Abstract
The mouse Igκ locus has three known transcriptional enhancers: the MAR/intronic enhancer (MEi), the 3′ enhancer (E3′) and the further downstream enhancer (Ed). Previous studies have shown that both MEi and E3′ enhancers are required for maximal gene rearrangement of the locus, and that E3′ is also required for maximal expression and somatic hypermutation (SHM). To functionally elucidate Ed in vivo, we generated knockout mice with a targeted germline deletion of Ed. Ed deleted homozygous mice (Ed−/−) have moderately reduced numbers of Igκ expressing B cells and correspondingly increased numbers of Igλ expressing B cells in spleen. Ed−/− mice also have decreased Igκ mRNA expression in resting and T-cell dependent activated splenic B cells and reduced Igκ chains in sera. However, our analysis indicates that Igκ gene rearrangement is normal in Ed−/− mice. In addition, our results show that Ed−/− mice exhibit reduced SHM in the Igκ gene J-C intronic region in germinal center B cells from Peyer's patches. We conclude that Ed positively regulates Igκ gene expression and SHM, but not gene rearrangement.
Keywords: B cells, gene regulation, knockout mice, enhancer, transcription, somatic hypermutation
Introduction
During B cell development the Ig heavy chain gene rearranges first, by sequential D-J and then by V-(D)J joining, leading to the pro- and pre-B cell stages of development, respectively (1). The Igκ locus is poised for rearrangement in pre-B cells, and upon appropriate signaling one of the 95 Vκ genes is semi-randomly selected for recombination to a J region (for review of recombination, see ref. 2), evidently by the RAG proteins first making single-stranded nicks at Vκ recombination signal sequences (RSS)3 followed by a capture model for synapsis (3). The Igκ locus thus offers the opportunity to visualize changes in chromatin structure that may precede gene rearrangement and transcriptional activation during B lymphocyte differentiation, as well as those remodeling events that may accompany or be a consequence of gene activation (4 and refs. within). In addition, the mouse Igκ gene locus has provided a paradigm for investigating site-specific recombination and tissue-specific transcriptional regulation (reviewed in ref. 5); somatic hypermutation (SHM) (reviewed in ref. 6); DNA methylation (7); higher-order chromatin organization (8); and nuclear organization and allelic exclusion (reviewed in ref. 9).
Several previous studies have identified a number of cis-acting regulatory elements in the mouse Igκ gene locus. All of these elements reside in a 32 kb segment near or within the Jκ-Cκ region toward the 3′ end of the 3.2 mega base locus, except for Vκ gene promoter elements and their RSSs (10) and the recombining sequence (RS) (11). These include: a recombination silencer (12), two germline promoter elements (13, 14), KI-KII sequences (15), RSSs associated with J regions (16), a nuclear matrix association region (MAR) (17), an intronic enhancer (Ei) (18), a transcription terminating region (19), a 3′ enhancer (E3′) (20), and further downstream enhancer (Ed) (4).
The functional significance of several of the above cis-acting sequences has been addressed by creating their targeted deletion from the native locus in cell lines or mice. Deletion of a germline promoter, or KI-KII sequences, or both, results in lower levels of gene rearrangement (15, 21, 22). Deletion of the MAR from the mouse germline downregulates somatic hypermutation (SHM) and mildly stimulates precocious Vκ-Jκ joining (23), whereas its deletion in a pre-B cell line results in hyper-recombination (24). Deletion of either MEi or E3′ severely reduces but does not abolish Igκ gene rearrangement (25, 26), whereas deletion of both enhancers nearly completely blocks recombination, indicating that each element has a redundant but critical role in regulating gene rearrangement in the locus (27). Deletion of E3′ also results in reduced SHM (28), in contrast to an earlier report (29), and also reduced Igκ gene expression (26, 28).
Several years ago our laboratory discovered a third enhancer in the mouse Igκ locus, initially from an analysis of B cell specific DNase I hypersensitive sites (HSs) in chromatin (4). This enhancer is in the far downstream region, some 8 kb 3′ of the E3′ element, and was coined Ed. Because the mouse Ed sequence is heavily conserved in the human genome and shares NF-κB and E2A binding sites, we decided to elucidate its function in the native locus by creating a targeted deletion in the mouse germline, and report here the phenotypes resulting from this deletion. We demonstrate that Ed plays no role in regulating recombination, a result consistent with our previous findings that the element exhibits HS in plasmacytoma cells but not in pre-B cells (4). Rather, we have found that Ed is required for maximal Igκ gene expression in resting and T-cell-dependent stimulated splenic B cells. We further demonstrate the SHM is downregulated in germinal center (GC) B cells from Peyer's patches of these Ed−/− knockout mice.
Materials and Methods
Generation of Ed knockout mice
Homology arms of the Ed targeting vector were genomic DNA fragments from a lambda phage clone containing the mouse Ed genomic locus, which was isolated by screening a 129SVJ mouse genomic library (Stratagene). The Ed targeting vector was constructed as follows: First, a PGK-neor gene fragment flanked by two loxP sites was excised from the NL-24 vector (30) and cloned into pBluescript II KS (Stratagene), and a NdeI site was introduced into the PBS vector by inserting a polylinker. Next, a 3.6 kb NdeI 129SVJ genomic DNA fragment as a 3′ homology arm was cloned into the NdeI site. And then, a 1.8 kb HindIII 129SVJ genomic DNA fragment corresponding to the 5′ homology arm was cloned into a HindIII site. Finally, the 1.2 kb HindIII/NdeI Ed enhancer containing fragment was PCR amplified using 129SVJ genomic DNA as template. A loxp site was introduced into this PCR fragment using the primers: Eddu:CATTAGCGGCCGCATAACTTCGTATAGCATACATTATACGAAGTTATCT G G G T G T A T C T G A T C G T T T T A G a n d E d d d : CATTAGCGGCCGCATGTATGCTATGGTACACTCAGG. The PCR reaction used 35 cycles, each cycle consisted of 1 min at 94°C, 1 min at 59°C and 2 min at 68°C. Then the PCR amplified Ed fragment was cloned into the Ed targeting vector. The SM-1 (129/SvEvTac) embryonic stem cell line was transformed by electroporation with SacII linearized targeting vector. G418 drug-resistant embryonic stem cell clones were first screened by PCR and correctly targeted clones were then verified by Southern blotting. Probe A or probe B (Fig. 1) residing outside of the targeting vector were used to hybridize NcoI digested ES cell genomic DNA. Three independent Ed targeted clones were injected into C57/Bl6 blastocysts. Chimeric mice were bred with C57/BL6 mice to obtain germline transmission. Then germline transmissible mice were bred with Cre recombinase expressing MORE mice (31) to obtain Ed and neor deletion mice. All mice were used in accordance with protocols approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee (IACUC).
Flow Cytometry
Single-cell suspensions were prepared from bone marrow and spleen. Cells (1 × 106) were stained with an optimal concentration of FITC-, PE- or biotin-conjugated antibodies in 100 μl of PBS/2%FCS for 20 min on 4°C. Stained cells were analyzed by a FACS Calibur (Becton-Dickinson). The following antibodies were used: FITC-anti-B220 (RA3-6B2), anti-λ (R26-46), anti-CD21 (7G6) (PharMingen); PE-anti-Igκ (187.1), anti-B220 (RA3-6B2) (PharMingen); biotinylated-anti-IgM (II/41), anti-CD138 (syndecan-1) (281-2) (PharMingen); FITC-anti-IgD (11-26), APC-anti-CD23 (2G8) (Southern Biotech). The biotin conjugates were revealed with allophycocyanin-streptavidin (Southern Biotech). Only cells residing in the lymphocyte gate were analyzed. Dead cells were excluded by size and forward scatter gating. Data were analyzed with CellQuest (Becton Dickinson) or FlowJo (Tree Star) software.
Northern blotting analysis of Igκexpression
Single-cell suspensions were prepared and 2 × 107 spleen cells from wild type (WT) or Ed−/− mice were incubated with an optimal concentration of biotinylated anti-κ Abs and purified by using MACS MS separation columns (Miltenyi Biotec) according to the manufacturer's instructions. κ+ cell populations were found to be more than 93% pure by FACS analysis (data not shown). Splenic activated and resting B cells were purified by Percoll (Sigma-Aldrich) gradient centrifugation as described (32). Total RNA was isolated from purified cells by extraction with Trizol reagent (Invitrogen) according to the manufacturer's instructions. Northern blotting was performed by using NorthernMax kits (Ambion) according to the manufacturer's protocol. The β-actin probe was supplied by (Ambion). The μ chain probe was a 300 bp PCR product from the mouse IgH Cμ exon1. The κ chain probe was a 410 bp XmnI fragment from the Cκ region from the pSPIg8 plasmid.
B cell culture
Single-cell suspensions of spleens from WT and Ed−/− mice were incubated with an optimal concentration of biotinylated-anti-CD43 (Ly-48, PharMingen) antibody and biotinylated-anti-λ (JC5-1, Southern Biotech) antibody. Then MACS MS separation columns (Miltenyi Biotec) were used to deplete CD43+ and λ+ B cells. κ+ enriched B cells were cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin/streptomycin, L-glutamine, and 50 μM β-mercaptoethanol at 106 cells/ml, and then were stimulated with 20 μg/ml LPS (Sigma-Aldrich), 2.5 μg/ml anti-CD40 Abs (HM40-3, BD Pharmingen) or 2.5 μg/ml anti-CD40 Abs and 10 ng/ml recombinant mouse IL-4 (Pharmingen) for 4 days.
ELISA analysis
Sera collected from 10-16 week old WT and Ed−/− mice were analyzed for Igκ and Igλ chain concentrations by sandwich ELISA. PVC microtiter plates (DYNEX technologies INC) were coated with polyclonal goat anti-mouse Ig (H+L) (Southern Biotech) at 10 μg/ml as the capturing antibody. After blocking for 2 h with 3% bovine serum albumin (BSA) in PBS at room temperature, diluted sera were added and incubated at 4°C overnight. Plates were subsequently washed three times and incubated with either goat anti-mouse kappa-HRP or goat anti-mouse lambda-HRP (Southern Biotech) for 2 h. After washing three times, plates were developed with the ABTS substrate (Southern Biotech) following the manufacturer's instructions. Purified mouse IgG3, κ isotype standard and purified mouse IgG2a, λ isotype standard (Pharmingen) were used to quantitate ELISA data.
Southern blotting and quantitative PCR analysis of Igκ V-J rearrangement
Single-cell suspensions containing 2 × 107 splenic or bone marrow cells from WT or Ed−/− mice were incubated with an optimal concentration of biotinylated-anti-κ antibody or biotinylated-anti-B220 antibody, and κ+ cells or B220+ cells were purified by using MACS MS separation columns. The purified cells were lysed in lysis buffer with proteinase K. Genomic DNA was then purified by phenol/chloroform extraction, followed by ethanol precipitation. 10 μg of genomic DNA was digested with BamHI/EcoRI, and transferred to Zeta-probe GT genomic membrane (Bio-Rad) after electrophoretic separation on agarose gels. Prehybridization and hybridization were performed at 65°C with ExpressHyb hybridization buffer (Clonetech). The membranes were hybridized with [α-32P] dCTP-labeled Cκ and c-myb probes (26). Membranes were exposed to PhosphorImaging screens, and images were analyzed using ImageQuant software (Molecular Dynamics). A quantitative Igκ V-J rearrangement PCR assay was performed as described (23, 27). Briefly, the VκJκ rearrangement products were PCR amplified by using the Platinum Taq high fidelity DNA polymerase (Invitrogen) with a degenerate Vκ primer (VκD) (33) and a MAR35 primer (27). PCR cycles were as follows: 94°C denaturation for 3 min, followed by 27 cycles at 94°C for 1min, 60°C for 1 min, 68°C for 3 min, and a final 5 min extension at 68°C. The amount of genomic DNA used in each PCR reaction was controlled by the c-myb PCR reaction, which specifically amplifies a 0.7-kb c-myb gene genomic fragment (26). PCR products were resolved by electrophoresis in 1% agarose gels and transferred to Zeta-probe GT genomic membrane (Bio-Rad). The membranes were hybridized with a probe within the Jκ to MEi region (Fig. 7D). Membranes were exposed to PhosphorImaging screens, and images were analyzed using ImageQuant software (Molecular Dynamics).
Cell sorting and SHM analyses
Single-cell suspensions prepared from Peyer's patches were stained with PE-anti-B220 and FITC-PNA (Vector Laboratories). B200+ PNAhigh germinal center cells and B220+ PNAlow B cells were sorted on a MoFlo machine (Dako Cytomation). For the Jκ-Cκ intronic region SHM analysis, genomic DNA was purified from sorted WT and Ed−/−germinal center B cells. The Jκ-Cκ intronic regions from rearranged genes were PCR amplified by using the Platinum Taq high fidelity DNA polymerase (Invitrogen) with a degenerate Vκ primer (33) and a reverse primer located approximately 600 bp downstream of the Jκ5: AGCGAATTCAACTTAGGAGACAAAAGAGAGAAC. The PCR reaction used 35 cycles, each cycle consisted of 30s at 94°C, 30s at 56°C and 1 min at 68°C. Gel purified Vκ-Jκ5 PCR products were cloned into the PGEM-T vector (Promega). Vκ-Jκ5 clones were identified and sequenced by use of a T7 primer. Sequences were aligned with the mouse Jκ5 downstream sequence using the Vector NT I (Invitrogen) AlignX program and mismatches were scored as mutations in the 500 bp region downstream of Jκ5.
Results
Generation of Ed−/− mice
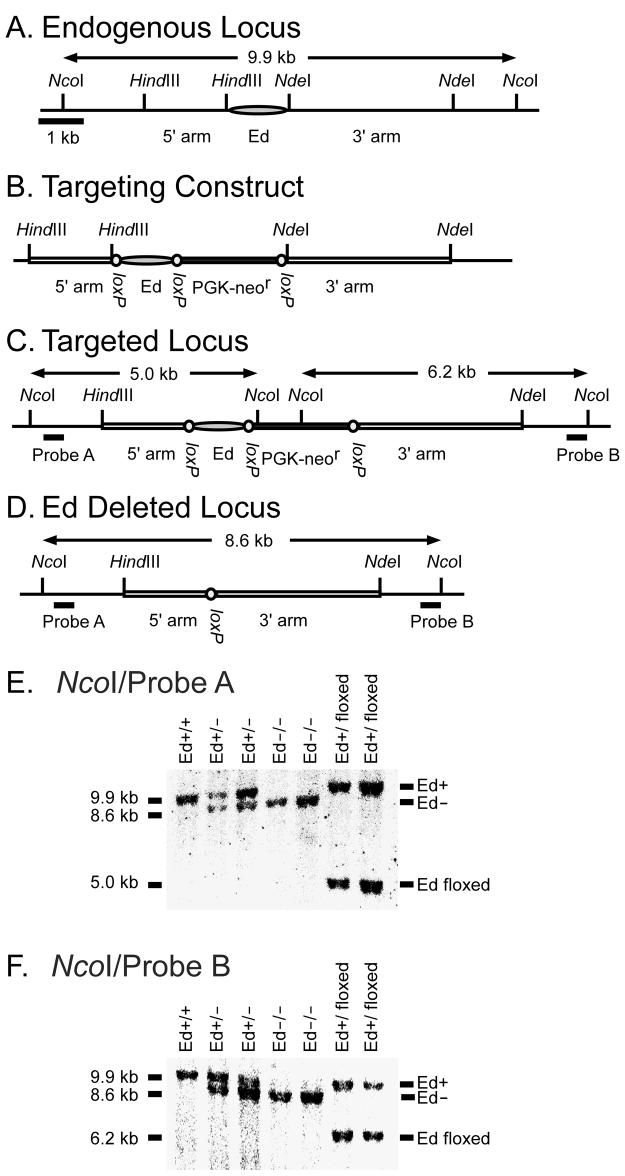
To delete Ed from the native locus we first constructed a targeting vector in which the entire 1.2 kb enhancer was replaced with Ed and a PGK-neor gene flanked by loxP sites (Fig. 1A, B). Embryonic stem (ES) cells were transfected with this linearized construct and several clones that exhibited site-directed integration were obtained after screening by PCR and Southern blotting (Fig. 1C). Three independent targeted ES cell clones were used to generate chimeric mice lines. Chimeric mice were bred with C57/BL6 mice to obtain germline transmission. To delete Ed and the PGK-neor gene (Fig. 1D), we bred germline transmissible mice with Cre recombinase expressing MORE mice (31). Ed deleted heterozygous mice were interbred to obtain Ed deleted homozygous mice, which are referred to hereafter as Ed−/− mice. Various stages of the targeting and Ed deletion were confirmed by Southern blotting. Genomic DNA was digested with NcoI and hybridized with either probe A or B, which generated a 9.9 kb WT band, a 8.6 kb Ed deleted band, and a 5.0 or 6.2 kb PGK-neor gene replacement band detected by probe A and B, respectively (Fig. 1E, F).
FIGURE 1.
Generation of Ed knockout mice. A, Genomic organization of the 3′ region of the Igκ locus. The 1.2 kb HindIII-NdeI fragment shown as a shaded oval contains the entire Ed region. B, Targeting construct, with Ed and the neor gene flanked by three loxP sites. The HindIII-HindIII and NdeI-NdeI fragments were used as 5′ and 3′ homology arms. C, Targeted locus. Endogenous Ed was replaced with Ed-PGK-neor flanked by loxP sites. Probes are represented with thick lines. D, Ed deleted locus. Ed and the neor gene were deleted in vivo by Cre-loxP mediated recombination. E and F, Southern blot analysis of tail DNA. Tail genomic DNA was digested with NcoI and hybridized with probe A (E) or probe B (F). A 9.9 kb band derived from the WT allele and a 8.6 kb band derived from the Ed deleted allele were detected by Probe A or Probe B. A 5.0 kb band and a 6.2 kb band derived from the floxed allele were detected by probe A and probe B, respectively. NEB DNA markers were used for gel calibrations to determine fragment sizes.
Ed−/− mice exhibit a modest decrease in Igκ expression in splenic B cells
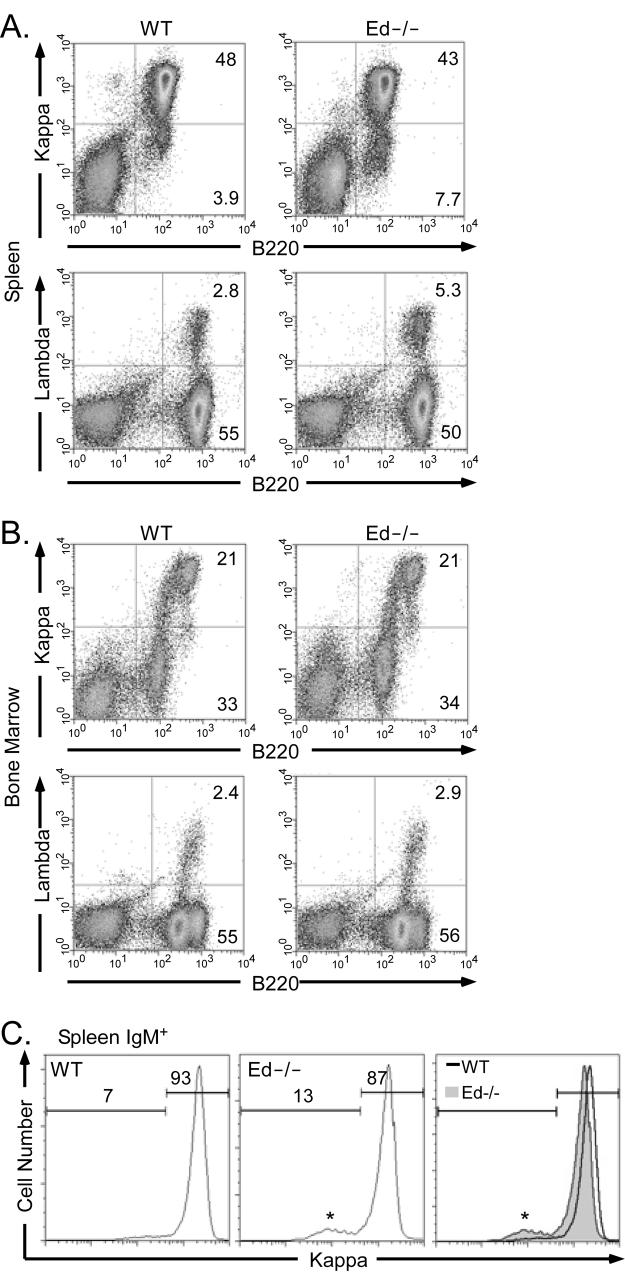
We found that Ed−/− mice exhibited no significant differences in bone marrow and spleen B cell numbers compared to those of their WT littermates or age-matched WT mice (data not shown). However, Ed−/− mice exhibited a moderate decrease of Igκ+ B cells in spleen compared with WT mice (Fig. 2A, 43±1% versus 48±2%, as percentages of Igκ+ B cells among total lymphocytes, n=6, P<0.01, Student's t test). In contrast, the percentage of Igλ+ B cells was increased by about 2-fold (Fig. 2A, 5.3±1.5% versus 2.8±0.5%, as percentages of Igλ+ B cells among total lymphocytes, n=5, P<0.01, Student's t test). These changes resulted in a moderate decrease of Igκ/λ ratio, approximately 8.1/1 in Ed−/− mice as opposed to 17.1/1 in WT mice. Because the percentages of Igκ+ and Igλ+ B cells in bone marrow were not different between Ed−/− and WT mice (Figure 2B), we conclude that the alteration in the distributions of Igκ+ and Igλ+ cells occurs in the periphery. Furthermore, FACS analysis showed that the percentages of pro- and pre-B cells (B220low/IgM−), newly generated immature B cells and recirculating B cells (B220+/IgM+) in bone marrow exhibited no significant differences between Ed−/− and WT mice (data not shown). We also observed a similar decrease of Igκ+ cells in gated splenic IgM+ cells in Ed−/− mice (Fig. 2C). And the mean fluorescence intensity (MFI) of Igκ expressing B cells in Ed−/− mice was decreased compared to those of WT (Fig. 2C, 1138±5.0 versus 1268±21.5, n=3, P<0.01, Student's t test). These results indicate that the decreased Igκ+ cells and altered Igκ/λ ratio in the spleen of Ed−/− mice must be primarily established during the maturation of B cells in the spleen, and not during the generation of B cells in bone marrow at the level of recombination.
FIGURE 2.
Flow-cytometric analysis of cell surface Ig expression in spleen and bone marrow. A, Single cell suspensions from spleen were simultaneously stained with PE-conjugated anti-Igκ and FITC-conjugated anti-B220 antibodies (upper); or PE-conjugated anti-B220 and FITC-conjugated anti-Igλ antibodies (lower). Stained cells were analyzed by FACS. Only cells residing in the lymphocyte gate were analyzed. Percentages of cells residing in various windows are shown in the figure sub-panels. Data are representative of independent FACS analyses from at least 5 mice of each genotype. B, Single cell suspensions from bone marrow were stained and analyzed as described in A. C, Splenic cells were stained with PE-conjugated anti-Igκ, FITC-conjugated anti-B220 and biotinylated anti-IgM. The biotinylated anti-IgM was revealed with allophycocyaninstreptavidin. IgM+ cells were analyzed for cell surface expression of Igκ. Histograms of Igκ fluorescence intensity and cell percentages are displayed for WT and Ed−/− splenic cells. Only cells residing in the lymphocyte gate were analyzed. The asterisks indicate small peaks of IgM+ λ+ cells, which disappear if samples are first depleted in λ+ B cells with biotinylated-anti-λ antibody and MACS columns prior to FACS (data not shown).
Ed−/− mice exhibit little differences from WT mice in marginal zone, follicular, and transitional splenic B cells and in plasma cell formation
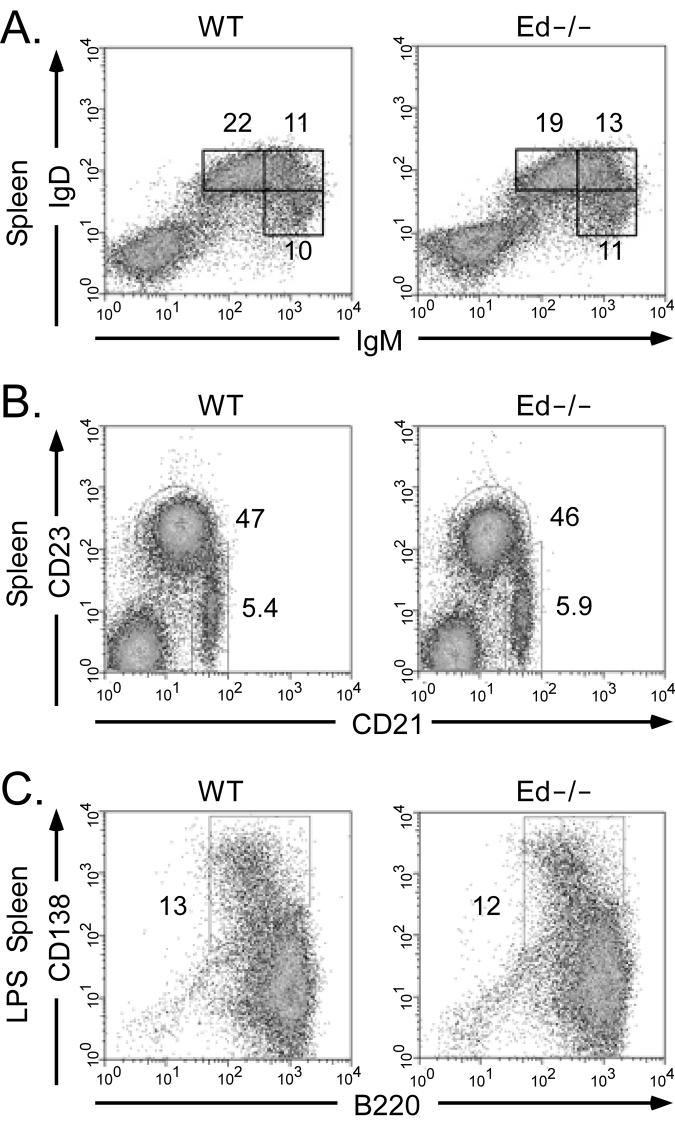
To further investigate the effect of Ed deletion on the development of B cells, we analyzed the percentages of transitional B cells, follicular mature B cells and marginal zone B cells in the spleen by FACS. Ed−/− mice exhibited a slight increase in T1 (IgMhiIgD−/lo) and T2 (IgMhiIgDhi) transitional B cells, and a slight decrease in follicular mature (IgMintIgDhi) B cells relative to the corresponding cell distributions exhibited by WT mice (Fig. 3A). We also found that the percentage of marginal zone B cells was normal in Ed−/− mice (Fig. 3B). Previous studies in our laboratory demonstrated that Ed exhibits a plasmacytoma cell-specific DNase I hypersensitive site (4). To address the question of whether plasma cell development is impaired in Ed−/− mice, we analyzed the percentages of B220+/−CD138+ plasma cells in spleen by FACS, and we found there was no significant difference between WT and Ed−/− mice (0.27±0.13% versus 0.29±0.05%, respectively; data not shown). When cultured with LPS in vitro, splenic B cells proliferate and differentiate into Ig-secreting cells (34). We found that the percentage of B220+/−CD138+ Ig-secreting cells was almost same between LPS treated WT and Ed−/− splenic B cells (Fig. 3C, 13±1.5% versus 12±0.6%). Northern blot analysis showed that the level of Igκ mRNA expression in these purified Ed−/− B220+/−CD138+ Ab-secreting cells were also normal (data not shown). In summary, these data indicate that the splenic B cell distributions and plasma cell formation are approximately normal in Ed−/− mice.
FIGURE 3.
Flow-cytometric analysis of B cell development and Ig-secreting cell differentiation in spleen. A, Single cell suspensions from spleen were simultaneously stained with FITC-conjugated anti-IgD and biotinylated anti-IgM. The biotinylated anti-IgM was revealed with allophycocyanin-streptavidin. Stained cells were analyzed by FACS. Only cells residing in the lymphocyte gate were analyzed. Gates of IgMloIgDhi,IgMhiIgDhi, IgMhiIgDlo are shown with the percentages of cells indicated. Data are representative of independent FACS analyses from at least 5 mice of each genotype. B, Single cell suspensions from spleen were simultaneously stained with FITC-conjugated anti-CD21 and APC-conjugated anti-CD23 antibodies and analyzed by FACS as described in A. Gates of follicular B cells (CD21+CD23+) and marginal zone B cells (CD21+CD23−/lo) are shown with the percentages of cells indicated. C, Analysis of Ig-secreting cell differentiation. Splenic cells were stimulated with LPS for 4 days and stained with PE-conjugated anti-B220 and biotinylated anti-CD138. The biotinylated anti-CD138 was revealed with allophycocyanin-streptavidin. Stained cells were analyzed by FACS. Gates of B220+/− CD138+ Ig-secreting cells are shown with the percentages of cells indicated.
Ed is required for normal Igκ expression
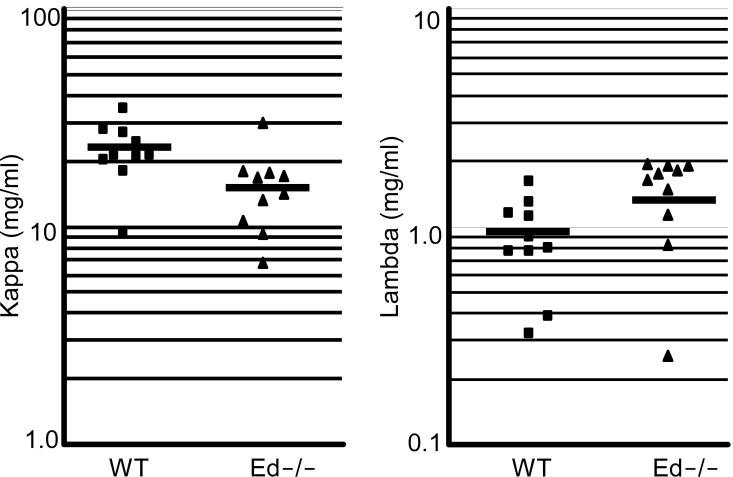
Because we found that cell surface Igκ expression levels were decreased in Ed−/− splenic Igκ+ B cells (Fig. 2C), we assayed for the steady-state levels of Igκ mRNA by Northern blot analysis. RNA samples from splenic Igκ+ cells were hybridized with Cκ, Cμ and β-actin probes. Then Igκ RNA levels were normalized to the total amount of β-actin RNA in the corresponding lanes. Igκ RNA levels of Ed−/− B cells were decreased compared to those of WT (Fig. 4A, C, 67±8.2% versus 100±5.6%, n=5, P<0.01, Student's t test). Furthermore, when we normalized Igκ RNA levels to those of Igμ RNA, we found a similar decrease in Ed−/− B cells compared to those of WT (Fig. 4A, 51±22.8% versus 100±7.7%, n=3, P<0.01, Student's t test). We also compared Igκ RNA levels of Ed−/− and WT B cells after 4 days of LPS stimulation in culture. We found Ed−/− and WT B cells expressed similar levels of Igκ RNA after LPS stimulation (Fig. 4B, D, 105±13% versus 100±8.7%, n=5, P=0.51, Student's t test). To further determine whether Igκ RNA levels were decreased in either activated or/and resting B cells, we purified activated and resting B cells from spleen by use of Percoll gradient centrifugation and analyzed the Igκ RNA levels by Northern blotting as above. We found Igκ RNA levels were decreased about 50% in Ed−/− resting B cells compared with those of WT. In contrast, when compared with resting B cells, the reduction of Igκ RNA levels in Ed−/− activated B cells was less dramatic (Fig. 4E). We also analyzed sera for Igκ chain protein levels by ELISA. The sera Igκ chain levels were decreased in Ed−/− mice compared to those of WT (Fig. 5, 15.3±6.4 versus 22.5±6.7 mg/ml, n=10, P=0.02, Student's t test). A corresponding increase in sera Igλ chain levels was observed in Ed−/− (Fig. 5, 1.46±0.56 versus 0.91±0.40 mg/ml, n=10, P=0.02, Student's t test). In summary, these data indicate that Igκ expression is impaired in splenic B cells lacking the Ed enhancer.
FIGURE 4.
Northern blot analysis of Igκ expression. A, Total RNA from splenic Igκ+ cells was blotted and probed first with a Cκ probe, and then sequentially hybridized with β-actin and Cμ probes. B, Total RNA from splenic Igκ+ cells cultured for 4 days with 20 μg/ml LPS was blotted and probed as described in A. C, The Northern blot signals of splenic Igκ+ cells were quantitated by PhosphorImager analysis. The levels of Igκ expression were normalized to those of β-actin (Cκ/β-actin) or Cμ (Cκ/Cμ). The Cκ/β-actin ratios of WT mice were set as 100% expression. The percentages of Igκ expression (Cκ (%), y axis) in Ed−/− mice were calculated as: [(Cκ/β-actin ratio of Ed−/− mice)/(Cκ/β-actin ratio of WT mice)] ×100%. Data are presented as means ±SD. Statistical significance was determined by a paired Student's t test. D, The Northern blot signals of LPS stimulated splenic Igκ+ cells RNA were quantitated by PhosphorImager analysis. The percentages of Igκ expression in Ed−/− mice were calculated as described in C. Data in A to D are representative of three independent experiments. E, Northern blot analysis of Igκ expression in activated B cells and resting B cells. Activated B cells and resting B cells were purified by Percoll gradient centrifugation from pooled WT and Ed−/− splenic cells. Total RNA from the purified cells was blotted and hybridized with Cκ and β-actin probes. The percentages of Igκ expression in Ed−/− mice were calculated as described in C.
FIGURE 5.
Analysis of sera Igκ and Igλ chain concentrations by ELISA. Sera from 10-16 week-old WT mice (n=10) and Ed−/− mice (n=10) were collected and analyzed by ELISA. The bar represents mean value of samples. Statistical significance was determined by a paired Student's t test.
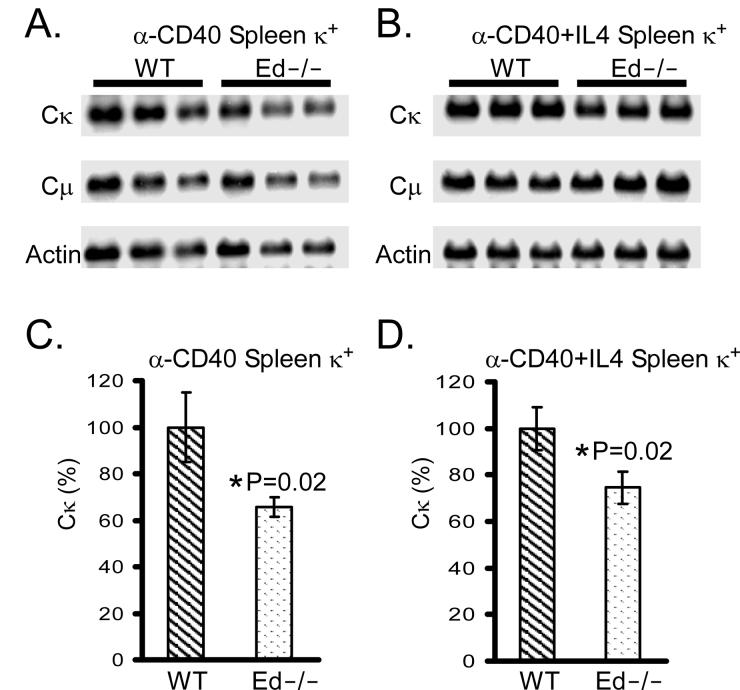
Igκ expression is impaired in T-cell-dependant stimulated Ed −/− splenic B cells
Previous studies have shown that Igκ expression is normal in E3′ enhancer knockout splenic B cells after stimulation with LPS (26). However, Igκ expression was decreased in these cells if activated by B cell receptor (BCR) or T-cell-dependant stimulation, such as anti-IgM or/and anti-CD40 antibody (28). To test whether Ed is required for normal Igκ expression in a T-cell-dependant simulated immune response, we stimulated splenic B cells with anti-CD40 antibody or anti-CD40 antibody plus IL4. Northern analyses revealed that Igκ mRNA levels were decreased in anti-CD40 activated Ed−/− splenic B cells compared with those of WT (Fig. 6A, C, 66±4% versus 100±15%, n=3, P=0.02, Student's t test). Similar results were found in anti-CD40 plus IL4 activated Ed−/− splenic B cells (Fig. 6B, D, 75±7% versus 100±9%, n=3, P=0.02, Student's t test). Furthermore, when we normalized the Igκ RNA levels to those of Igμ, we found a similar decrease in Ed−/− B cells compared to those of WT (Fig. 6A, B and data not shown). We also found that the percentages of B220+/−CD138+ Ig-secreting cells were similar between these T-cell-dependant stimulated Ed−/− and WT B cells (data not shown), indicating that the stimulation efficiencies were similar between Ed−/− and WT B cells.
FIGURE 6.
Northern blot analysis of Igκ expression in T-cell-dependent stimulated splenic B cells. A and B, Total RNA from splenic Igκ+ cells cultured for 4 days with anti-CD40 (A) or anti-CD40+IL4 (B) was blotted and hybridized first with a Cκ probe and then sequentially with β-actin and Cμ probes. C, The Northern blot signals of anti-CD40 stimulated splenic Igκ+ cells were quantitated by PhosphorImager analysis. The levels of Igκ expression were normalized by β-actin (Cκ/β-actin) or Cμ (Cκ/Cμ) expression. The Cκ/β-actin ratios of WT mice were set as 100% expression. The percentages of κ expression (Cκ (%), y axis) in Ed−/− mice were calculated as: [(Cκ/β-actin ratio of Ed−/− mice)/(Cκ/β-actin ratio of WT mice)] ×100%. Statistical significance was determined by a paired Student's t test. D, The Northern blot signals of anti-CD40+IL4 stimulated splenic Igκ+ cells were quantitated by PhosphorImager analysis. The percentages of Igκ expression in Ed−/− mice were calculated as described in C.
Igκ locus rearrangement is normal in Ed−/− splenic and bone marrow B cells
To further investigate the mechanism responsible for decreased splenic Igκ+ cells in Ed−/−mice, Southern blotting was used to assay for VκJκ rearrangement levels in purified splenic Igκ+ and bone marrow B cells. Electrophoretically resolved genomic DNA from purified cells that had been digested with EcoRI-BamHI was transferred and hybridized with a Cκ probe. In agreement with a previous report (35), the level of germline fragments was approximately 29% in Igκ+ splenic B cells (Fig. 7B, WT), very similar to the levels seen in the corresponding cells from Ed−/− mice (Fig. 7B, 27±3% versus 29±9%, n=3, P=0.65, Student's t test). Related assays on bone marrow B cells also revealed very similar levels of germline fragments from WT and Ed−/− mice that were not statistically different (Fig. 7C, 60±11% versus 51±11%, n=3, P=0.33, Student's t test). To examine the possibility that Jκ region usage and secondary VκJκ rearrangement levels might be altered in Ed−/− mice, we used a quantitative PCR assay to analyze VκJκ rearrangement products in purified splenic Igκ+ and bone marrow B cells. The results revealed that the relative frequencies of Jκ region usage were similar in WT and Ed−/− samples (Fig. 7E). We conclude that Igκ gene rearrangement is unaffected in Ed−/− mice.
FIGURE 7.
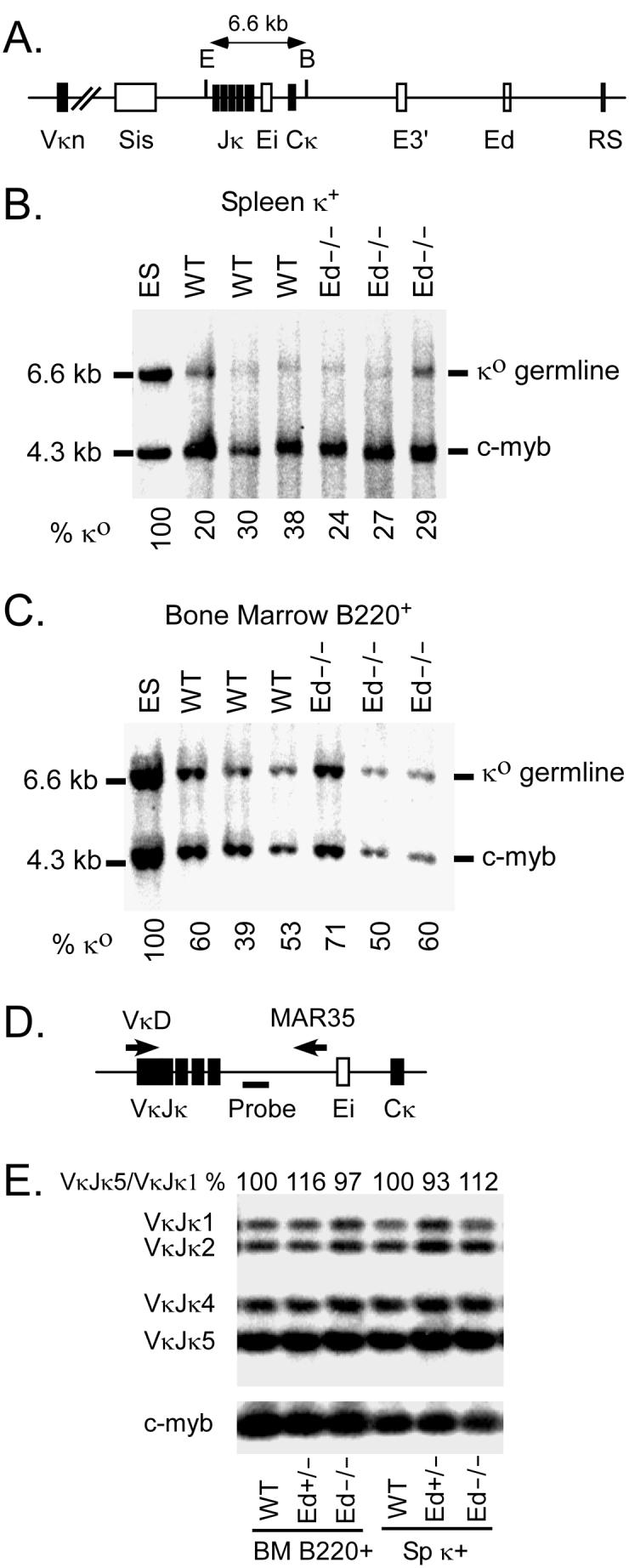
Analysis of Vκ-Jκ rearrangement levels by Southern blotting and quantitative PCR. A, Schematic diagram of the Southern blot assay for Vκ-Jκ rearrangement. E=EcoRI; B=BamHI. B , Electrophorectically resolved genomic DNA from splenic Igκ+ cells that had been digested with EcoRI + BamHI and transferred was hybridized with Cκ and c-myb probes. The amount of EcoRI + BamHI digested DNA was normalized by the c-myb signal. Igκ germline (κo)/c-myb ratio in ES cells was set as 100%. The % κo for each sample were calculated as: [(κo /c-myb in samples)/(κo/c-myb in ES cells)] ×100%. C, Bone marrow B220+ cells genomic DNA was analyzed as described in Panel B. Data in B and C are representative of at least two independent experiments. D, Schematic diagram of the quantitative PCR assay used for VκJκ rearrangement. The positions of VκD and MAR35 primers are indicated by the arrows, and the probe used for the Southern blot is indicated by a thick line. E, Analysis of Jκ regions usage in splenic κ+ and bone marrow B cells. VκJκ rearrangement PCR products were electrophoretically separated on agarose gels and the intensities of VκJκ1 to VκJκ5 bands were quantitated by PhosphorImager analysis of the resulting Southern blot. The VκJκ5/VκJκ1 ratio of WT samples were set as 100%, and the ratio of Ed+/− and Ed−/− samples are shown in the figure. The Southern blot results of PCR amplification of c-myb are shown at the bottom, which were used to control for the amount of genomic DNA template in PCR reactions.
SHM is decreased in the J-C intronic region of Ed−/− GC B cells
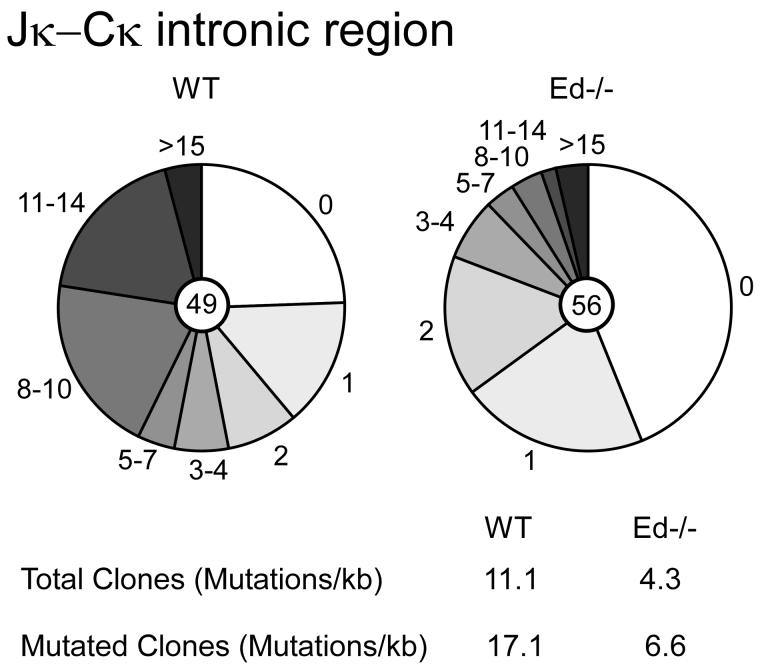
The E box motif has been reported to enhance SHM (36, 37). In addition, the Igκ transcription efficiency has also been correlated with levels of SHM (6). Because Ed contains two E box sites and we also observed reduced Igκ expression in Ed−/− B cells and T-cell-dependant activated B cells, we investigated the possibility that SHM might be decreased in Ed−/− GC B cells. We purified B220+PNAhigh GC B cells from Peyer's patches of Ed−/− and WT mice by flow cytometry. Genomic DNA was isolated from these GC B cells and VκJκ rearrangements were amplified using high fidelity PCR. VκJκ5 rearrangement products were gel purified, cloned and sequenced. We found that the mutation frequency of a 500 bp intronic region downstream of Jκ5 was 11.3 × 10−3 mutations per base in WT GC B cells. In contrast, only 4.3 × 10−3 mutations per base were detected in Ed−/− GC B cells (Fig. 8, P<0.01). The overall mutation frequency in Ed−/− GC B cells was 2.5-fold reduced compared with that of WT GC B cells. Among clones bearing mutations, the mutation frequency was 17.1 × 10−3 mutations per base in WT GC B cells, and only 6.6 × 10−3 mutations per base in Ed−/− GC B cells (Fig. 8). The mutation frequency was also about 2.5-fold reduced in Ed−/− GC B cells. Control experiments revealed that error frequency of the high fidelity PCR was only 0.13 mutations per kb, and the mutation frequency of WT B220+PNAlow B cells was less than 1 mutation per kb (data not shown). Furthermore, we found that the percentage of clones with no mutations was 43% in Ed−/− GC B cells, but only 25% in WT GC B cells. In summary, these data indicate that Igκ SHM is decreased in Ed−/− GC B cells from Peyer's patches.
FIGURE 8.
Analysis of Igκ SHM in GC B cells. B220+PNAhigh GC B cells were sorted from 4-6-month-old WT (left) and Ed−/− (right) mice, and SHM in a 500 bp Jκ-Cκ region that is immediately 3′ of Vκ-Jκ5 recombination products was analyzed. Each pie slice represents the proportions of sequences having the indicated numbers of mutations. The numbers of plasmid clones that were sequenced are shown in pie centers. The mutation frequencies from the total or mutated clones are shown at the bottom.
Discussion
The results of our study complete the picture on the individual roles of the three Igκ transcriptional enhancers in the locus with respect to gene rearrangement, transcription of rearranged genes, and SHM. Unlike MEi and E3′, which each serve a redundant role in triggering V-J joining in the locus at the appropriate stage of B cell development (25-27), our results on Ed−/− mice reveal no defect in gene rearrangement, both by the numbers of Igκ+ cells in bone marrow and germline DNA content in bone marrow and spleen (Fig. 2 & Fig. 7). In addition, the fact that Igκ V-J rearrangement was essentially abolished in MEi and E3′ double knockout mice also supports the conclusion that Ed by itself cannot trigger Igκ gene rearrangement (27). We have also found no defects in RS recombination in Ed−/− mice (data not shown). We conclude that the deficit of Igκ chains in sera (Fig. 5) and in the fraction of Igκ+ cells in the periphery (Fig. 2) in Ed−/− mice must be due to defects in B cell behavior after the cells have left the bone marrow.
We have previously noted that Ed forms a DNase I hypersensitive site in B cell chromatin in plasmacytoma but not pre-B cell lines (4), an observation also consistent with a functional role for Ed later in B cell maturation. These findings prompted us to measure the percentages of plasma cells in spleen and the production of Ig-secreting cells upon stimulating splenic B cells with LPS. However, we found that there was no difference in plasma cell number between WT and Ed−/− cells (Fig. 3C). The deficit of Igκ chains in Ed−/− mice sera can't be explained only by decreased Igκ+ cells in the periphery, because MEi−/− and E3′−/− knockout mice have more dramatic decreases in Igκ+ cells in the periphery than do Ed−/− mice, but Igκ chains in sera of MEi−/− and E3′−/− mice are at concentrations similar to those of WT (25-27). In conclusion, we still cannot rule out the possibility that Ed plays roles in antibody secretion in vivo. T-cell-dependent and independent immunization experiments are required to further elucidate the function of Ed in immune responses and/or antibody secretion.
With respect to transcription levels of rearranged genes in unstimulated B cells, previous studies have shown the MEi−/− mice have no defect in rearranged Igκ gene expression (25, 28). By contrast, lower levels of rearranged gene transcripts exist in splenic B cells of E3′−/− mice (26, 27), or upon their BCR or T-cell-dependent stimulation (28), but not after LPS stimulation (26, 28). Ed−/− mice Igκ gene transcription levels closely fit the same patterns as those exhibited by E3′−/− mice, with moderately reduced cell surface Igκ chains and Igκ gene mRNA levels in splenic κ+ cells, serious deficits in resting splenic B cells transcript levels (Fig. 2C & Fig. 4C, E) and in responsiveness to transcriptional activation by a T-cell-dependent pathway (Fig. 6C, D), but not by the LPS activation pathway (Fig. 4D). We found that in vivo activated Ed−/− splenic B cells only exhibited slightly reduced Igκ gene transcript levels, which can be explained by the fact that part of splenic B cells, maginal zone cells, can be activated by T-cell-independent antigens (38). Our previous studies have shown that the Ed enhancer synergistically activates transcription in combination with other enhancers, like Ei (4), and that Ed forms complexes with Ei and E3′ with the looping out of the intervening DNA in stimulated splenic B cells as well as plasmacytoma cell lines (8). Taken together with the results of the present investigation, it seems clear that E3′ and Ed in particular must work together to optimally set up active chromatin loop domains and to drive high level Igκ gene expression during resting and activated B cell states.
We also studied Igμ transcript levels relative to those of Igκ in WT and Ed−/− mice in various B cell populations. Surprisingly, in cell populations where Igκ transcript levels were compromised in Ed−/− mice, no corresponding down regulation was observed for Igμ transcript levels (Fig. 4 and Fig. 6). This raises the possibilities that overabundant Igμ chains may be cytoplasmic, or associated with up-regulated surrogate light chains on the cell surface, or that Igμ protein levels are stoichiometrically adjusted to be equimolar with Igκ chains by regulation of their translational efficiency.
Studies on cis-acting elements that are involved in specifying SHM in mice were first performed on rearranged Igκ transgenes that lacked Ed (39, 40). These studies found that both MEi and E3′ were required for SHM and that the level of SHM correlated with the level of expression. Another more recent transgenic study, again with a construct lacking Ed, was able to disconnect the correlation between expression level and SHM, because upon mutation of E2A binding sites in the transgene, no affect on transcription was seen but SHM went down by 4-fold (36). This result points to a role for certain transcription factors, and not simply to the levels of RNA polymerase II traversal, in the targeting of the mutator apparatus. The first experiments on SHM in Igκ enhancer knockouts in the native locus were performed in the Alt laboratory. By contrast to the transgenic studies and unexpectedly, E3′−/− mice were found to exhibit no defects in the production of high-affinity Ab when immunized against 2-phenyl-5-oxazolone and these Ab possessed the normal levels of SHM (29). Recognizing that Ag-driven affinity maturation may have selected for survival of this B cell subset, Inlay et al. (28) recently reinvestigated the role of E3′ in the process of SHM under physiological conditions without continuous challenge with a single Ag and did find that E3′ was necessary to achieve WT levels of SHM in the 5′ end of the Igκ gene J-C intron. They further showed that MEi in the native locus played no role in determining the level of SHM. Taking a similar experimental approach to that of Inlay et al. (28), we have found that Ed−/− mice show about a 2.5-fold decrease in the level of SHM relative to those of WT mice (Fig. 8). This fold decline is in the same range as that seen in E3′−/− mice (28). We conclude that both E3′ and Ed contribute to SHM. Notably, both these enhancers contribute to Igκ gene transcription levels and each also have E2A binding sites, making it difficult to judge which of these defects or both might be responsible for a decline in SHM.
In conclusion, our results together with other published studies on endogenous Igκ locus enhancer knockout mice permit the conclusions that MEi and E3′ but not Ed are required for maximal gene rearrangement (25-27), that E3′ and Ed, but not MEi are required for maximal Igκ rearranged gene transcription (25-28), and that E3′ and Ed, but not MEi are required for maximal SHM in rearranged Igκ genes (28). Since E3′ and Ed might have redundant functions to drive Igκ rearranged gene transcription and SHM, one might predict from these combined results that E3′ and Ed double knockout mice might be severely impaired for high level expression of rearranged Igκ genes, and totally compromised with respect to the process of SHM.
Footnotes
This investigation was supported by Grants GM29935 and AI067906 from the National Institutes of Health and Grant I-0823 from the Robert A. Welch Foundation to WTG.
Abbreviation: Ed, downstream enhancer; GC, germinal centers; RS, recombination sequence; RSS, recombination signal sequence; SHM, somatic hypermutation.
References
- 1.Yancopoulos GD, Alt FW. Developmental controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 2.Gellert M. Recombination: RAG proteins, repair factors, and regulation. Ann. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 3.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nature Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 4.Liu ZM, George-Raizen JB, Li S, Meyers KC, Chang MY, Garrard WT. Chromatin structural analyses of the mouse Igκ gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J. Biol. Chem. 2002;277:32640–32649. doi: 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- 5.Schlissel MS. Regulation of activation and recombination of the murine Igκ locus. Immunol. Rev. 2004;200:215–223. doi: 10.1111/j.0105-2896.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 6.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 7.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Garrard WT. Long range interactions between three transcriptional enhancers, active Vκ gene promoters and a 3' boundary sequence spanning 46 kb. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuxa M, Skok JA. Transcriptional regulation in early B cell development. Curr. Opin. Immunol. 2007;19:1–8. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Brekke KM, Garrard WT. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 2004;56:490–505. doi: 10.1007/s00251-004-0659-0. [DOI] [PubMed] [Google Scholar]
- 11.Durdik JM, Moore MW, Selsing E. Novel κ light chain rearrangements in mouse λ light chain producing B lymphocytes. Nature. 1984;307:749–750. doi: 10.1038/307749a0. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Widlak P, Zou Y, Xiao F, Oh M, Li S, Chang MY, Shay JW, Garrard WT. A recombination silencer that specifies heterochromatin positioning and Ikaros association in the immunoglobulin κ locus. Immunity. 2006;24:405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 13.VanNess B, Weigert M, Coleclough C, Mather EL, Kelle DE, Perry RP. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa - C kappa gene. Cell. 1981;27:593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- 14.Martin D, VanNess B. Initiation and processing of two kappa immunoglobulin germline transcripts in mouse B cell. Mol. Cell. Biol. 1990;10:1950–1958. doi: 10.1128/mcb.10.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferradini L, Gu H, Smet AD, Rajewsky K, Reynaud C-A, Weill J-C. Rearrangement-enhancing element upstream of the mouse immunoglobulin kappa chain J cluster. Science. 1996;271:1416–1420. doi: 10.1126/science.271.5254.1416. [DOI] [PubMed] [Google Scholar]
- 16.Max EE, Maizel JV, Jr., Leder P. The nucleotide sequence of a 5.5 kilobase DNA segment containing the mouse κ immunoglobulin J and C region genes. J. Biol. Chem. 1981;256:5116–5120. [PubMed] [Google Scholar]
- 17.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 18.Queen C, Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983;33:741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 19.Xu M, Barnard MB, Rose SM, Cockerill PN, Huang S-Y, Garrard WT. Transcription termination and chromatin structure of the active immunoglobulin κ gene locus. J. Biol. Chem. 1986;261:3838–3845. [PubMed] [Google Scholar]
- 20.Meyer KB, Neuberger MS. The immunoglobulin κ locus contains a second, stronger B cell specific enhancer which is located downstram of the constant region. EMBO J. 1989;8:1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cocea L, DeSmet A, Saghatchian M, Fillatreau S, Ferradini L, Schurmans S, Weill J-C, Reynaud C-A. A targeted deletion of a region upstream from the Jκ cluster impairs κ chain rearrangement in cis in mice and in the 103/bcl2 cell line. J. Exp. Med. 1999;189:1443–1449. doi: 10.1084/jem.189.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Van Ness B. Gene targeting of the KI-KII sequence elements in a model pre-B cell line:effects on germline transcription and rearrangement of the κ locus. Mol. Immunol. 1999;36:464–469. doi: 10.1016/s0161-5890(99)00065-6. [DOI] [PubMed] [Google Scholar]
- 23.Yi M, Wu P, Trevorrow KW, Claflin L, Garrard WT. Evidence that the Igκ gene MAR regulates the probability of premature V-J joining and somatic hypermutation. J. Immunol. 1999;162:6029–6039. [PubMed] [Google Scholar]
- 24.Hale MA, Garrard WT. A targeted κ immunoglobulin gene containing a deletion of the nuclear matrix association region exhibits spontaneous hyper- recombination in pre-B cells. Mol. Immunol. 1998;35:609–620. doi: 10.1016/s0161-5890(98)00060-1. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matix attachment region impairs but does not abolish Vκ-Jκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 26.Gorman JR, Van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Igκ 3′ enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 27.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the κ light chain intronic enhancer and 3′ enhancer in κ rearrangement and demethylation. Nat. Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 28.Inlay MA, Gao HH, Odegard VH, Lin T, Schatz DG, Xu Y. Roles of the Ig kappa light chain intronic and 3′ enhancers in Igκ somatic hypermutation. J. Immunol. 2006;177:1146–1151. doi: 10.4049/jimmunol.177.2.1146. [DOI] [PubMed] [Google Scholar]
- 29.van der Stoep N, Gorman JR, Alt FW. Reevaluation of 3′ Eκ function in stage- and lineage-specific rearrangement and somatic hypermutation. Immunity. 1998;8:743–750. doi: 10.1016/s1074-7613(00)80579-8. [DOI] [PubMed] [Google Scholar]
- 30.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 31.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Bondada S, Robertson DA. Current Protocols in Immunology. John Wiley and Sons, Inc.; New York: 2003. Purification of resting B cells by Percoll gradient centrifugation; pp. 3.8.15–3.8.17. [Google Scholar]
- 33.Schlissel MS, Baltimore D. Activation of immunoglobulin κ gene rearrangement correlates with induction of germline κ gene-transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 35.Zou YR, Takeda S, Rajewsky K. Gene targeting in the Ig kappa locus: efficient generation of lambda chain-expressing B cells, independent of gene rearrangements in Ig kappa. EMBO J. 1993;12:811–820. doi: 10.1002/j.1460-2075.1993.tb05721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 37.Schoetz U, Cervelli M, Wang YD, Fiedler P, Buerstedde JM. E2A expression stimulates Ig hypermutation. J. Immunol. 2006;177:395–400. doi: 10.4049/jimmunol.177.1.395. [DOI] [PubMed] [Google Scholar]
- 38.Fagarasan S, Honjo T. T-independent immune response: new aspects of B cell biology. Science. 2000;290:89–92. doi: 10.1126/science.290.5489.89. [DOI] [PubMed] [Google Scholar]
- 39.Sharpe MJ, Neuberger M, Pannell R, Surani MA, Milstein C. Lack of somatic mutation in a κ light chain transgene. Eur. J. Immunol. 1990;20:1379–1385. doi: 10.1002/eji.1830200625. [DOI] [PubMed] [Google Scholar]
- 40.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger M. Elements regulating somatic mutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]