Abstract
Auxin regulates a host of plant developmental and physiological processes, including embryogenesis, vascular differentiation, organogenesis, tropic growth, and root and shoot architecture. Genetic and biochemical studies carried out over the past decade have revealed that much of this regulation involves the SCFTIR1/AFB-mediated proteolysis of the Aux/IAA family of transcriptional regulators. With the recent finding that the TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) proteins also function as auxin receptors, a potentially complete, and surprisingly simple, signaling pathway from perception to transcriptional response is now before us. However, understanding how this seemingly simple pathway controls the myriad of specific auxin responses remains a daunting challenge, and compelling evidence exists for SCFTIR1/AFB-independent auxin signaling pathways.
Introduction
In the plant life cycle, few developmental processes occur without the involvement of the phytohormone auxin. Initially described by Charles and Francis Darwin as a mobile substance that mediates phototropism in grass coleoptiles, auxin was chemically identified in the 1930s as indole-3-acetic acid [1]. Subsequent experiments involving the application of auxin to plant tissues implicated this hormone in a plethora of physiological and developmental processes. In the 1980s, it was recognized that auxin regulates the transcription of many genes, thereby linking the auxin signal to changes in gene expression [2•]. However, the nature of the molecular mechanisms of auxin perception and signal transduction underlying this regulation remained a mystery. In this review, we focus on these central questions and the impressive progress that has been made toward illuminating this fundamental area of plant biology.
Auxin-responsive gene expression
Early molecular approaches to elucidate auxin action identified several genes that exhibit a rapid and specific increase in expression in response to exogenous auxin. Among the most thoroughly characterized are three gene families (SAURs [Small Auxin-Up RNAs], GH3s and Aux/IAAs), which are induced within minutes of auxin application. The promoter regions of several auxin-responsive genes contain one or more auxin-responsive elements (AuxRE), which are capable of conferring auxin-regulated gene expression to reporter constructs [3]. The identification of the AuxRE sequence led to the isolation of the Arabidopsis AUXIN RESPONSE FACTOR 1 (ARF1) gene [4], and subsequent genetic, genomic, and molecular studies have identified 23 ARF genes in Arabidopsis [5]. In addition to a conserved amino-terminal (N-terminal) domain that mediates AuxRE binding [6], most ARF transcription factors also contain carboxyl-terminal (C-terminal) dimerization elements (domains III and IV). In between is a variable middle region (MR) that generally is either glutamine (Q)-rich or serine (S)-rich. This difference is apparently a major determinant of ARF function, with S-rich ARFs acting as transcriptional repressors and Q-rich ARFs as transcriptional activators in protoplast transfection assays [6].
Genetic studies have implicated various ARFs in distinct developmental processes, including embryogenesis (ARF5/MONOPTEROS [MP] [7] and ARF17 [8•]), root development (ARF7, ARF19 [9•], ARF10 and ARF16 [8•,10•]), flower development (ARF2 [11], ARF3/ETTIN [ETT] [12], ARF6 and ARF8 [13,14]), and senescence (ARF1 and ARF2 [13]). Owing to some functional redundancy, several arf mutants lack obvious phenotypes, but exacerbated developmental defects are seen in several double mutant combinations [9•,14].
Although the transcription of most ARF genes is not auxin-regulated, hormonal control of at least the Q-rich ARFs is achieved through their interactions with Aux/IAA proteins (Figure 1a). The 29 members of the Arabidopsis Aux/IAA family of auxin-inducible genes encode nuclear proteins, most of which share four conserved domains [5], including the C-terminal domains III and IV that are found in most ARF proteins. These domains mediate Aux/IAA-ARF heterodimerization [15], which has been shown to repress transcriptional activation by Q-rich ARFs in protoplast assays [6]. Aux/IAA domain I has recently been shown to mediate this transcriptional repression, although the mechanistic details remain to be elucidated [16].
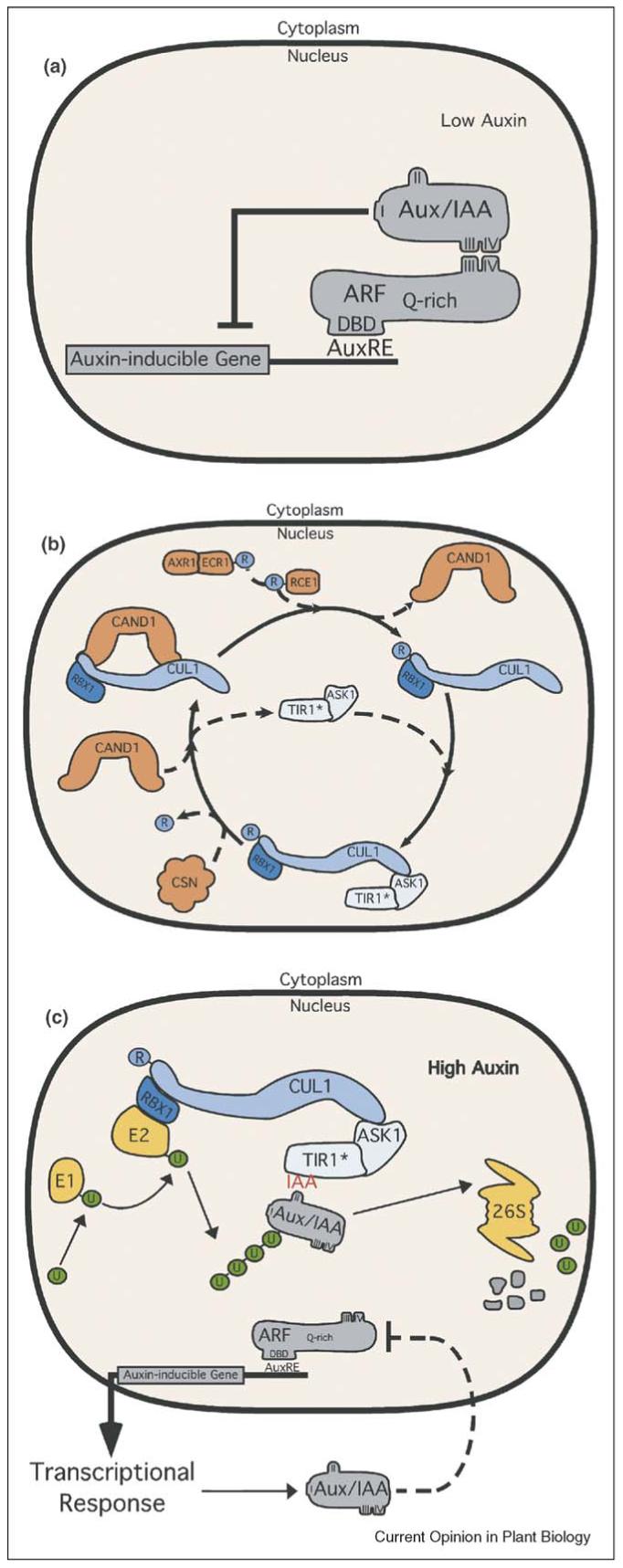
Auxin regulation of gene expression. (a) Under sub-threshold auxin concentrations the Aux/IAA proteins heterodimerize with the ARF transcription factors, thereby repressing auxin-inducible gene expression. (b) The active SCFTIR1 complex, containing RUB-modified CUL1, is shown at the bottom. The CSN complex can cleave the RUB modifier from CUL1, thus facilitating CAND1 binding to CUL1 and SCF disassembly. Conjugation of RUB to CUL1 by the AXR1-ECR1 and RCE1 enzymes might free CUL1 from CAND1, promoting re-assembly of the active complex. Genetic studies have shown that all of the components depicted in this figure are required for optimal SCFTIR1 activity in vivo. (c) Auxin binding to the TIR1/AFB receptors promotes the recruitment of Aux/IAA proteins to the SCF complex. Subsequent Aux/IAA ubiquitinylation and proteasome-mediated degradation results in a decline in Aux/IAA protein levels, thus de-repressing auxin-inducible gene expression. In addition to genes responsible for specific auxin responses, the Aux/IAA genes themselves are auxin-inducible. This might represent a negative feedback loop that ensures a transient response, with the nascent Aux/IAA proteins attenuating the signaling pathway as auxin levels fall by restoring repression of the ARF transcription factors. DBD, DNA-binding domain; E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; U, ubiquitin; R, RUB. *,AFB1, AFB2, or AFB3.
A number of loss-of-function mutations in Aux/IAA genes have been characterized but they confer no apparent mutant phenotypes, suggesting genetic redundancy within this large gene family [17•]. Most of the insight into the biological function of Aux/IAA proteins gathered to date comes from the characterization of dominant, gain-of-function mutations [2•] in several Aux/IAA genes that confer dramatic phenotypes characteristic of auxin signaling defects. Each of these mutations alters the highly conserved core of domain II. Most Aux/IAAs are extremely short-lived proteins, whose half-lives decrease further in the presence of auxin [18]. The 13-amino-acid domain II sequence functions as a degron that targets the Aux/IAA for ubiquitin-mediated proteolysis [19,20], and these dominant mutations disrupt this regulation, thereby stabilizing the affected protein. Recent findings suggest that sequences outside of domain II also contribute to Aux/IAA proteolysis, and it is interesting to note that degradation rates vary considerably among different Aux/IAA family members [21•].
Although genetic studies suggest considerable functional redundancy within the ARF and Aux/IAA gene families [9•,17•], the fact that several arf mutants exhibit specific auxin-related defects clearly indicates some functional specialization. Likewise, the phenotypes of the different gain-of-function Aux/IAA mutants that have been characterized vary considerably. Recent results suggest that auxin signals are converted into specific responses by matching pairs of co-expressed ARF and Aux/IAA proteins. For example, gain-of-function mutations in the Aux/IAA gene IAA12/BODENLOS [BDL] confer a dramatic embryo- or seedling-lethal phenotype similar to those associated with loss-of-function mutations in arf5/mp [22]. These findings suggest an antagonistic relationship between IAA12/BDL and ARF5/MP, and indeed, IAA12 was found to interact with ARF5 and to negatively regulate its activity [23]. Although yeast two-hybrid studies have not revealed substantial specificity in the interactions between individual ARF and Aux/IAA proteins, the above findings suggest that such specificity might exist in planta. The spatiotemporal transcriptional control of Aux/IAA gene expression is also a key determinant in the regulation of specific ARFs: promoter-swap experiments have suggested that IAA12/BDL and IAA3/SHORT HYPOCOTYL 2 [SHY2] are largely functionally interchangeable [24••]. The precise control of expression pattern might explain why several ARF mRNAs are under the additional control of microRNA (miRNA)- and trans-acting small interfering RNAs (tasiRNA)-mediated degradation [8•,10•,25].
Proteolysis and auxin signaling
The genetic identification of Arabidopsis auxin-resistant mutants has provided crucial insight into the molecular mechanisms underlying auxin signaling. Molecular and biochemical studies of the gene products that are affected by these mutations have positioned the SCFTIR1 ubiquitin-ligase complex as a central regulator of auxin signaling. E3 ubiquitin ligases catalyze the conjugation of ubiquitin to substrate proteins. Once ubiquitinylated, these substrates are targeted to the 26S proteasome for degradation. SCF complexes are the largest family of ubiquitin ligases in plants. They are composed of Skp1, cullin, an F-box protein, and the small RING protein Rbx1 (for RING-BOX 1). The cullin subunit acts as a scaffold, binding Skp1 at its N-terminus and Rbx1 at its C-terminus. Skp1 binds the F-box domain of the F-box protein, which functions as a substrate-specific adaptor, recruiting specific proteins to the complex for ubiquitinylation [26,27].
Mutations in the TRANSPORT INHIBITOR RESPONSE1 (TIR1) gene confer reduced auxin response [28]. TIR1 encodes a nuclear protein belonging to the F-box protein family of Arabidopsis, which has approximately 700 members. TIR1 interacts with the core SCF subunits [29,30], and additional support for SCF-mediated regulation of auxin signaling was rapidly obtained through genetic studies of CULLIN 1 (CUL1), ARABIDOPSIS SKP1-LIKE (ASK1) and RBX1 [29-32]. These findings established the SCFTIR1 complex as a positive regulator of auxin response and suggested a model invoking the SCFTIR1-mediated ubiquitinylation of a repressor of auxin signaling. Given the short-lived nature of Aux/IAA proteins, together with the results of molecular and genetic studies implicating these proteins as negative regulators of auxin response, Aux/IAA proteins are attractive candidates for substrates of the SCFTIR1 complex. Indeed, Aux/IAA stability is increased in mutants that have defects in SCFTIR1 activity, and biochemical studies have confirmed that Aux/IAA proteins interact with TIR1 [20]. Furthermore, this interaction requires the Aux/IAA domain II degron and is rapidly promoted by auxin in a concentration-dependent manner.
The study of several additional auxin-resistant mutants has provided insight into the regulation of SCFTIR1 activity. Proper SCFTIR1 function requires the covalent modification of the CUL1 subunit by conjugation of the RELATED TO UBIQUITIN protein (RUB, known as NEDD8 in mammals). RUB conjugation requires RUB-specific E1 (AUXIN RESISTANT 1 [AXR1]-E1 C-TERMINAL RELATED1 [ECR1]) and E2 enzymes (RUB1 CONJUGATING ENZYME1 [RCE1]), as well as RBX1, which appears to function as a RUB E3 ligase. Defects in any of these enzymes severely impair SCFTIR1 activity, conferring a dramatic reduction in auxin response [20,30,33,34].
RUB modification of CUL1 is a dynamic process. The SCF interacts with the COP9 SIGNALOSOME (CSN) [35], which possesses a RUB-isopeptidase activity that cleaves the RUB modifier off of CUL1 [26]. Surprisingly, plants that have impaired CSN function exhibit diminished auxin response and increased Aux/IAA stability. Thus, like RUB conjugation, RUB cleavage off of CUL1 is also required for optimal SCFTIR1 activity [35]. While the molecular function of RUB modification remains unclear, an interesting avenue has emerged with the identification of CAND1 (CULLIN-ASSOCIATED and NEDD8-DISSOCIATED1). CAND1 specifically interacts with unmodified cullin, and can be dissociated by the RUB modification of CUL1 in vitro [26]. Furthermore, CAND1 and SKP1 binding to CUL1 are mutually exclusive, suggesting a model whereby CAND1 sequesters a fraction of the CUL1 pool and thus negatively regulates ubiquitin-ligase activity by preventing SCF assembly [26]. Genetic studies of Arabidopsis cand1 mutants, however, suggest a more complicated scenario. Rather than exhibiting increased SCFTIR1 activity as might be expected, cand1 mutants are defective in Aux/IAA proteolysis and display diminished auxin-response phenotypes similar to those of SCF mutants [36-38]. To reconcile these observations, it has been suggested that CAND1 is required to sustain SCF activity in vivo by promoting cycles of SCF assembly and disassembly ([26]; Figure 1b). Such a cycle might be essential in a cellular context in which many F-box proteins compete for access to the common core SCF subunits. The cyclical conjugation and cleavage of the RUB modifier has been proposed to drive this cycle. The finding that auxin response in cand1 mutants is still highly dependent on the RUB conjugation pathway is, however, difficult to reconcile with this model, and at least suggests that this is not the primary means by which RUB regulates SCF function [37].
Auxin receptors
Amid an ever-clearer picture of how auxin triggers changes in gene expression, the auxin receptor remained a conspicuous omission from models of auxin action. Early biochemical efforts to isolate auxin receptors identified AUXIN-BINDING PROTEIN1 (ABP1) [39]. The majority of ABP1 protein localizes to the endoplasmic reticulum, where the pH is too high for auxin binding. However, some ABP1 is also found on the plasma membrane, and ABP1 antibody experiments have implicated this pool in auxin-mediated cell expansion. Overexpression studies also support a role for ABP1 in cell expansion but no evidence has been obtained to link ABP1 to auxin-regulated transcriptional changes, which are clearly responsible for many auxin responses.
An important breakthrough in the auxin receptor hunt came when auxin action was demonstrated in a cell-free system [40]. Specifically, the addition of auxin to crude plant extracts was shown to promote the interaction between Aux/IAA proteins and the SCFTIR1 complex, indicating the presence of a soluble auxin receptor. SCF-substrate interactions frequently require the post-translational modification of the substrate [26]. This did not appear to be the case in this instance, however, because Aux/IAA domain II, which was previously shown to be sufficient for auxin-induced TIR1 binding, is not modified when incubated with auxin-supplemented extracts [41•].
Finally, two recent reports revealed the remarkable simplicity of the TIR1 pathway by showing that radiolabeled auxin that was added to crude extracts co-purified with the SCFTIR1-Aux/IAA complex [42••,43••]. This binding activity is saturable, specific for active auxins, and possesses the affinity expected of an auxin receptor. Although this evidence alone suggests only that a receptor is associated with SCFTIR1, the finding that TIR1 that is expressed in animal cells also interacts with Aux/IAAs (or a domain II peptide) in an auxin-dependent manner strongly suggests that TIR1 is an auxin receptor. To date, it has not been possible to separate the auxin binding and Aux/IAA binding activities of TIR1, raising the possibility that auxin might coordinate the complex, with both TIR1 and domain II contacting the hormone.
The fact that tir1 null mutants exhibit a relatively weak auxin response defect suggested that TIR1 might not be the only auxin receptor. Indeed, three additional AUXIN SIGNALING F-BOX proteins, AFB1-AFB3, are highly related to TIR1 and were recently shown to exhibit auxin-dependent binding to Aux/IAA proteins [44••]. Correspondingly, the introduction of afb mutations into tir1 plants results in a progressive reduction in auxin response, culminating with tir1 afb1 afb2 afb3 quadruple mutants exhibiting a seedling lethal phenotype similar to that of mp and bdl mutants. Curiously, a fraction of these quadruple mutants do survive and are able to complete development, suggesting that auxin signaling is not completely abolished in these mutants.
Conclusions
The discovery that the TIR1/AFB F-box proteins function as auxin receptors is a surprising development that fills in a crucial piece of the auxin puzzle and might well serve as a new paradigm for the regulation of SCF-mediated ubiquitinylation by small ligands. Many details remain to be established, including a definition of the auxin-binding site and a comparison of the binding affinities of the receptors, but we can now follow the auxin signal from perception through to changes in gene expression (Figures 1a,c). But can this apparently simple signaling pathway account for the diversity of specific auxin responses? The presence of a small receptor family suggests the potential for functional specialization. However, the relatively ubiquitous expression patterns of the TIR1/AFB genes, their biochemical interactions with common Aux/IAA proteins and the results of genetic studies all suggest that the receptors act in a largely redundant fashion [44••]. This would again suggest that the control of specific auxin responses might largely occur downstream in the web of potential Aux/IAA-ARF interactions. The recent finding that Aux/IAA degradation kinetics vary considerably among different family members is an exciting development [21•], particularly if future structural studies reveal that Aux/IAA proteins contribute to auxin binding. If so, one intriguing possibility is that these stability differences reflect varying affinities for IAA, which could facilitate Aux/IAA proteins serving as sensors of auxin concentration.
The SCFTIR1/AFB-mediated proteolysis of Aux/IAA proteins is clearly responsible for many of the effects of auxin, but the resulting changes in gene expression occur too slowly to account for the most rapid auxin responses, including ion fluxes across the plasma membrane [45], mitogen-activated protein (MAP) kinase activation [46], and vesicle trafficking [47••]. The success in elucidating the SCFTIR1/AFB pathway has resulted in other potential signaling mechanisms receiving significantly less attention than they probably deserve. There is evidence that implicates a cell-surface auxin receptor in auxin-mediated cell expansion [48]. This receptor might very likely be ABP1 [49], but the mechanism of ABP1 action and the identities of other components of this pathway await discovery. Furthermore, several factors, including the repressing ARFs, SAUR proteins, MAP kinase pathway components, and the INDOLE-3-BUTYRIC ACID-RESPONSE5 (IBR5) [50] dual-specificity phosphatase, have been implicated in auxin signaling but are presently without a home in current models. Clearly, much remains to be learned about the mechanics of auxin signaling.
Acknowledgements
We thank P Overvoorde for helpful comments on the manuscript and apologize to colleagues whose research could not be included in the available space. Research in the authors’ lab is supported by a US National Institutes of Health grant (GM067203 to WMG), and a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (to MQ).
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Went FW, Thimann KV. Phytohormones. Macmillan; New York: 1937. [Google Scholar]
- 2•.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An excellent detailed overview of auxin biology.
- 3.Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 5.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- 6.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis auxin response factor17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The expression of three ARF genes is shown to be regulated by the miR160 microRNA. Expression of a miR160-resistant form of ARF17 conferred dramatic auxin-related defects, demonstrating the importance of this regulation.
- 9•.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. Functional genomic analysis of the auxin response factor gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This paper describes the identification and characterization of loss-of-function mutations in 18 Arabidopsis ARF genes, revealing unique and redundant functions of different ARF proteins.
- 10•.Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors find that miR160 plays an important role in regulating ARF10 and ARF16 expression during root cap development.
- 11.Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 12.Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 13.Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 14.Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. Functional genomic analysis of the auxin/indole-3-acetic acid gene family members in Arabidopsis thaliana. Plant Cell. 2005;17:3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Characterization of twelve Arabidopsis Aux/IAA loss-of-function mutants and some double and triple mutant combinations reveals extensive functional redundancy in this gene family.
- 18.Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of aux/IAA proteins. Proc Natl Acad Sci USA. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 21•.Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis AUX/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors show that regions outside the domain II degron affect Aux/IAA stability and demonstrate distinct degradation kinetics for different Aux/IAA family members.
- 22.Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 24••.Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. Developmental specificity of auxin response by pairs of ARF and aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aux/IAA and ARF specificities are examined in planta using a promoter-swap strategy. The authors find that both expression patterns and optimized pairs of interacting ARF and Aux/IAA proteins contribute to the specificity of response.
- 25.Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petroski MD, Deshaies RJ. Function and regulation of Cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 27.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F box-Skp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 28.Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray WM, Hellmann H, Dharmasiri S, Estelle M. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell. 2002;14:2137–2144. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, del Pozo C, Reinhardt D, Estelle M. Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 2003;22:3314–3325. doi: 10.1093/emboj/cdg335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quint M, Ito H, Zhang W, Gray WM. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 2005;43:371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Pozo JC, Estelle M. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA. 1999;96:15342–15347. doi: 10.1073/pnas.96.26.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M. The RUB/ Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 2003;22:1762–1770. doi: 10.1093/emboj/cdg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Dai X, Zhao Y. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 2004;135:1020–1026. doi: 10.1104/pp.104.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuang HW, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCFTIR1 ubiquitin ligase. Plant Cell. 2004;16:1883–1897. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell. 2004;16:1870–1882. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Napier RM, David KM, Perrot-Rechenmann C. A short history of auxin-binding proteins. Plant Mol Biol. 2002;49:339–348. [PubMed] [Google Scholar]
- 40.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13:1418–1422. doi: 10.1016/s0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- 41•.Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors demonstrate that the auxin-dependent TIR1-Aux/IAA interaction probably doesnotrequire post-translational modificationof the Aux/IAA protein. This strongly suggests that this interaction is regulated very differently than previously characterized F-box protein-substrate interactions.
- 42••.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Radiolabeled IAA co-purifies with SCFTIR1 and TIR1 that is expressed in animal cells interacts with Aux/IAA proteins in an auxin-dependent manner, strongly suggesting that TIR1 is an auxin receptor.
- 43••.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- See annotation to [42••].
- 44••.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- This paper demonstrates that the three additional members of the TIR1 family, AFB1-AFB3, also function as auxin receptors and collectively mediate auxin responses throughout plant development. Quadruple tir1 afb1 afb2 afb3 mutants are auxin-insensitive, exhibiting a bdl-like phenotype, although a considerable fraction reaches the adult stage. This suggests either that an unknown auxin signaling pathway that is independent of TIR1/AFB is still intact in the quadruple mutant or that Aux/IAA proteins such as BDL can be degraded by an alternative pathway.
- 45.Hager A. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res. 2003;116:483–505. doi: 10.1007/s10265-003-0110-x. [DOI] [PubMed] [Google Scholar]
- 46.Mockaitis K, Howell SH. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 2000;24:785–796. doi: 10.1046/j.1365-313x.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- 47••.Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- Auxin is shown to inhibit the endocytosis of plasma membrane proteins. This previously undescribed rapid auxin response provides a feedback mechanism for regulating auxin transport.
- 48.Napier RM, David KM, Perrot-Rechenmann C. A short history of auxin-binding proteins. Plant Mol Biol. 2002;49:339–348. [PubMed] [Google Scholar]
- 49.Steffens B, Feckler C, Palme K, Christian M, Bottger M, Luthen H. The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J. 2001;27:591–599. doi: 10.1046/j.1365-313x.2001.01103.x. [DOI] [PubMed] [Google Scholar]
- 50.Monroe-Augustus M, Zolman BK, Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15:2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]


