Abstract
Background
A novel non-invasive imaging technique, the heart-model-based 3-dimensional cardiac electrical imaging (3DCEI) approach was previously developed and validated to estimate the initiation site (IS) of cardiac activity and the activation sequence (AS) from body surface potential maps (BSPMs) in a rabbit model. The aim of the present study was to validate the 3DCEI in an intact large mammalian model (swine) during acute ventricular pacing.
Methods and results
The heart-torso geometries were constructed from pre-operative MR images acquired from each animal. Body surface potential mapping and intracavitary non-contact mapping (NCM) were performed simultaneously during pacing from both right ventricular (intramural) and left ventricular sites (endocardial). Subsequent 3DCEI analyses were performed from the measured BSPMs. The estimated ISs were compared to the precise pacing locations and estimated ASs were compared to those recorded by the NCM system. In total, 5 RV and 5 LV sites from control and heart failure animals were paced and sequences of 100 paced beats were analyzed (10 for each site). The averaged localization error of the RV and LV sites were 7.3 ± 1.8 mm (n=50) and 7.0 ± 2.2 mm (n=50), respectively. The global 3-dimensional activation sequences throughout the ventricular myocardium were also derived. The endocardial ASs as a subset of the estimated 3-dimensional ASs were consistent with those reconstructed from the NCM system.
Conclusion
The present experimental results demonstrate that the noninvasive 3DCEI approach can localize the IS and estimate AS with good accuracy in an in vivo setting; under control, paced and/or diseased conditions.
Keywords: 3D Cardiac Electrical Imaging, Pacing, Electrocardiophysiology, body surface potential mapping, Noncontact mapping
Introduction
Radiofrequency catheter ablation has become commonly employed strategies for the treatment of various types of cardiac arrhythmias1. Recently, advanced cardiac mapping systems have been developed to guide such ablation procedures2–4. In general, these invasive mapping methods often require considerable procedural and fluoroscopic time in the electrophysiology (EP) laboratory5. Furthermore, a successful ablation may become compromised by a prolonged procedure due to underlying patient’s hemodynamic instability and/or the inability to simultaneously acquire maps due to aberrant heart rhythms. As such, major research efforts have been put forth towards developing clinically applicable, noninvasive mapping techniques of the arrhythmic activities by analyzing the thoracic electrocardiogram (ECG) recordings. Specifically, by solving the so called “inverse problem”, the various noninvasive cardiac imaging techniques allow for the ability to estimate and localize the arrhythmogenic substrate before the intervention. The heart surface (epicardium and/or endocardium) mapping technique has been developed to noninvasively estimate the surface potentials and/or activation sequences by analyzing the body surface potential maps (BSPMs)6–11, and the validation on human subjects has been recently reported12–13. Although the intramural myocardial activation could be inferred from the information on the heart surface, the direct three-dimensional (3D) imaging of the intramural cardiac activation is highly expected so to provide important patho-anatomic information: e.g., ventricular arrhythmias may arise from transmural regions within the myocardial tissue.
Recently, He and co-workers proposed and developed the 3D cardiac electrical imaging (3DCEI) approach for noninvasively imaging of 3D cardiac electrical activity employing BSPMs14–20. More recently, this 3DCEI approach was validated using 3D intracardiac mapping in a rabbit model21–22. In the present study, we aimed to validate 3DCEI by using swine, whose cardiac size, heart-torso geometry and electrophysiological characteristics were chosen to best approximate those of an adult human. In addition to the control animal investigations, a chronically induced heart failure (HF) model was also used to test the performance of 3DCEI under diseased conditions.
Methods
Control and Chronic Heart Failure Swine Models
Control swine (n=2) and a single swine with chronically induced heart failure (HF) were employed in these investigations. The induced HF was achieved by high-rate pacing (VVI mode, 200 beats/min) for four week period. The surgical preparation of these animals for hemodynamic and electrical monitoring has been previously reported23. During the comparative mapping studies, each animal was anesthetized with a fentanyl infused at 0.75 mcg/kg/min; all were intubated and mechanically ventilated with 65% air and 35% O2 to maintain a PaCO2 of 40 ± 2 mmHg.
Active-fixation pacing leads were implanted in both the right ventricular apex (RVA) and the RV septum (RVS) (Model 3830, Medtronic, Inc., USA); intramural pacing. Endocardial surface pacing from either the left ventricular (LV) apex (LVA), LV lateral wall (LVL), or LV anterior (LVAn) sites was accomplished by a quadripolar EP catheter (MarinR, Medtronic, Inc.). The 3D locations of the endocardial pacing sites were recorded on the reconstructed LV geometry acquired with the NCM system.
For each animal, the pre-operative magnetic resonance imaging (MRI; ECG gated to end diastole) was acquired approximately 5–7 days before the in vivo mapping experiment so to obtain the needed anatomical geometry information. On the mapping study day, body surface potential mapping and intracavitary mapping (by the EnSite® NCM system) were simultaneously performed during the various pacing protocols. For the body surface potential mapping, up to 100 disposable electrodes were placed on the anterolateral chest (see Figure 1). The body surface electrodes were referenced to the Wilson central terminal. The 3D locations of the body surface electrodes and the fiducial points were digitized using a radio frequency localizer (Fastrak, Polhemus Inc.). After the completion of the data collection protocol, each heart was removed and fixed in formalin with the intramural pacing leads remaining in place. These isolated hearts were again MRI scanned to record the precise locations of the RV pacing sites.
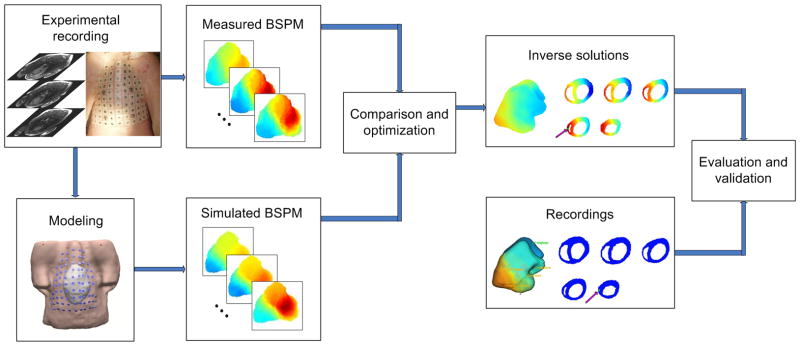
Figure 1.
The schematic diagram of the noninvasive three-dimensional electrical imaging approach.
Principles of the 3D cardiac electrical imaging
The heart-model-based 3DCEI methodology was previously described in detail15,17,24. A schematic diagram of 3DCEI is shown in Figure 1. In brief, for each animal, a heart-excitation model and the heart-torso volume conductor model were constructed based on the pre-operative MRI scans and the prior known physiological knowledge, specifically, of the swine heart. The MR images were segmented to obtain the detailed cardiac geometry and the cellular-automaton heart model. The entire heart excitation process could be simulated and the corresponding BSPMs were calculated by using the boundary element method. A preliminary classification system was employed to initialize the parameters of the heart-excitation model, and then the model parameters were iteratively adjusted in an attempt to minimize the dissimilarity between the measured and the heart-model-generated BSPMs until the convergent criteria were satisfied. In this study, for each pacing site, both the 3D location of the initiation site for electrical activation and the corresponding activation sequence throughout the ventricles were noninvasively estimated by the above procedure.
Evaluation of the 3DCEI solutions
The relative performance of this 3DCEI approach was further validated by evaluating the localization errors of the initiation sites of activation and comparing the estimated activation sequences with that reconstructed by the endocardial NCM system.
The locations of the initiation sites of activation were inversely estimated in the heart-excitation models, which were obtained from the pre-operative MR images. The precise locations of the pacing sites in RV were obtained by locating the distal end of the pacing leads in the post-operative MR images of the isolated hearts; note that each heart was fixed in an end diastolic state. For the purposes of determining the precise 3D locations of the initiation sites in the resultant heart-excitation models, an image registration procedure between the isolated, fixed hearts and the beating hearts were performed. The epicardial surfaces of the fixed heart and the beating heart were extracted from the postoperative MR images and the pre-operative MR images, respectively, and then were registered by applying a multidimensional registration approach based on the Euclidean distance transform and the Marquardt-Levenberg optimization algorithm25. The primary principle of this approach was to minimize the averaged distance between the two sets of special points. After the registration, the precise locations of the initial activation sites in RV were determined in the heart-excitation model and could be directly compared with the estimated locations.
Each heart was also paced at endocardial locations within the LV with an EP catheter. The endocardial geometries of the LV and the precise locations of the endocardial pacing sites were recorded by the NCM system. In order to determine the precise locations of the pacing sites reflected in the resultant heart-excitation models, the endocardial surface maps were extracted from the NCM recordings and then registered with the corresponding surface of the beating heart segmented from the pre-operative MR images. The registration approach used here was identical to that used on the epicardial surface registration described above. We defined the precise locations of the endocardial pacing sites within each heart-excitation model as the closest point on the endocardial surface of the beating heart to the corresponding pacing sites on the endocardial surface recorded by the NCM system.
In addition to the localization of the initiation sites of activation, the ability of the 3DCEI approach to estimate global, 3D activation sequences was also evaluated. The NCM system recorded the endocardial activation sequences of the LV, while the 3D activation sequences throughout the myocardium were noninvasively estimated by the 3DCEI approach. The endocardial activation sequences were extracted from the 3D solutions, and then projected from the endocardial surfaces of the heart models to the endocardial surface maps recorded by the NCM system; so that we could compare our results with the output of the NCM system based on the same geometrical surfaces.
Results
Experimentation and modeling
In these in vivo studies, the heart-model-based 3DCEI analyses were conducted on sets of data obtained from three swine (healthy controls, n=2, and another with chronically induced HF). The computational heart-torso geometries were constructed for each animal basis of their prior obtained individual MR images. Each model contained over 100,000 cellular units with the spatial resolution of 1.5 mm. Due to the large number of cellular units of these heart models, each heart was further sub-divided into 500–650 myocardial segments. Cardiac electrical activity at each myocardial segment was represented by a regional current dipole for evaluating the BSPM during ventricular activations. Pacing was conducted in each animal’s heart from a number of intramural (RVA and RVS) and endocardial ventricular sites (LVA, LVL, LVAn). Although the HF animal was paced from the RVS, because the pacing lead shifted before the post-operative MR scan was performed, this pacing site was not included in the subsequent validation analyses. A summary of the modeling and experimental parameters can be found in Table 1.
Intramural RV Pacing
Intramural pacing was successfully completed in the RV in all three animals. Figure 2 demonstrates an example of the BSPM results during RVS pacing in a control animal. In panel (a), six beats are displayed, including two beats under sinus rhythm (the first two) and three beats when the heart is captured by pacing (the last three). In panel (b), the BSPMs are shown from the onset of pacing to the end of activation. The 3DCEI analysis was applied on the BSPMs collected (corresponding to the fifth beat in panel (a)) and the estimated location of the initiation site and the detailed 3D activation sequence are shown in panel (c). Figure 3(a) depicts an example of the imaging results when the HF swine was paced from RVA.
Figure 2.
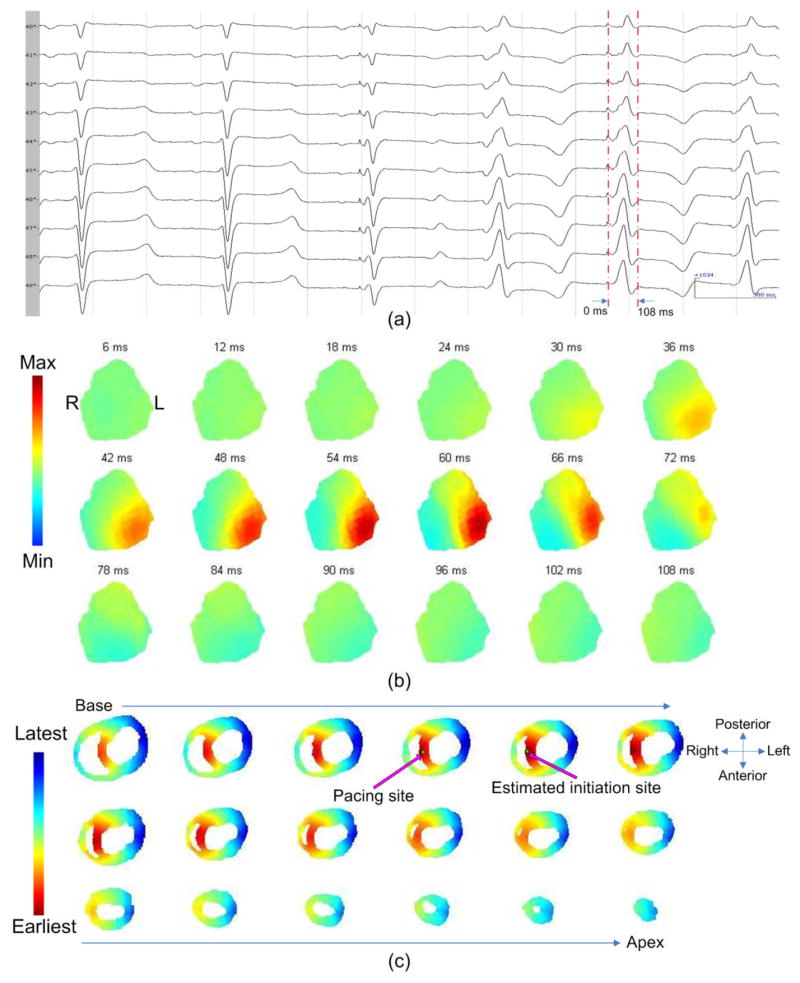
(a): The ECG waveforms recorded from the body surface electrodes with corresponding time intervals. (b): The body surface potential maps. (c): The location of the initiation site and the 3D activation sequence (AS). The relative geometry of the heart is shown by horizontal sections, arranged from base to apex. The precise location of the initiation site and the estimated location of the initiation site are indicated. The 3D AS is shown on the horizontal sections.
Figure 3.
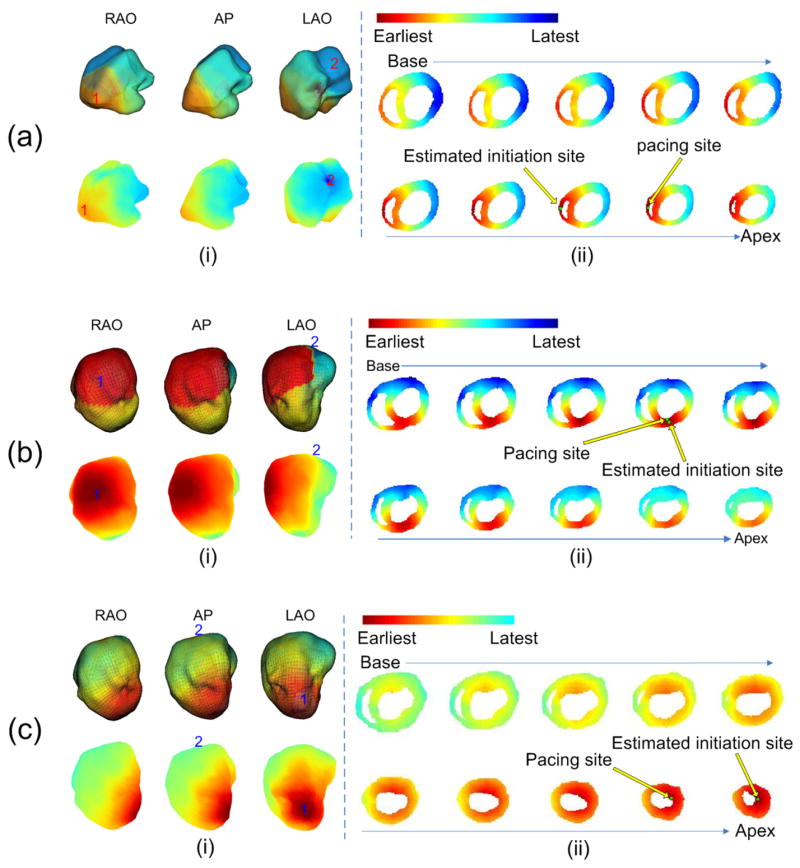
The evaluation of the inverse results when: (a) the HF animal was paced from the RV apex; (b) a control animal was paced from the endocardial LV anterior; (c) a control animal was paced from the endocardial LV apex. In each panel, (i): the activation sequence (AS) on the LV endocardial surface. The AS reconstructed by the NCM system is shown in the upper row in 3 views: RAO, AP and LAO. RED represents the earliest activated area and BLUE represents the latest activated area. The earliest activated site is indicated by the number “1” and the latest activated site is indicated by “2” on the figure. The corresponding AS estimated by 3DCEI is shown in the lower row. The earliest activated site and the latest activated site are also indicated by “1” and “2”, respectively. (ii): the location of the initiation site and the 3D AS. The geometry of the heart is shown by 10 horizontal sections, arranged from base to apex. The precise location of the initiation site and the estimated location of the initiation site are indicated. The 3D AS is shown on the 10 sections.
In total, 5 intramural sites in RV were paced, and 50 paced beats were analyzed (10 for each site). The evaluation of the estimation results are summarized in Table 2. Over the 50 beats analyzed, the averaged localization error (LE) of the initiation site of activation was 7.3±1.8 mm. It was observed that the depolarization propagation was similar between the estimated activation sequence by 3DCEI and those recorded on the NCM system for all pacing sites.
Endocardial LV Pacing
Stable endocardial pacing within LV was successfully obtained in one control animal and in the HF animal (Table 1). Figure 3(b) represents an example of the imaging results that was observed when pacing a control animal from the LV anterior endocardial site, and Figure 3(c) represents another example when pacing the same animal from the LV apical endocardial site. In total, 5 endocardial sites in LV were paced, and 50 paced beats were analyzed (10 for each site). The evaluation of the estimation results are summarized in Table 3. Over the 50 beats analyzed, the averaged LE of the initiation site of activation was 7.0 ± 2.2 mm. Similar to the results when intramural pacing was conducted, the estimated endocardial activation sequence by 3DCEI demonstrated a similar depolarization wavefront to the recorded endocardial activation sequence generated by the NCM system.
Discussion
The present study has experimentally demonstrated the capability of our noninvasive 3D electrical imaging approach to localize the electrical initiation sites and estimate the resultant activation sequences. To our knowledge, the present study represents the first reported effort in literature to validate electrocardiographic inverse solutions using simultaneous body surface and intracavitary recordings in an in vivo animal, under both control and diseased conditions. The animal model chosen for these studies was the swine due to their close approximation to humans in their cardiac sizes, heart-torso geometries and electrophysiological characteristics. Swine have a non-compliant thorax which is more similar to the human (versus canine) condition and further has a relative thoracic surface mimicking an average adult. Therefore, we believe the validation employing a swine model provides stronger clinical significance than the previous validation in a rabbit model where an open-chest procedure was used22. Importantly, promising results were also demonstrated when investigating a disease heart model, which suggests the 3DCEI approach may be robust enough to account for complex, clinical conditions that may be found in a clinical setting.
The localization of the initiation site and/or arrhythmogenic substrate of various cardiac arrhythmias has direct clinical benefit for the guidance of catheter ablation procedures. Efforts have been made to find the earliest activated site(s) of cardiac events on the epicardium6, endocardium3 or the entire heart surface (endocardium and epicardium)7,11. These methods define their solutions on the heart surface and beneficial results have been reported. However, often times, the cardiac event occurs within the myocardial wall, thereby increasing the difficulty in locating the accurate location and depth for targeting the ablative therapy. The noninvasive 3D imaging methods provides for greater anatomical resolution and thus may provide a greater clinical benefit and efficacious therapy. In the present study, we consider that the successful localization of the initiation sites induced by pacing suggests the potential feasibility of localizing the initiation sites of focal ventricular tachycardia and/or other cardiac arrhythmias throughout the 3D myocardium. Pacing was performed within both ventricular chambers, from the endocardial surfaces and with implanted electrodes (intramural activation). We obtained minimal to relatively small localization errors for such pacing employing this 3DCEI approach. The averaged localization error of all pacing trials was approximately 7 mm, which suggests the potential of guiding catheter ablation procedures by applying 3DCEI.
In addition to localizing the arrhythmogenic focal trigger and/or substrate, importantly the 3DCEI approach can further estimate the global 3D electrical activation sequence in multiple cardiac chambers simultaneously. In the present study, we evaluated our estimated AS by comparing with the AS reconstructed by the NCM system. It was observed that the endocardial AS estimated by 3DCEI demonstrated a consistent propagation pattern to that reconstructed by the NCM system (examples shown in Figure 3), which is currently routinely used in the clinical setting for the guidance and navigation of atrial ablation procedures. The similarity in activation sequence provides evidence that the estimated endocardial AS extracted from the estimated 3D AS could prove clinically valuable and efficacious.
The present study further investigated an animal with high-rate pacing induced HF. Thereby, the heart was observed to have a larger geometry than the control animal (nearly a doubled diastolic volume), and associated increased durations of the QRS complexes. It should be noted that there were no regional cardiac dysfunctions in this animal (such as that due to an infarction). Yet, the high correlation between the simulated BSPMs and the recorded BSPMs provided further validation of our forward model. Specifically, in this animal, we successfully recorded the simultaneous mapping data when pacing from three different sites. The analyses of the obtained data proved that on this diseased swine model, the 3DCEI approach could generate adequate inverse solutions; as with the controls (shown in Table 2 and Table 3). This robust performance of 3DCEI indicates its ability to afford strong clinical utility in complex procedures.
To date, we have experimentally demonstrated the feasibility of localizing the initiation sites and image maps the global electrical activation sequences using 3DCEI techniques for single-site pacing. Future investigations will need to focus on more complex arrhythmic events, such as reentrant ventricular tachycardias, so to provide further clinical utility of such an approach.
Limitations
Although we have obtained the 3D activation sequence, we can only evaluate its endocardial propagation pattern under the present experimental protocol without using 3D intracardiac mapping procedure. Although employing non-contact mapping within the protocol provides many benefits for the testing of the 3DCEI methodology in a setting similar to clinical studies in human patients, it also represents a limitation in such that we do not have direct measurements within the 3D ventricular volume. We used non-contact endocardial mapping results, as practiced in clinical cardiology, to validate our 3DCEI results and its ability to display activation sequence propagation. The NCM system does not provide the quantitative output of the activation sequence on the endocardial surface, so the quantitative comparison between our estimated activation sequence and the NCM results was not done in the present study. However, the present comparison study, although qualitative in nature, should still provide important experimental evidence to assess the performance of the 3DCEI, as the present protocol does not need an open-chest procedure.
As described in the Methods session, in order to determine the precise locations of the endocardial pacing sites in the heart-excitation model, the endocardial surface recorded by the NCM system was registered with the corresponding surface of the beating heart segmented from the pre-operative MR images. Although both of the endocardial surfaces correspond to diastole, they can not exactly match due to the complex positional and size changes that may occur during the cardiac and respiratory cycles. Because of this problem, error might be introduced when the registration procedure was applied. Such error is not unique to this Ensite/MRI registration. A similar case was reported to register the surface recorded by CARTO® system with the CT images26. Their results showed that the registration accuracy could be significantly increased when multiple surfaces were involved in the registration procedure. For instance, we might detect the geometries of both LV and superior vena cava for the purpose of registration in future study.
Conclusions
We have demonstrated the feasibility of localizing 3D initiation sites of activation and estimating the activation sequences within both healthy and diseased large mammalian models from noninvasive body surface potential measurements by using 3DCEI. These promising results suggest that the 3DCEI approach may provide a useful tool not only for basic cardiovascular research, but may have future utility for the clinical diagnosis and management of arrhythmias.
Acknowledgments
The authors would like to thank Bill Gallagher, Michael Eggen, Jason Quill, Sara Anderson, Eric Richardson, Christopher Wilke, Zhongming Liu, and Chengzong Han for technical assistance of the data acquisition. This work was supported in part by NIH RO1 HL080093-01A2, NSF BES-0411480, NSF BES-0602957, and by the Institute of Engineering in Medicine of the University of Minnesota. Chenguang Liu was supported in part by the Predoctoral Fellowship #0515472Z from the American Heart Association, Midwest Affiliate.
List of abbreviations
- 3D
three dimension
- 3DCEI
three-dimensional cardiac electrical imaging
- AS
activation sequence
- BSPM
body surface potential map
- ECG
electrocardiogram
- EP
electrophysiology
- HF
heart failure
- IS
initiation site
- LE
localization error
- LV
left ventricle
- MCG
magnetocardiogram
- MRI
magnetic resonance imaging
- NCM
non-contact mapping
- RV
right ventricle
Footnotes
This work was conducted at the University of Minnesota.
References
- 1.Morady F. Radio-frequency ablation as treatment for cardiac arrhythmias. N Engl J Med. 1999;340:534–544. doi: 10.1056/NEJM199902183400707. [DOI] [PubMed] [Google Scholar]
- 2.Gepstein L, Hayam G, Ben-Haim SA. A novel method for nonfluoroscopic catheter-based electroanatomical mapping of the heart: in vitro and in vivo accuracy results. Circ. 1997;95:1611–1622. doi: 10.1161/01.cir.95.6.1611. [DOI] [PubMed] [Google Scholar]
- 3.Gornick CC, Adler SW, Pederson B, Hauck J, Budd J, Schweitzer J. Validation of a new noncontact catheter system for electroanatomic mapping of left ventricular endocardium. Circ. 1999;99:829–835. doi: 10.1161/01.cir.99.6.829. [DOI] [PubMed] [Google Scholar]
- 4.Gurevitz OT, Glikson M, Asirvatham S, Kester TA, Grice SK, Munger TM, Rea RF, Shen WK, Jahangir A, Packer DL, Hammill SC, Friedman PA. Use of advanced mapping systems to guide ablation in complex cases: experience with noncontact mapping and electroanatomic mapping systems. Pacing Clin Electrophysiol. 2005;28:316–323. doi: 10.1111/j.1540-8159.2005.09477.x. [DOI] [PubMed] [Google Scholar]
- 5.Varanasi S, Dhala A, Blanck Z, Deshpande S, Akhtar M, Sra J. Electroanatomic mapping for radiofrequency ablation of cardiac arrhythmias. J Cardiovasc Electrophysiol. 1999;10:538–544. doi: 10.1111/j.1540-8167.1999.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 6.Oster HS, Taccardi B, Lux RL, Ershler PR, Rudy Y. Noninvasive electrocardiographic imaging: reconstruction of epicardial potentials, electrograms, and isochrones and localization of single and multiple electrocardiac events. Circ. 1997;96:1012–1024. doi: 10.1161/01.cir.96.3.1012. [DOI] [PubMed] [Google Scholar]
- 7.Huiskamp G, Greensite F. A new method for myocardial activation imaging. IEEE Trans Biomed Eng. 1997;44:433–446. doi: 10.1109/10.581930. [DOI] [PubMed] [Google Scholar]
- 8.Greensite F, Huiskamp G. An improved method for estimating epicardial potentials from the body surface. IEEE Trans Biomed Eng. 1998;45:98–104. doi: 10.1109/10.650360. [DOI] [PubMed] [Google Scholar]
- 9.Brooks DH, Ahmad GF, MacLeod RS, Maratos GM. Inverse electrocardiography by simultaneous imposition of multiple constraints. IEEE Trans Biomed Eng. 1999;46:3–18. doi: 10.1109/10.736746. [DOI] [PubMed] [Google Scholar]
- 10.Pullan AJ, Cheng LK, Nash MP, Bradley CP, Paterson DJ. Noninvasive electrical imaging of the heart: theory and model development. Ann Biomed Eng. 2001;29:817–836. doi: 10.1114/1.1408921. [DOI] [PubMed] [Google Scholar]
- 11.Tilg B, Fischer G, Modre R, Hanser F, Messnarz B, Schocke M, Kremser C, Berger T, Hintringer F, Roithinger FX. Model-based imaging of cardiac electrical excitation in humans. IEEE Trans Med Imaging. 2002;21:1031–1039. doi: 10.1109/TMI.2002.804438. [DOI] [PubMed] [Google Scholar]
- 12.Ramanathan C, Raja NG, Jia P, Ryu K, Rudy Y. Noninvasive electrocardiographic imaging for cardiac electrophysiology and arrhythmia. Nature Medicine. 2004;10:422–428. doi: 10.1038/nm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger T, Fischer G, Pfeifer B, Modre R, Hanser F, Trieb T, Roithinger FX, Stuehlinger M, Pachinger O, Tilg B, Hintringer F. Single-beat noninvasive imaging of cardiac electrophysiology of ventricular pre-excitation. J Am Coll Cardiol. 2006;48:2045–2052. doi: 10.1016/j.jacc.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 14.He B, Wu D. Imaging and Visualization of 3D Cardiac Electric Activity. IEEE Transactions on Information Technology in Biomedicine. 2001;5:181–186. doi: 10.1109/4233.945288. [DOI] [PubMed] [Google Scholar]
- 15.He B, Li G, Zhang X. Noninvasive three-dimensional activation time imaging of ventricular excitation by means of a heart-excitation model. Phys Med Bio. 2002;47:4063–4078. doi: 10.1088/0031-9155/47/22/310. [DOI] [PubMed] [Google Scholar]
- 16.He B, Li G, Zhang X. Noninvasive imaging of cardiac transmembrane potentials within three-dimensional myocardium by means of a realistic geometry anisotropic heart model. IEEE Trans Biomed Eng. 2003;50:1190–1202. doi: 10.1109/TBME.2003.817637. [DOI] [PubMed] [Google Scholar]
- 17.Li G, He B. Localization of the site of origin of cardiac activation by means of a heart-model-based electrocardiographic imaging approach. IEEE Trans Biomed Eng. 2001;48:660–669. doi: 10.1109/10.923784. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Zhang X, Lian J, He B. Noninvasive localization of the site of origin of paced cardiac activation in human by means of a 3-D heart model. IEEE Trans Biomed Eng. 2003;50:1117–1120. doi: 10.1109/TBME.2003.816068. [DOI] [PubMed] [Google Scholar]
- 19.Li G, He B. Noninvasive estimation of myocardial infarction by means of a heart-model-based imaging approach – a simulation study. Med Biol Eng Comput. 2004;42:128–136. doi: 10.1007/BF02351022. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Li G, He B. Localization of Site of Origin of Reentrant Arrhythmia from BSPMs: A Model Study. Phys Med Bio. 2005;50:1421–1432. doi: 10.1088/0031-9155/50/7/006. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Zhang X, Liu Z, Pogwizd SM, He B. Three-dimensional myocardial activation imaging in a rabbit model. IEEE Trans Biomed Eng. 2006;53:1813–1820. doi: 10.1109/TBME.2006.873701. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Ramachandra I, Liu Z, Muneer B, Pogwizd SM, He B. Noninvasive three-dimensional electrocardiographic imaging of ventricular activation sequence. Am J Physiol Heart Circ Physiol. 2005;289:H2724–2732. doi: 10.1152/ajpheart.00639.2005. [DOI] [PubMed] [Google Scholar]
- 23.Laske TG, Skadsberg ND, Hill AJ, Klein GJ, Iaizzo PA. Excitation of the intrinsic conduction system through his and intraventricular septal pacing. Pacing Clin Electrophysiol. 2006;29:397–405. doi: 10.1111/j.1540-8159.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 24.He B. Electrocardiographic Tomographic Imaging. In: He B, editor. Modeling and Imaging of Bioelectric Activity: Principles and Applications. Kluwer Academic/Plenum Publishers; 2004. pp. 161–182. [Google Scholar]
- 25.Kozinska D, Tretiak OJ, Nissanov J, Ozturk C. Multidimensional alignment using the euclidean distance transform. Graphical Models and Image Processing. 1997;59:373–387. [Google Scholar]
- 26.Dong J, Calkins H, Solomon SB, Lai S, Dalal D, Lardo AC, Brem E, Preiss A, Berger RD, Halperin H, Dickfeld T. Integrated electroanatomic mapping with three-dimensional computed tomographic images for real-time guided ablations. Circ. 2006;113:186–194. doi: 10.1161/CIRCULATIONAHA.105.565200. [DOI] [PubMed] [Google Scholar]