore, Bedford, MA). Blots were blocked in 0.5% skim milk in PBS for 30 min and labeled with the primary antibody in 0.5% skim milk in PBS containing 0.05% Tween 20 (PBS-T) at the recommended dilution. The blots were washed in PBS-T and labeled with a secondary anti-mouse or anti-rabbit antibody conjugated to Alexa Fluor 680 (1:40,000) (Molecular Probes, Eugene, OR) or LI-COR IRDye 800 (1:10,000) (Rockland, Gilvertsville, PA) for analysis on a LI-COR Odyssey imager (LI-COR Biosciences, Lincoln, NE). Quantification of the band intensities from three different ROS and disk preparations were determined from the integrated peak area using the LI-COR imaging analysis program. For MS analysis, samples (50 μg of protein) were denatured in lithium dodecyl sulfate denaturing buffer containing 50 mm DTT and separated on 4–12% Tris/glycine NuPAGE gels (Invitrogen).
In-solution and In-gel Digestion of Proteins for Proteomics Analysis—
For in-solution digests, protein samples were prepared as described previously (26). Briefly samples (5 μg of protein) were solubilized with 6 m urea, 2 m thiourea in 10 mm HEPES, pH 8.0; reduced with 1 μg of DTT/50 μg of protein for 30 min; alkylated with 5 μg of iodoacetamide/50 μg of protein for 30 min; digested with 1 μg of endopeptidase Lys-C/50 μg of protein for 3 h; and diluted 4× with 50 mm NH4HCO3. The proteins were subsequently digested overnight at 37 °C with 1 μg of porcine modified trypsin (Promega, Nepean, Ontario, Canada)/50 μg of protein.
For in-gel digests, each lane of an SDS gel was cut into 15 pieces and digested exactly as described previously (27). Peptide mixtures were desalted and concentrated using stop and go extraction (STAGE) tips (28). To gain better coverage when comparing disks versus ROS membranes, dimethylated peptides (see below) were further resolved between pH 3 and 10 by OFFGEL in-solution isoelectric focusing (Agilent Technologies, Mississauga, Ontario, Canada) according to the manufacturer's instructions.
Formaldehyde Labeling—
Formaldehyde isotopologues (29) were used to quantify relative abundances of proteins in disks versus total ROS membranes. Briefly peptides purified with C18 stop and go extraction (STAGE) tips (28) were resuspended in 5 μl of 200 mm formaldehyde or deuterated formaldehyde (Cambridge Isotope Laboratories) and 0.5 μl of 1 m sodium cyanoborohydride and incubated for 30 min at room temperature away from light. The reaction mixture was then adjusted to pH 7.5, and a further 5 μl of the respective formaldehyde isotopologue plus 1 μl of 1 m cyanoborohydride was added. The reaction was allowed to continue for a further 30 min before being quenched by addition of 6 μl of 2.5 m NH4Cl for 10 min at room temperature.
LC-MS/MS—
Peptides were resolved by reverse phase chromatography on 15-cm-long, 75-μm-inner diameter fused silica emitters with an 8-μm-diameter opening (Polymicro, Phoenix, AZ) packed with 3-μm-diameter ReproSil-Pur C18 beads (Dr. Maisch, Ammerbuch-Entringen, Germany) with an Agilent 1100 Series nanoflow HPLC instrument coupled on line to LTQ-FT and LTQ-Orbitrap systems (ThermoFisher, Bremen, Germany) using nanospray ionization sources (Proxeon Biosystems, Odense, Denmark). Running buffer A consisted of 0.5% acetic acid, and running buffer B consisted of 0.5% acetic acid, 80% acetonitrile. Gradients were run from 6 to 30% B over 60 min, 30 to 80% B for 10 min, held at 80% B for 5 min, and then dropped to 6% B for 15 min to recondition the column. The LTQ-FT system was set to acquire a full-range scan at 25,000 resolution in FT from which the three most intense multiply charged ions per cycle were isolated for fragmentation in the LTQ. Selected ion monitoring scans in FT were also carried out on each of the three precursor ions as described previously (30). The LTQ-Orbitrap was set to acquire a full-range scan at 60,000 resolution from 350 to 1500 thomson in the Orbitrap and to simultaneously fragment the top five peptide ions in each cycle in the LTQ (31). Typically LC-MS/MS was carried out on at least three separate ROS and subcellular fraction preparations.
MS Data Analysis—
Fragment peak lists were generated by ExtractMSN (version 3.2, ThermoFisher) using the default parameters, and monoisotopic peak and charge state assignments were checked by DTA Supercharge, part of the MSQuant suite of software (SourceForge, Inc.). Fragment spectra were searched against the bovine IPI database (version 3.15, 33,270 sequences) using Mascot Server version 2.2 with the following parameters: trypsin specificity allowing up to one missed cleavage, cysteine carbamidomethylation as a fixed modification, ESI-trap fragmentation, 5-ppm mass tolerance for precursor ion mass, and 0.8-Da mass tolerance for fragment ions. For dimethylated samples four variable modifications were also considered (Unimod names): dimethyl:2H(4) (Lys), dimethyl:2H(4) (N terminus), dimethyl (N terminus), and dimethyl (Lys). Sequences of human keratins, trypsin, and Lys-C were added to the bovine IPI database prior to searching. The final, non-redundant list of proteins was generated using finaList.pl, an in-house script available by request that finds the smallest set of proteins that explains the observed peptides. Acceptance criteria for protein identifications were set to require two or more peptides of seven or more amino acids and each with scores greater than 24, corresponding to a false discovery rate of less than 1% based on the number of hits observed with the same conditions when the data were searched against the reversed IPI bovine database. Multiple isoforms were included in the final list when at least one peptide uniquely identifying each isoform was observed. Using a fixed score cutoff when the 95% identify score calculated by Mascot for each peptide can vary widely could potentially lead to some individual peptides having a higher false discovery rate, but this is balanced out overall by lower false discovery rates for those peptides with low identity scores and anyhow is taken into account by the reversed database searching. Quantitative ratios of dimethylated peptides based on extracted ion chromatograms were extracted from the raw MS data using MSQuant version 1.4.1, and peptides common to multiple proteins contributed to the quantitative measures for each protein. All peptides meeting the false discovery rate criteria were used for quantification. Absolute protein expression measurement (APEX) ratios were calculated as described previously (32) using 0.99 for the pi value for each hit and approximating the Oi term as the number of tryptic peptides (zero and one missed cleavages considered) with between 6 and 30 amino acids in each protein.
Immunofluorescence Microscopy—
For tissue labeling, rat or mouse eyes were fixed with 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.0 (PB) for 4 h. Cryosections were permeabilized and blocked in PB containing 0.2% Triton X-100 and 10% normal goat serum for 20 min. The sections were labeled overnight with the primary antibody diluted in PB containing 0.1% Triton X-100 and 2.5% normal goat serum according to the manufacturer's specifications. Sections were rinsed in PB, labeled for 1 h with the secondary antibody conjugated with Cy3, and counterstained with DAPI nuclear stain for analysis with a Zeiss LSM510 Meta confocal microscope equipped with a Zeiss LSM5 Image Browser.
RESULTS
Purity of ROS Preparations—
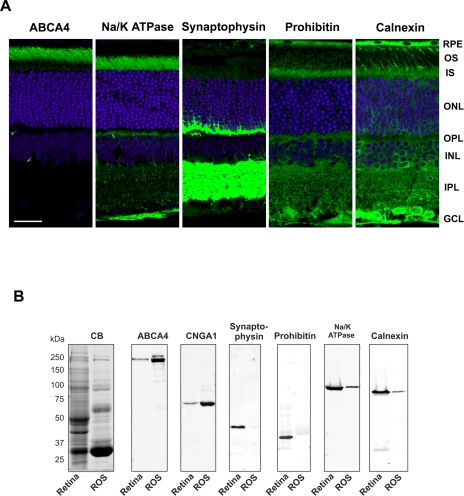
ROSs isolated from bovine retina by sucrose density centrifugation have been extensively studied by electron microscopy and SDS gel electrophoresis (25, 33). These preparations consist of stacks of disks enclosed by a plasma membrane with rhodopsin estimated to make up over 75% of the total protein content. For the current studies, the purity of these ROS preparations was further evaluated using a number of subcellular marker proteins together with immunofluorescence and Western blotting. The photoreceptor ABC transporter (ABCA4) and the cyclic nucleotide-gated channel α subunit (CNGA1) were used as markers for ROS disk and plasma membrane, respectively (6, 23); synaptophysin was used for synaptic vesicles (34), Na/K-ATPase was used for the inner segment plasma membrane (35), prohibitin was used for mitochondria (36), and calnexin was used for ER (37).
Immunofluorescence microscopy was first used to validate the use of these proteins as markers for retinal subcellular compartments (Fig. 1A). As previously reported ABCA4 and CNGA1 (not shown) were localized to the photoreceptor outer segment layer of the retina (6, 23, 38). In contrast, the Na/K-ATPase, synaptophysin, and prohibitin were not detected in the outer segment layer but were abundantly present in other retinal layers. Na/K-ATPase was primarily localized to the inner segment and outer plexiform layers and to a lesser extent the outer nuclear layer, synaptophysin was present in the outer and inner synaptic (plexiform) layers, and prohibitin was found in the photoreceptor inner segment layer as well as other retinal cell layers in agreement with its reported localization in mitochondria. Calnexin localized primarily to the inner segments of photoreceptors and the cell bodies of other retinal cells consistent with its role as an ER chaperone (37). However, more limited immunostaining was also observed in the outer segment layer suggesting that small amounts of calnexin translocate to this compartment.
Fig. 1.
Subcellular markers used for analysis of the purity of isolated bovine ROSs. A, localization of subcellular markers in rat retinal sections by confocal scanning microscopy. Rat retinal cryosections were labeled with primary antibodies for ABCA4 (outer segment marker), Na/K-ATPase (inner segment plasma membrane marker), synaptophysin (synaptic vesicle marker), prohibitin (mitochondrial marker), and calnexin (endoplasmic reticulum marker) followed by a secondary antibody tagged with Cy3 fluorescent dye (green). The sections were counterstained with DAPI nuclear stain. Retinal layers are: OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; and GCL, ganglion cell layer. Bar, 20 μm. B, SDS gels and Western blots of retina homogenate (Retina) and isolated ROSs stained with Coomassie Blue (CB) or labeled with antibodies to ABCA4, CNGA1 (cyclic nucleotide-gated channel A1 subunit), synaptophysin, prohibitin, Na/K-ATPase (α3 subunit), and calnexin.
SDS gel electrophoresis and Western blotting were used to determine whether the various subcellular markers were detectable in isolated ROS preparations as well as crude retinal extracts (Fig. 1B). A complex Coomassie Blue-stained protein banding pattern was evident in the retinal membrane extracts. In contrast, isolated ROSs showed a simpler protein pattern dominated by the intensely stained rhodopsin monomer (∼36 kDa) and more moderately stained rhodopsin dimer (∼70 kDa). Western blots revealed a marked increase in the level of ABCA4 and CNGA1 in isolated ROSs compared with the retinal membrane extracts as expected (Fig. 1B). In contrast, synaptophysin and prohibitin were detected in the retinal membranes but not isolated ROSs. A relatively low amount of Na/K-ATPase was observed in isolated ROSs suggesting that inner segment plasma membrane vesicles are a minor contaminant of ROS preparations. A low level of calnexin was also observed in the ROS fraction consistent with the detection of small amounts of calnexin in outer segments by immunofluorescence microscopy (Fig. 1A). Together these studies support the high degree of purity of these preparations.
Subfractionation of ROSs—
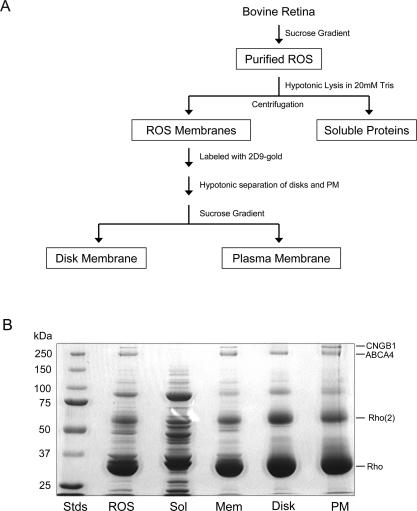
To obtain a comprehensive list of photoreceptor outer segment proteins, purified bovine ROSs were separated into membrane and soluble fractions (Fig. 2A). ROS membranes were further separated into a highly pure disk membrane fraction and an enriched plasma membrane fraction using an immunogold density perturbation procedure (25). The enriched plasma membrane preparations contain some disk vesicles that remain attached to the plasma membrane because it has been shown previously that hypotonic treatment of ROSs does not completely dissociate the rim regions of disk membranes from the plasma membrane (25). SDS gels of the various fractions are shown in Fig. 2B. The membrane fractions were dominated by monomeric and multimeric forms of rhodopsin (25, 33). The 250-kDa ABCA4 transporter was readily seen in all membrane preparations (6), whereas the β subunit of the cGMP-gated channel (CNGB1) was present in ROS membrane and plasma membrane fractions but absent in the disk membrane fraction (39) consistent with its localization to the plasma membrane of ROSs (23). The soluble fraction showed a more complex banding pattern. The dominant doublets at 37–39 and 89–91 kDa corresponded to the α and β subunits of transducin and phosphodiesterase of rod photoreceptors, respectively (40, 41). Numerous less intense proteins were visible in the range of 20–150 kDa.
Fig. 2.
A, flow diagram showing the main steps in the purification and fractionation of bovine ROSs. B, Coomassie Blue-stained SDS-polyacrylamide gels of the various ROS fractions used in our proteomics studies. From left to right are molecular weight standards (Stds), ROS, soluble fraction (Sol), rod outer segment membranes (Mem), disk fraction (Disk), and enriched plasma membrane fraction (PM). Each lane was loaded with 30 μg of proteins. Rho, rhodopsin monomer; Rho(2), rhodopsin dimer, CNGB1, cyclic nucleotide-gated channel (B1 or β subunit).
Proteomics Analysis of Bovine ROSs—
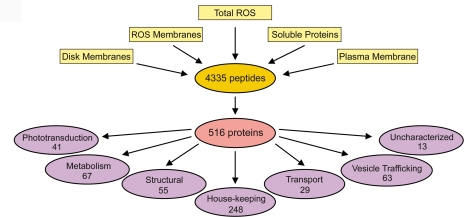
In-gel and in-solution tryptic digestion was carried out on five fractions: total ROSs, ROS membranes, soluble, disk, and plasma membrane. The resulting peptides were analyzed by LC-MS/MS. Database searching of the fragmentation spectra resulted in the identification of a combined total of 516 proteins (primary data set) from 4335 peptides (supplemental Tables S1 and S2) using a selection criterion of two or more peptides having a length of seven or more amino acids (Fig. 3). Reducing the criteria to one peptide and/or shorter peptide lengths resulted in a secondary data set of 500 additional proteins. However, this data set was not included in our proteomics analysis except where noted because these reduced criteria are known to generate a high level of false positives (21, 42).
Fig. 3.
Diagram depicting the proteomics analysis of rod outer segment fractions. MS/MS analysis of five fractions (total ROSs, ROS membranes, soluble proteins, disk membranes, and enriched plasma membranes) resulted in 4335 peptides from which 516 unique proteins (primary data set) were identified. These proteins were classified into seven broad categories according to their known or predicted functions.
To facilitate our analysis, the identified proteins were separated into six broad classes based on their principal known or predicted function (Fig. 3). A seventh class was added to segregate “hypothetical” proteins annotated in the genomic database for which no information on their putative function was available. Our complete primary data set of proteins is given in supplemental Table S3 along with the ROS fractions in which the proteins were detected.
Rod and Cone Phototransduction Proteins—
Proteins that function in phototransduction and the visual cycle comprise a prominent class of proteins (Table I and supplemental Table S3). All proteins known to play a direct role in phototransduction and retinoid processing in ROSs were detected in our primary data set with the exception of the relatively small proteins GCAP1, GCAP2, and the phosphodiesterase γ subunit. However, these three proteins were present in our secondary data set generated from a single peptide or peptides of less than seven amino acids in length. Apparently trypsin digestion of these relatively small proteins did not yield a sufficient number of long peptides detected by LC-MS/MS for inclusion into our primary data set. RPE65 and cellular retinaldehyde-binding protein (CRABLP), two proteins known to function in the visual cycle and retinoid binding, were found in our proteomics studies. However, because these proteins have been reported to reside in RPE and/or Mueller cells (43, 44), they most likely represent minor contaminants of our ROS preparations.
Table I.
Selected proteins involved in phototransduction and visual cycle in rods and cones
The number of unique peptides used for the identification is given. The protein to rhodopsin ratio was estimated by APEX analysis. NF, not found in the APEX list of 150 most abundant proteins from the primary data set.
| Protein | Unique peptides | Protein to rhodopsin ratio (APEX) |
|---|---|---|
| Phototransduction | ||
| Rods | ||
| Rhodopsin (Rho) | 10 | 1 |
| Transducin, α-1 subunit | 28 | 0.250 |
| Transducin, β-1 subunit | 15 | 0.106 |
| Transducin, γ-T1 subunit | 7 | 0.039 |
| Rod cGMP-specific 3′,5′-cyclic phosphodiesterase α subunit | 44 | 0.070 |
| Rod cGMP-specific 3′,5′-cyclic phosphodiesterase β subunit | 51 | 0.077 |
| cGMP-gated cation channel α subunit (CNGA1) | 24 | 0.049 |
| cGMP-gated cation channel 240-kDa β subunit (CNGB1) | 37 | 0.060 |
| Recoverin | 13 | 0.028 |
| Regulator of G protein signaling 9 (RGS9) | 33 | 0.021 |
| Guanine nucleotide-binding protein, β-5 subunit | 15 | 0.017 |
| Regulator of G protein signaling 9-binding protein (R9AP) | 12 | 0.010 |
| Retinal guanylyl cyclase 1 (GC1) | 39 | 0.047 |
| Retinal guanylyl cyclase 2 (GC2) | 23 | 0.008 |
| Rhodopsin kinase | 22 | 0.047 |
| Sodium/calcium-potassium exchanger 1 (NCKX1) | 16 | 0.030 |
| Splice isoform B of S-arrestin | 36 | 0.120 |
| Calmodulin 1 | 3 | NF |
| Protein phosphatase 2 α | 4 | 0.004 |
| Cones | ||
| Arrestin 3 (X-arrestin) | 4 | 0.007 |
| Blue-sensitive opsin | 2 | 0.023 |
| cGMP-gated cation channel α3 (CNGA3) | 6 | NF |
| Cone cGMP-specific 3′,5′-cyclic phosphodiesterase α subunit | 27 | 0.016 |
| Red opsin | 8 | 0.057 |
| Retina G protein-coupled receptor kinase 7 | 4 | 0.005 |
| Sodium/calcium-potassium exchanger 2 | 3 | NF |
| Transducin α-2 subunit | 16 | 0.054 |
| Transducin γ-T2 subunit | 2 | NF |
| Visual/retinoid cycle | ||
| ABCA4 | 95 | 0.047 |
| All-trans-retinol dehydrogenase | 7 | 0.021 |
| Cellular retinaldehyde-binding protein | 12 | 0.031 |
| Interphotoreceptor retinoid-binding protein (IRBP) | 46 | 0.061 |
| Retinal pigment epithelium-specific 65-kDa protein | 3 | NF |
In addition to rod phototransduction proteins, we also detected essentially all proteins known to function in cone phototransduction including red and blue cone opsin, cone transducin, cone arrestin, cone opsin kinase, the cone cGMP-gated channel α subunit (CNGA3), and the cone sodium/calcium-potassium exchanger (Table I). One protein that was not detected was the β subunit of the cone cGMP-gated channel (CNGB3). These results indicate that COSs co-purify with ROSs by the sucrose density centrifugation method used here.
Metabolic, Structural, Transport, and Housekeeping Proteins—
A significant number of metabolic, transport, and structural proteins were present in our data set as exemplified in Table II (see supplemental Table S3 for a complete listing). Essentially all the glycolytic enzymes, the GLUT-1 glucose transporter, and many enzymes that function in the hexose monophosphate shunt and creatine phosphate shuttle pathways were detected confirming earlier studies (10, 11, 45). Peripherin/rds, rom-1, prominin-1, and RP1 implicated in outer segment structure and morphogenesis and associated with various retinal degenerative diseases were also observed (14–16, 46). A large fraction of the proteins were classified as housekeeping proteins. These include chaperone proteins, nucleotide-processing proteins, transcription factors, and other proteins typically found in most cells.
Table II.
Selected proteins involved in outer segment structure, metabolism, and vesicular trafficking
The number of unique peptides used for the identification is given. The protein to rhodopsin ratio was estimated by APEX analysis. NF, not found in the APEX list of 150 most abundant proteins in the primary data set.
| Protein | Unique peptides | Protein to rhodopsin ratio (APEX) |
|---|---|---|
| Structure | ||
| Peripherin/rds | 14 | 0.141 |
| rom-1 | 11 | 0.248 |
| Prominin-1 | 18 | NF |
| Retinitis pigmentosa 1 protein | 24 | 0.006 |
| Actin 2 | 16 | 0.026 |
| Tubulin, β2 | 20 | 0.128 |
| Metabolism/transport | ||
| Brain creatine kinase | 10 | 0.039 |
| Glucose transporter 1 (GLUT-1) | 7 | 0.038 |
| Glyceraldehyde-3-phosphate dehydrogenase | 25 | 0.109 |
| Hexokinase 1 | 19 | 0.012 |
| Phosphoglycerate kinase 1 | 15 | 0.025 |
| Sodium/potassium-transporting ATPase | 34 | 0.029 |
| Transketolase | 5 | 0.004 |
| Vesicular trafficking | ||
| NSF | 16 | 0.008 |
| Rab 11b | 15 | 0.010 |
| Rab 1b | 10 | 0.030 |
| Rab 18 | 9 | 0.009 |
| Rab 3a | 11 | 0.049 |
| RabGDI (α) | 3 | NF |
| Syntaxin 3 | 8 | 0.013 |
| Syntaxin-binding protein 1 (Munc18-1) | 22 | 0.017 |
| VAMP2/3 | 3 | NF |
Vesicle-trafficking and Membrane Fusion Proteins—
Proteins implicated in vesicle trafficking and membrane fusion comprised a large class (Table II and supplemental Table S3). Of the 63 proteins in this category, 29 were members of the Rab subclass of small GTPase proteins (47) known to function as regulators of membrane trafficking and fusion. Both the α and β subunits of the Rab accessory protein RabGDI were also identified. In addition, numerous SNARE proteins implicated in membrane fusion were detected (48). These included syntaxin 3, VAMP2 or -3 (VAMP2/3 also known as synaptobrevin), NSF, syntaxin-binding protein (Munc 18-1), SNAP25, and others. In agreement with Western blots of ROSs and immunofluorescence studies, synaptophysin, an abundant synaptic protein, was not detected by MS/MS confirming the absence of synaptic vesicles in our outer segment preparations.
APEX—
APEX is a recently developed method that estimates protein abundance from the proportionality of the peptides expected and peptides observed in an LC-MS/MS experiment (32). This procedure was used to determine the 150 most abundant proteins in our isolated ROS preparation (Tables I and II and supplemental Table S4). As expected rhodopsin was the most abundant ROS protein. Most other phototransduction/visual cycle proteins and structural proteins previously shown to be in outer segments were represented within the 50 most abundant proteins. Using rhodopsin as a reference, the 150 most abundant proteins in our ROS preparations ranged from transducin at ∼25% the level of rhodopsin to a member of the kinesin family of proteins at 0.11% (Table I and supplemental Table S4).
Protein Profiling of ROS and Disk Membranes by LC-MS/MS and Western Blotting—
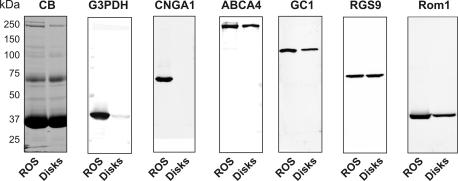
Protein profiling of isotope-labeled formaldehyde-derivatized tryptic peptides was used to determine the relative amounts of proteins in disks versus ROS membranes. The data from the LC-MS/MS experiments were first compared with results obtained by Western blotting for a number of proteins for which antibodies were available (Fig. 4). As summarized in Table III, the two methods are in close agreement. Proteins such as CNGA1, CNGB1, sodium/potassium-calcium exchanger, and glyceraldehyde-3-phosphate dehydrogenase previously shown to be localized in the plasma membrane of ROSs resulted in very high ROS:disk ratios (>20:1). In some cases, values could not be determined because the proteins were not detected in the disk fraction. In contrast proteins in the disk rim such as rom-1, ABCA4, peripherin, and GC1 had ratios just above 1, whereas rhodopsin, which is present in both the disk and plasma membrane but more abundant in the former (25), had a ratio just below 1. A complete protein profiling list obtained by LC-MS/MS is given in supplemental Table S5.
Fig. 4.
SDS gel and Western blots of total ROSs and isolated disks used for protein profiling. SDS gel was stained with Coomassie Blue (CB), and Western blots were labeled with antibodies to proteins known to be present in ROSs including glyceraldehyde-3-phosphate dehydrogenase (G3PDH), CNGA1 (cyclic nucleotide-gated channel A1 subunit, ABCA4, guanylate cyclase 1 (GC1), RGS9, and rom-1. The labeled proteins were quantified by densitometry measurements (see “Experimental Procedures”), and the protein profiling data obtained by Western blotting were compared with protein profiling determined by LC-MS/MS (see Table III and supplemental Table S5).
Table III.
Protein profiling in ROS versus disk membranes by mass spectrometry and Western blotting for selected proteins in the primary data set
n, number of experiments; values, ±S.D. for n = 3; av, average value for n = 2; high, >20.
| Protein | ROS:disk
|
|
|---|---|---|
| Mass spectrometry profiling | Western blotting | |
| Cyclic nucleotide-gated channel α1 (CNGA1) | High (n = 3) | High (n = 3) |
| Cyclic nucleotide-gated channel β1 (CNGB1) | High (n = 3) | High (n = 3) |
| Sodium/calcium-potassium exchanger 1 (NCKX1) | High (n = 1) | High (n = 3) |
| Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) | High (n = 3) | High (n = 3) |
| Rod outer segment membrane protein 1 (rom-1) | 3.84 ± 1.31 (n = 3) | 1.90 ± 0.53 (n = 3) |
| ABC transporter (ABCA4, ABCR, or rim protein) | 1.52 ± 0.91 (n = 3) | 1.12 ± 0.46 (n = 3) |
| Regulator of G protein signaling 9 (RGS9) | 1.08 ± 1.05 (n = 3) | 0.99 ± 0.20 (n = 3) |
| Retinal guanylyl cyclase 1 (GC1) | 1.70 ± 0.42 (n = 3) | 1.90 ± 0.75 (n = 3) |
| Rhodopsin (Rho) | 0.72 ± 0.36 (n = 3) | 1.09 ± 0.095 (n = 3) |
| Rab 11b | 0.77 av (n = 2) | 0.73 av (n = 2) |
| Synaptotagmin | 3.25 ± 0.75 (n = 3) | 5.01 ± 2.03 (n = 3) |
| Syntaxin 3 | 5.42 ± 2.03 (n = 3) | 5.13 (n = 1) |
Immunolocalization of Vesicle-trafficking Proteins—
A novel and interesting result of our proteomics analysis is the existence of Rab and SNARE vesicle-trafficking proteins in ROS preparations (Table II and supplemental Table S3). Rab and SNARE proteins have been localized to the inner segments and synaptic regions of photoreceptors (49–51), but their presence in outer segments has not been reported. Therefore, it was important to determine whether some of the Rab and SNARE proteins found in our proteomics analysis are true components of photoreceptor outer segments. This was investigated using a combination of Western blotting and confocal scanning microscopy.
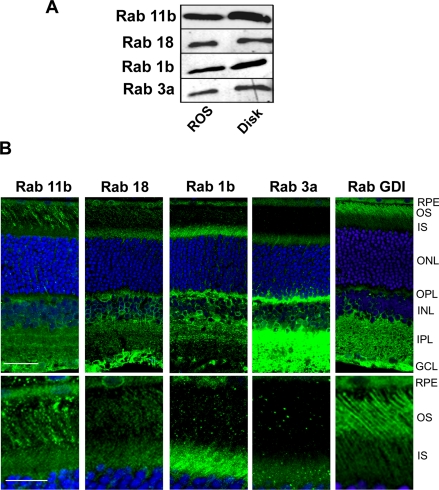
Of the 29 Rab proteins detected in our proteomics studies, we investigated four Rab proteins (Rab 3a, Rab 1b, Rab 11b, and Rab 18) present in our APEX list of the 150 most abundant ROS proteins. Western blots labeled with specific anti-Rab antibodies confirmed the presence of these Rab proteins in both ROSs and isolated disks (Fig. 5A), results consistent with MS/MS protein profiling studies (supplemental Table S5). Rab 11a was not detected in ROS preparations by Western blotting (data not shown) in agreement with its absence in our MS proteomics data set.
Fig. 5.
Detection and localization of Rab proteins in the ROS fractions and retina. A, Western blots of ROSs and disks labeled for Rab 11b, Rab 18, Rab 1b, and Rab 3a. B, top panel, confocal scanning microscopy of rat retinal cryosections labeled with primary antibodies to Rab 11b, Rab 18, Rab 1b, Rab 3a, and RabGDI (α subunit) followed by a secondary antibody tagged with the Cy3 fluorescent dye (green) and counterstained with the DAPI nuclear dye (blue). Bar, 20 μm. Bottom panel, same as top panel except showing a restricted region of the outer and inner segments of photoreceptor cells at higher magnification. Bar, 10 μm. Retinal layers are: OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; and GCL, ganglion cell layer.
To further determine whether these Rab proteins localize to outer segments, rat retinal cryosections were labeled with the Rab-specific antibodies for analysis by confocal scanning microscopy. As shown in Fig. 5B, the Rab antibodies labeled various layers in the retina including the inner segment and outer plexiform layer of photoreceptor cells and the inner plexiform, inner nuclear, and ganglion cell layers of the inner retina. Rab 11b and Rab 18 also displayed a relatively strong patchy staining pattern in the outer segment layer. Rab 1b staining was weaker, and Rab 3a staining was close to background. Retinal sections were also labeled for RabGDI, a Rab accessory protein observed in our proteomics data set. Strong staining was observed in the outer segment layer as shown in Fig. 5B. In control experiments no labeling was observed when the primary antibodies were omitted (data not shown).
Taken together, these studies provide strong evidence that Rab 11b, Rab 18, Rab 1b, and RabGDI are present in outer segments in significant amounts consistent with the MS results. Rab 3a detected in ROS and disk preparations by LC-MS/MS and Western blotting but not observed in outer segments by immunofluorescence labeling may be a contaminant from other retinal layers that co-purifies with ROSs and disks. Alternatively the limited labeling of Rab 3a observed by confocal scanning microscopy may result from the inaccessibility of the Rab 3a epitope for immunofluorescence labeling in outer segments possibly due to interaction with another outer segment protein. Additional studies are needed to determine whether Rab 3a is a true constituent of outer segments.
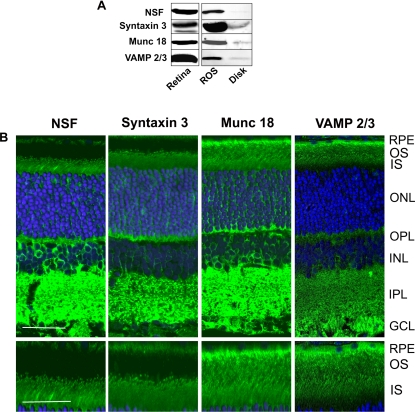
The distribution of several SNARE proteins detected in our proteomics screen was also examined by Western blotting and immunofluorescence microscopy. VAMP2/3, Munc 18-1, syntaxin 3, NSF, and synaptotagmin (not shown) exhibited strong staining in both the retinal extracts and ROS fractions but only weak staining in the disk membranes by Western blotting (Fig. 6A). By confocal scanning microscopy, VAMP2/3 and Munc 18-1 were observed throughout the outer segment layer of rat and mouse retina (Fig. 6B). Syntaxin 3 and NSF showed a striking punctate pattern of labeling at the base of the outer segment and strong labeling in the distal region of the outer segment layer adjacent to the RPE layer. The latter may reflect the labeling of aged disks within the outer segments or apical processes of RPE cells that penetrate into the outer segment layer. SNARE proteins were also observed in the synaptic regions consistent with their role in neurotransmitter release at ribbon synapses (49). Synaptotagmin exhibited strong immunostaining in the inner and outer plexiform layers, but unlike synaptophysin, weak staining was evident in the inner and outer segments (data not shown).
Fig. 6.
Detection and localization of SNARE proteins in the retina and ROSs. A, Western blots of retina, ROSs, and disks labeled with antibodies to NSF, syntaxin 3, Munc 18-1, and VAMP2/3. B, top panel, confocal scanning microscopy of rat retinal cryosections labeled with primary antibodies to NSF, syntaxin 3, Munc 18-1, and VAMP2/3 followed by a secondary antibody tagged with the Cy3 fluorescent dye (green) and counterstained with the DAPI nuclear dye (blue). Bar, 20 μm. Bottom panel, same as top panel except showing a restricted region of the outer and inner segments of photoreceptor cells at higher magnification. Bar, 10 μm. Similar labeling results were obtained with mouse retina. Retinal layers are: OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; and GCL, ganglion cell layer.
DISCUSSION
Outer Segment Proteome—
In this study we used sensitive tandem MS together with subcellular fractionation procedures to identify proteins in highly purified photoreceptor outer segment preparations. Most ROS proteins known to function in phototransduction, retinoid cycling, metabolic pathways, and outer segment structure and proteins associated with retinal degenerative diseases were detected using our LC-MS/MS approach. Furthermore although cone photoreceptors comprise less than 5% of the photoreceptors in bovine retina, all proteins known to be involved in cone phototransduction were present in our primary data set with the exception of the cone cGMP-gated channel β subunit (CNGB3). These results provide strong support for the high sensitivity of the MS approach used here. This study also shows that COSs co-purify with ROSs on sucrose density gradients. On this basis our primary data set most likely contains most of the resident proteins in photoreceptor outer segments. However, a small fraction of proteins could have been missed because of 1) the inability of trypsin digestion to produce multiple peptides of seven or more amino acids for inclusion into our primary data set as in the case of GCAP1, GCAP2, and phosphodiesterase (γ), 2) the failure to detect or identify hydrophobic or posttranslationally modified peptides, 3) the use of an incomplete bovine genomic database for mining of proteins, or 4) extremely low abundance proteins.
The 516 proteins in our primary data set most likely represent an overestimation of the number of proteins in photoreceptor outer segments. Although the outer segment preparations used in this study are highly pure based on electron microscopic and biochemical analyses as reported previously (25, 33) and further demonstrated here using subcellular markers, low levels of contaminating subcellular membranes are present as is typically the case in all subcellular fractionations. Tissue homogenization can result in the association of various subcellular membranes with outer segment membranes. These membrane aggregates cannot be readily removed because they co-sediment with isolated outer segments on sucrose gradient. Examples of protein from contaminating subcellular compartments include: the Na/K-ATPase, an abundantly expressed protein in the plasma membrane of photoreceptor inner segments (35, 52, 53); RPE 65, cellular retinaldehyde-binding protein (CRABLP), and ezrin, abundant proteins localized to subcellular organelles of RPE and Müller cells (43, 44); and calcium-dependent ATPase and myosin VI, proteins associated with other photoreceptor membranes (54, 55). In addition, hemoglobin and serum albumin appear as contaminants. These abundant soluble proteins most likely adsorbed nonspecifically to outer segment membranes when the retinal tissue was subjected to freeze-thawing and/or hypotonic lysis and as a result co-purify with outer segments on sucrose gradients. Accordingly it is important to validate MS proteomics results with other biochemical and immunocytochemical techniques to more accurately define the photoreceptor outer segment proteome. Interestingly we did not detect many photoreceptor cilium-specific components such as retinitis pigmentosa GTPase regulator (RPGR), retinitis pigmentosa GTPase regulator interacting protein (RPGRIP), and intraflagellar transport proteins suggesting that the connecting cilium is not a significant contaminant in our ROS preparations. A recent proteomics study of mouse photoreceptor sensory cilium preparations catalogued close to 2000 distinct proteins (56). These preparations contain the connecting cilium, basal body, and inner segments along with the outer segment. The larger number of proteins observed in these photoreceptor sensory cilium preparations results in part from proteins located in these subcellular compartments. Use of the complete mouse genome database for mining of proteins also may have contributed to the identification of additional proteins not found in our studies of bovine outer segments.
APEX analysis enabled us to identify the 150 most abundant proteins in ROS preparations (supplemental Table S4). Over a third of these proteins have been shown previously to be abundantly expressed in photoreceptor outer segment proteins with 32 being present in the top 50 most abundant proteins. The rank order of abundance of outer segment proteins obtained by APEX is in very good agreement with previous quantitative results obtained by more conventional biochemical analysis (57). Rhodopsin stands out as the most abundant protein with the α subunit of rod transducin as the next most abundant protein. Other relatively abundant proteins include rom-1 and peripherin/rds known to form homo- and heterotetrameric complexes (58), tubulin, arrestin, glyceraldehyde-3-phosphate dehydrogenase, transducin β subunit, rod phosphodiesterase α and β subunits known to form a 1:1 complex, interphotoreceptor retinoid-binding protein, cGMP-gated channel, retinal guanylyl cyclase (RetGC1), ABCA4 (rim protein), and others. The ability of APEX to reliably estimate the relative abundance of proteins in our data set is further demonstrated by the finding that red cone opsin is 6% as abundant as rhodopsin in outer segments, a result that is in general agreement with the known ratio of rod to cone photoreceptors in rod-dominant mammalian retinas. There are some apparent anomalies. For example, the level of rom-1 and peripherin/rds is higher than determined previously (59), and the β subunit of the rod cGMP-gated channel appears to be more abundant than the α subunit, although the stoichiometry of the native tetrameric channel is 3α:1β (60, 61). However, the latter can be explained on the basis of abundant glutamic acid rich protein splice variants of the β subunit that contribute to the peptide counting of the β subunit (39, 62). Finally the actual abundance of the proteins in the APEX list relative to rhodopsin may be an overestimation by ∼2–3-fold because it has been reported that the amount of transducin in mammalian rod cells is only 10–15%, and phosphodiesterase is about 2–3% the level of rhodopsin (57). The underestimation of rhodopsin levels may result from a deficiency in peptide counting of this exceptionally high abundance protein. Nonetheless the accuracy of the APEX-derived protein abundant measurements obtained in our studies are within the 2–3-fold range of reliability estimated in APEX analysis of yeast and Escherichia coli cell lysates by Lu et al. (32).
MS and Western blotting protein profiling studies provided additional information on the relative abundance of proteins in ROS versus disk membrane preparations. For the known ROS proteins, the ROS:disk ratios ranged from very high (>20) for plasma membrane-specific proteins (cyclic nucleotide-gated channel, Na/Ca-K exchanger, and glyceraldehyde 3-phosphate dehydrogenase) to below 1 for proteins that preferentially reside in the lamellar region of the disks or proteins that are in both the disk and plasma membrane (rhodopsin, all-trans-retinol dehydrogenase, RGS9, and R9AP). Outer segment proteins that localize to the rim region of disks typically had ratios greater than 1 (rom-1, peripherin/rds, guanylate cyclase 1, and ABCA4) consistent with the fact that the rim regions are only partially dissociated from the ROS plasma membrane by hypotonic lysis (25). Although protein profiling provides a useful framework for defining the localization of proteins within ROSs, the data have to be evaluated in context to proteins conclusively shown to reside in outer segments. In particular, high ROS:disk ratios were also obtained for contaminating proteins in ROSs that do not co-purify with disk membrane such as Na/K-ATPase.
Rab and SNARE Proteins and Their Role in Vesicle Trafficking and Outer Segment Renewal—
A novel aspect of this study is the finding that outer segments contain a subset of SNARE and Rab proteins. These proteins are known to function in membrane trafficking and fusion in such dynamic cellular processes as neurotransmitter release, protein secretion, membrane recycling, phagocytosis, and membrane protein trafficking (47, 48). Photoreceptor membrane renewal is a highly dynamic process involving vesicle trafficking and membrane fusion within the inner segment, protein transport through the connecting cilium, formation of new disk membrane at the base of the outer segment, and eventually the removal of aged membranes at the distal end. Membrane proteins such as rhodopsin that are destined for the outer segments are synthesized in the ER of the inner segments and targeted to the base of the connecting cilium on post-Golgi vesicles, a process shown to be regulated by Rab 6 and Rab 8 proteins (50, 51, 63, 64). The post-Golgi vesicles appear to fuse with the plasma membrane of the inner segment near the connecting cilium enabling proteins to be transported through the connecting cilium to the base of the outer segment by a microtubule-based motor involving kinesin-2 (65). Our results showing the presence of SNARE proteins in the distal region of the inner segment suggests that post-Golgi vesicle trafficking and membrane fusion are mediated by SNARE proteins.
Outer segments are maintained at a constant length by two distinct processes. At the base of the outer segment new disks are formed. This process is generally thought to occur by evagination of the ciliary plasma membrane followed by joining of adjacent membranes through the outgrowth of the specialized disk rim region to generate mature, closed disks in the ROS (66). At the distal end of the outer segment, packets of aged disks are removed every 24 h through a process of shedding and phagocytosis (1). Molecular mechanisms underlying outer segment renewal are not well understood, but membrane fusion must play a central role in both disk morphogenesis and outer segment shedding. Our finding that SNARE and Rab proteins are present in photoreceptor outer segments suggests a novel mechanism by which SNARE and Rab proteins mediate membrane fusion as part of the outer segment renewal process. Syntaxin 3 and NSF are localized in membrane vesicular structures at the base of the outer segment where disk morphogenesis takes place. We propose that these proteins along with other SNARE and Rab proteins mediate membrane vesicle targeting and fusion at the base of the outer segment as part of disk morphogenesis. The absence of syntaxin 3 and NSF in the more distal part of the outer segment containing mature disks is consistent with the absence of membrane fusion in this region of the outer segment. VAMP2/3 and Munc 18 exhibit a more uniform pattern of distribution throughout the outer segment. This suggests that VAMP2/3 and Munc 18 are present in different membranes than syntaxin 3 and NSF and are not subjected to the restricted localization observed for syntaxin 3 and NSF. Studies are now in progress to more clearly define the cellular distribution of these proteins at the level of electron microscopy. While this manuscript was under review, Chuang et al. (67) also reported the localization of syntaxin 3 to vesicular structures at the base of the outer segment. Our proteomics studies are consistent with their findings and provide support for the concept that SNARE proteins play a crucial role in disk morphogenesis. Rab proteins cycle between their active GTP-bound state and inactive GDP-bound state (47). RabGDI observed throughout the outer segments may play a role in regulating the functional state of Rab proteins such that Rab proteins may be in their active state at the base of the outer segments and an inactive state over the remainder of the outer segment. The possible role of Rab proteins in regulating outer segment renewal is currently under investigation.
In summary, we identified most photoreceptor proteins known to function in phototransduction, the visual cycle, outer segment structure, and metabolic pathways and photoreceptor proteins linked to retinal degenerative diseases. In addition a large number of proteins were detected that have not been shown previously to be present in photoreceptor outer segments including Rab and SNARE proteins. The localization of SNARE proteins to the base of the outer segment provides novel mechanistic insight into their potential role in membrane vesicle trafficking and fusion events critical for outer segment renewal. Finally the data set of proteins generated in this study should serve as a valuable resource not only to further define the molecular and cellular basis of outer segment structure, function, and renewal but also for the identification and characterization of proteins linked to various inherited retinal degenerative diseases including retinitis pigmentosa and macular degeneration.
Supplementary Material
Footnotes
Published, MCP Papers in Press, January 31, 2008, DOI 10.1074/mcp.M700571-MCP200
The abbreviations used are: ROS, rod outer segment; COS, cone outer segment; RPE, retinal pigment epithelium, pAb, polyclonal antibody; mAb, monoclonal antibody, ER, endoplasmic reticulum, NSF, N-ethylmaleimide-sensitive factor; VAMP, vesicle-associated membrane protein; SNARE, soluble NSF attachment protein receptor; GDI, GDP dissociation inhibitor; ABC, ATP-binding cassette; IPI, International Protein Index; APEX, absolute protein expression measurement(s); DAPI, 4′,6-diamidino-2-phenylindole; LTQ, linear trapping quadrupole.
This work was supported, in whole or in part, by National Institutes of Health Grant EY 02422, NEI (to R. S. M.). This work was also supported by Canadian Institutes of Health Grant MOP-77688 (to L. J. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Young, R. W., and Bok, D. ( 1969) Participation of the retinal pigment epithelium in the rod outer segment renewal process. J. Cell Biol. 42, 392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young, R. W. ( 1967) The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns, M. E., and Baylor, D. A. ( 2001) Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu. Rev. Neurosci. 24, 779–805 [DOI] [PubMed] [Google Scholar]
- 4.Arshavsky, V. Y., Lamb, T. D., and Pugh, E. N., Jr. ( 2002) G proteins and phototransduction. Annu. Rev. Physiol. 64, 153–187 [DOI] [PubMed] [Google Scholar]
- 5.Lamb, T. D., and Pugh, E. N., Jr. ( 2004) Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res. 23, 307–380 [DOI] [PubMed] [Google Scholar]
- 6.Illing, M., Molday, L. L., and Molday, R. S. ( 1997) The 220-kDa rim protein of retinal rod outer segments is a member of the ABC transporter superfamily. J. Biol. Chem. 272, 10303–10310 [DOI] [PubMed] [Google Scholar]
- 7.Rattner, A., Sun, H., and Nathans, J. ( 1999) Molecular genetics of human retinal disease. Annu. Rev. Genet. 33, 89–131 [DOI] [PubMed] [Google Scholar]
- 8.Weng, J., Mata, N. L., Azarian, S. M., Tzekov, R. T., Birch, D. G., and Travis, G. H. ( 1999) Insights into the function of Rim protein in photoreceptors and etiology of Stargardt's disease from the phenotype in abcr knockout mice. Cell 98, 13–23 [DOI] [PubMed] [Google Scholar]
- 9.Sun, H., Molday, R. S., and Nathans, J. ( 1999) Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J. Biol. Chem. 274, 8269–8281 [DOI] [PubMed] [Google Scholar]
- 10.Wallimann, T., Wegmann, G., Moser, H., Huber, R., and Eppenberger, H. M. ( 1986) High content of creatine kinase in chicken retina: compartmentalized localization of creatine kinase isoenzymes in photoreceptor cells. Proc. Natl. Acad. Sci. U. S. A. 83, 3816–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, S. C., and Molday, R. S. ( 1991) Glycolytic enzymes and a GLUT-1 glucose transporter in the outer segments of rod and cone photoreceptor cells. J. Biol. Chem. 266, 21745–21752 [PubMed] [Google Scholar]
- 12.Hsu, S. C., and Molday, R. S. ( 1994) Glucose metabolism in photoreceptor outer segments. Its role in phototransduction and in NADPH-requiring reactions. J. Biol. Chem. 269, 17954–17959 [PubMed] [Google Scholar]
- 13.Molday, R. S., Hicks, D., and Molday, L. ( 1987) Peripherin. A rim-specific membrane protein of rod outer segment discs. Investig. Ophthalmol. Vis. Sci. 28, 50–61 [PubMed] [Google Scholar]
- 14.Bascom, R. A., Manara, S., Collins, L., Molday, R. S., Kalnins, V. I., and McInnes, R. R. ( 1992) Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron 8, 1171–1184 [DOI] [PubMed] [Google Scholar]
- 15.Arikawa, K., Molday, L. L., Molday, R. S., and Williams, D. S. ( 1992) Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J. Cell Biol. 116, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, Q., Lyubarsky, A., Skalet, J. H., Pugh, E. N., Jr., and Pierce, E. A. ( 2003) RP1 is required for the correct stacking of outer segment discs. Investig. Ophthalmol. Vis. Sci. 44, 4171–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poetsch, A., Molday, L. L., and Molday, R. S. ( 2001) The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 276, 48009–48016 [DOI] [PubMed] [Google Scholar]
- 18.Pacione, L. R., Szego, M. J., Ikeda, S., Nishina, P. M., and McInnes, R. R. ( 2003) Progress toward understanding the genetic and biochemical mechanisms of inherited photoreceptor degenerations. Annu. Rev. Neurosci. 26, 657–700 [DOI] [PubMed] [Google Scholar]
- 19.Sullivan, L. S., Bowne, S. J., Birch, D. G., Hughbanks-Wheaton, D., Heckenlively, J. R., Lewis, R. A., Garcia, C. A., Ruiz, R. S., Blanton, S. H., Northrup, H., Gire, A. I., Seaman, R., Duzkale, H., Spellicy, C. J., Zhu, J., Shankar, S. P., and Daiger, S. P. ( 2006) Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Investig. Ophthalmol. Vis. Sci. 47, 3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 21.Foster, L. J., de Hoog, C. L., Zhang, Y., Xie, X., Mootha, V. K., and Mann, M. ( 2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 22.Kim, T. S., Reid, D. M., and Molday, R. S. ( 1998) Structure-function relationships and localization of the Na/Ca-K exchanger in rod photoreceptors. J. Biol. Chem. 273, 16561–16567 [DOI] [PubMed] [Google Scholar]
- 23.Cook, N. J., Molday, L. L., Reid, D., Kaupp, U. B., and Molday, R. S. ( 1989) The cGMP-gated channel of bovine rod photoreceptors is localized exclusively in the plasma membrane. J. Biol. Chem. 264, 6996–6999 [PubMed] [Google Scholar]
- 24.Moritz, O. L., and Molday, R. S. ( 1996) Molecular cloning, membrane topology, and localization of bovine rom-1 in rod and cone photoreceptor cells. Investig. Ophthalmol. Vis. Sci. 37, 352–362 [PubMed] [Google Scholar]
- 25.Molday, R. S., and Molday, L. L. ( 1987) Differences in the protein composition of bovine retinal rod outer segment disk and plasma membranes isolated by a ricin-gold-dextran density perturbation method. J. Cell Biol. 105, 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster, L. J., De Hoog, C. L., and Mann, M. ( 2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U. S. A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 28.Rappsilber, J., Ishihama, Y., and Mann, M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 29.Hsu, J.-L., Huang, S.-Y., Chow, N.-H., and Chen, S.-H. ( 2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 30.Olsen, J. V., and Mann, M. ( 2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U. S. A., 101, 13417–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan, Q. W., Howes, C. G., and Foster, L. J. ( 2006) Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics 5, 2252–2262 [DOI] [PubMed] [Google Scholar]
- 32.Lu, P., Vogel, C., Wang, R., Yao, X., and Marcotte, E. M. ( 2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 33.Papermaster, D. S., and Dreyer, W. J. ( 1974) Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry. 13, 2438–2444 [DOI] [PubMed] [Google Scholar]
- 34.Brandstatter, J. H., Lohrke, S., Morgans, C. W., and Wassle, H. ( 1996) Distributions of two homologous synaptic vesicle proteins, synaptoporin and synaptophysin, in the mammalian retina. J. Comp. Neurol. 370, 1–10 [DOI] [PubMed] [Google Scholar]
- 35.Wetzel, R. K., Arystarkhova, E., and Sweadner, K. J. ( 1999) Cellular and subcellular specification of Na,K-ATPase α and β isoforms in the postnatal development of mouse retina. J. Neurosci. 19, 9878–9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh, S. Y., Shih, T. C., Yeh, C. Y., Lin, C. J., Chou, Y. Y., and Lee, Y. S. ( 2006) Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics 6, 5322–5331 [DOI] [PubMed] [Google Scholar]
- 37.Hammond, C., and Helenius, A. ( 1994) Folding of VSV G protein: sequential interaction with BiP and calnexin. Science 266, 456–458 [DOI] [PubMed] [Google Scholar]
- 38.Papermaster, D. S., Schneider, B. G., Zorn, M. A., and Kraehenbuhl, J. P. ( 1978) Immunocytochemical localization of a large intrinsic membrane protein to the incisures and margins of frog rod outer segment disks. J. Cell Biol. 78, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korschen, H. G., Illing, M., Seifert, R., Sesti, F., Williams, A., Gotzes, S., Colville, C., Muller, F., Dose, A., Godde, M., Molday, L. L., Kaupp, U. B., and Molday, R. S. ( 1995) A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15, 627–636 [DOI] [PubMed] [Google Scholar]
- 40.Molday, R. S. ( 1998) Photoreceptor membrane proteins, phototransduction, and retinal degenerative diseases. The Friedenwald Lecture. Investig. Ophthalmol. Vis. Sci. 39, 2491–2513 [PubMed] [Google Scholar]
- 41.Kuhn, H. ( 1980) Light- and GTP-regulated interaction of GTPase and other proteins with bovine photoreceptor membranes. Nature 283, 587–589 [DOI] [PubMed] [Google Scholar]
- 42.Cargile, B. J., Bundy, J. L., and Stephenson, J. L., Jr. ( 2004) Potential for false positive identifications from large databases through tandem mass spectrometry. J. Proteome Res. 3, 1082–1085 [DOI] [PubMed] [Google Scholar]
- 43.Bunt-Milam, A. H., and Saari, J. C. ( 1983) Immunocytochemical localization of two retinoid-binding proteins in vertebrate retina. J. Cell Biol. 97, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamel, C. P., Tsilou, E., Pfeffer, B. A., Hooks, J. J., Detrick, B., and Redmond, T. M. ( 1993) Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268, 15751–15757 [PubMed] [Google Scholar]
- 45.Hsu, S. C., and Molday, R. S. ( 1990) Glyceraldehyde-3-phosphate dehydrogenase is a major protein associated with the plasma membrane of retinal photoreceptor outer segments. J. Biol. Chem. 265, 13308–13313 [PubMed] [Google Scholar]
- 46.Maw, M. A., Corbeil, D., Koch, J., Hellwig, A., Wilson-Wheeler, J. C., Bridges, R. J., Kumaramanickavel, G., John, S., Nancarrow, D., Roper, K., Weigmann, A., Huttner, W. B., and Denton, M. J. ( 2000) A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum. Mol. Genet. 9, 27–34 [DOI] [PubMed] [Google Scholar]
- 47.Grosshans, B. L., Ortiz, D., and Novick, P. ( 2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103, 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jahn, R., and Scheller, R. H. ( 2006) SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell. Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 49.Von Kriegstein, K., Schmitz, F., Link, E., and Sudhof, T. C. ( 1999) Distribution of synaptic vesicle proteins in the mammalian retina identifies obligatory and facultative components of ribbon synapses. Eur. J. Neurosci. 11, 1335–1348 [DOI] [PubMed] [Google Scholar]
- 50.Deretic, D. ( 1997) Rab proteins and post-Golgi trafficking of rhodopsin in photoreceptor cells. Electrophoresis 18, 2537–2541 [DOI] [PubMed] [Google Scholar]
- 51.Moritz, O. L., Tam, B. M., Hurd, L. L., Peranen, J., Deretic, D., and Papermaster, D. S. ( 2001) Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol. Biol. Cell 12, 2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider, B. G., and Kraig, E. ( 1990) Na+,K+-ATPase of the photoreceptor: selective expression of α3 and β2 isoforms. Exp. Eye Res. 51, 553–564 [DOI] [PubMed] [Google Scholar]
- 53.Molday, L. L., Wu, W. W., and Molday, R. S. ( 2007) Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J. Biol. Chem. 282, 32792–32801 [DOI] [PubMed] [Google Scholar]
- 54.Krizaj, D., Demarco, S. J., Johnson, J., Strehler, E. E., and Copenhagen, D. R. ( 2002) Cell-specific expression of plasma membrane calcium ATPase isoforms in retinal neurons. J. Comp. Neurol. 451, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitamoto, J., Libby, R. T., Gibbs, D., Steel, K. P., and Williams, D. S. ( 2005) Myosin VI is required for normal retinal function. Exp. Eye Res. 81, 116–120 [DOI] [PubMed] [Google Scholar]
- 56.Liu, Q., Tan, G., Levenkova, N., Li, T., Pugh, E. N., Jr., Rux, J. J., Speicher, D. W., and Pierce, E. A. ( 2007) The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 6, 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pugh, E. N., and Lamb, T. D. ( 2000) Phototransduction in vertebrate rods and cones: molecular mechanisms of amplification, recovery, and light adaptation, in Handbook of Biological Physics (Stavenga, D. G., de Grip, W. J., and Pugh, E. N., eds) Vol. 3, pp. 183–255, Elsevier, Amsterdam
- 58.Loewen, C. J., and Molday, R. S. ( 2000) Disulfide-mediated oligomerization of Peripherin/Rds and Rom-1 in photoreceptor disk membranes. Implications for photoreceptor outer segment morphogenesis and degeneration. J. Biol. Chem. 275, 5370–5378 [DOI] [PubMed] [Google Scholar]
- 59.Goldberg, A. F., and Molday, R. S. ( 1996) Subunit composition of the peripherin/rds-rom-1 disk rim complex from rod photoreceptors: hydrodynamic evidence for a tetrameric quaternary structure. Biochemistry 35, 6144–6149 [DOI] [PubMed] [Google Scholar]
- 60.Zhong, H., Molday, L. L., Molday, R. S., and Yau, K. W. ( 2002) The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 420, 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weitz, D., Ficek, N., Kremmer, E., Bauer, P. J., and Kaupp, U. B. ( 2002) Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 36, 881–889 [DOI] [PubMed] [Google Scholar]
- 62.Colville, C. A., and Molday, R. S. ( 1996) Primary structure and expression of the human β-subunit and related proteins of the rod photoreceptor cGMP-gated channel. J. Biol. Chem. 271, 32968–32974 [DOI] [PubMed] [Google Scholar]
- 63.Deretic, D., Huber, L. A., Ransom, N., Mancini, M., Simons, K., and Papermaster, D. S. ( 1995) rab8 in retinal photoreceptors may participate in rhodopsin transport and in rod outer segment disk morphogenesis. J. Cell Sci. 108, 215–224 [DOI] [PubMed] [Google Scholar]
- 64.Deretic, D., and Papermaster, D. S. ( 1993) Rab6 is associated with a compartment that transports rhodopsin from the trans-Golgi to the site of rod outer segment disk formation in frog retinal photoreceptors. J. Cell Sci. 106, 803–813 [DOI] [PubMed] [Google Scholar]
- 65.Marszalek, J. R., Liu, X., Roberts, E. A., Chui, D., Marth, J. D., Williams, D. S., and Goldstein, L. S. ( 2000) Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell 102, 175–187 [DOI] [PubMed] [Google Scholar]
- 66.Steinberg, R. H., Fisher, S. K., and Anderson, D. H. ( 1980) Disc morphogenesis in vertebrate photoreceptors. J. Comp. Neurol. 190, 501–508 [DOI] [PubMed] [Google Scholar]
- 67.Chuang, J. Z., Zhao, Y., and Sung, C. H. ( 2007) SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell 130, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.