Abstract
Under conditions of oxidative stress, the formamidopyrimidine lesions (FapyG and FapyA) are formed in competition with the corresponding 8-oxopurines (OG and OA) from a common intermediate. In order to reveal features of the repair of these lesions, and the potential contribution of repair in mitigating or exacerbating the mutagenic properties of Fapy lesions, their excision by three glycosylases, Fpg, hOGG1 and Ntg1, was examined in various base pair contexts under single-turnover conditions. FapyG was removed at least as efficiently as OG by all three glycosylases. In addition, the rates of removal of FapyG by Fpg and hOGG1 were influenced by their base pair partner, with preference for removal when base paired with the correct Watson–Crick partner C. With the FapyA lesion, Fpg and Ntg1 catalyze its removal more readily than OG opposite all four natural bases. In contrast, the removal of FapyA by hOGG1 was not as robust as FapyG or OG, and was only significant when the lesion was paired with C. The discrimination by the various glycosylases with respect to the opposing base was highly dependent on the identity of the lesion. OG induced the greatest selectivity against its removal when part of a promutagenic base pair. The superb activity of the various OG glycosylases toward removal of FapyG and FapyA in vitro suggests that these enzymes may act upon these oxidized lesions in vivo. The differences in the activity of the various glycosylases for removal of FapyG and FapyA compared to OG in nonmutagenic versus promutagenic base pair contexts may serve to alter the mutagenic profiles of these lesions in vivo.
A plethora of modifications of the heterocyclic DNA bases are produced when the biopolymer is exposed to oxidative conditions (1–4). Oxidative DNA base damage and inefficient remediation via repair are intimately linked to the initiation and progression of carcinogenesis (4, 5). The oxidation product of guanine, 7,8-dihydro-8-oxoguanine (OG)1,2 has been the focus of investigations on the effects of oxidative damage (4, 6–9). However, ring-opened formamidopyrimidine lesions, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 4,6-diamino-5-formamidopyrimidine (FapyA) are formed in vitro in yields comparable to the respective 8-oxopurines from common radical intermediates (Figure 1) (10–13). Oxygen-rich conditions favor the 8-oxopurines, while reductive conditions favor production of Fapy lesions (12, 13). For example, FapyG was observed to be the major product in mammalian chromatin or a human leukemia cell line exposed to γ-radiolysis under anoxic conditions (13, 14). In addition, FapyG and OG are formed endogenously in DNA, with the relative amounts highly dependent on the cell type (15). Similarly, oxidation of adenine in mammalian chromatin by γ-radiolysis under O2 deficient conditions produces FapyA in greater amounts than 7,8-dihyro-8-oxoadenine (OA). However, the levels of OA present in cells are an order of magnitude less than OG (14).
Figure 1.

Formation of FapyG and OG from a common intermediate. FapyA and OA are also formed from a common intermediate in an analogous mechanism.
The wealth of attention paid to OG stems in part from the ease with which oligonucleotides with OG at defined positions may be obtained. This is due to the well-established synthetic procedures available to make the phosphoramidite monomer for automated DNA synthesis (16). Information regarding the mutagenic potential of FapyG and FapyA lesions is beginning to emerge due to the recent development of synthetic methods for preparing oligonucleotides containing these lesions at a defined location (17–19). In vitro replication studies using Escherichia coli DNA polymerase I Klenow fragment (KF) show that FapyG is bypassed less efficiently than OG (20, 21). Similar to OG, the presence of FapyG results in misinsertion of dAMP as well as correct insertion of dCMP by KF (20, 22). The KF misinsertion frequency opposite FapyG is also reduced compared to OG (21). Experiments using a template containing FapyA at a defined site show low levels of misincorporation of dAMP and dGMP opposite the lesion by KF exo− (23). The studies with isolated enzymes are consistent with the mutational response to OG and FapyG observed in cellular experiments. Numerous investigations have shown that the presence of OG in bacterial, yeast and mammalian cells mediates G → T transversion mutations (2, 3). The mutagenic potential of the formamidopyrimidine lesions relative to oxopurine lesions was studied in simian kidney cells using a shuttle phagemid vector containing the lesions at defined positions (24). These experiments revealed that FapyG induces G →T mutations at a higher frequency than OG, while FapyA is only weakly mutagenic and induces A → C mutations. In contrast, in E. coli, the frequency of G → T transversions induced by FapyG were minimal, and clearly below the amount observed for OG (21).
Important features that influence the mutagenic potential of a given lesion are the efficiency of its repair and the sensitivity of the repair enzymes to the correct base-pairing context. In E. coli, the mutagenic effects of OG are significantly reduced by the synergistic activities of three enzymes that constitute the “GO” repair pathway (25). Two of these are base excision repair (BER) glycosylases that act upon DNA lesions within duplex DNA. Any OG present in OG:C base pairs is targeted by the formamidopyrimidine-DNA glycosylase (Fpg/MutM). Fpg carries out the first step of BER by catalyzing removal of the damaged OG base. The BER machinery then acts appropriately to reconstitute the original undamaged DNA sequence (26–28). If OG is not removed prior to formation of an OG:A base pair, the MutY adenine glycosylase removes the misincorporated A, providing another opportunity to recreate an appropriate OG:C substrate for Fpg. Functionally similar enzymes are present in most organisms (28).
Fpg was originally discovered on the basis of its ability to mediate the release of ring-opened 7-methylguanine (MeFapyG) from DNA treated with alkylating agents followed by alkali (29). Thus, many reports of BER glycosylases, such as Fpg, have used MeFapy as a convenient model for FapyG (30–33). Other studies utilized DNA that had been treated with ionizing radiation or chemical agents to randomly produce FapyG and FapyA (and many other oxidized bases) in high-molecular-weight DNA (34–37). In addition to the bacterial enzyme Fpg, such studies have revealed a number of glycosylases capable of releasing FapyG or MeFapyG, including the human OG glycosylase (hOGG1) (38), the yeast OG glycosylases, yOGG1 and Ntg1 (yOGG2) (39–41) and the mammalian “Nei-like” enzymes, NEIL1 and NEIL2 (37, 42). Previous work showed that Fpg excises FapyG and FapyA localized at a specific site in a synthetic oligonucleotide duplex (43). While Fpg excises FapyG opposite C approximately 20-fold more efficiently than when opposite A, it indiscriminately removes FapyA opposite the four native nucleotides. A more recent investigation of Fapy lesion repair by mammalian cell extracts using synthetic oligonucleotides revealed that OGG1 and NTH1 are the major DNA glycosylases responsible for removing FapyG and FapyA, respectively (44).
In an effort to further understand features influencing recognition and removal of FapyG and FapyA lesions relative to OG, we examined the base excision activity of three BER enzymes (Fpg, hOGG1 and Ntg1) on synthetic duplexes containing these lesions in various base-pairing contexts. By carrying out experiments under single-turnover conditions, the intrinsic properties of the base recognition and removal process were revealed because complications associated with product release are removed. Indeed, with most BER glycosylases, product release is rate limiting. Therefore, kcat measured under steady-state conditions is primarily a reflection of the rate of product release rather than the steps involved in base excision. The quantitative measurements demonstrate that Fapy lesions are excellent substrates for these repair enzymes. Indeed, FapyG was found to be as good or better as a substrate than OG for all three enzymes. Moreover, these studies revealed subtle differences between the various glycosylases with respect to the selectivity for the base opposite the FapyG and FapyA lesions. Based on the results reported herein, the action of the glycosylase enzymes would be anticipated to alter the mutagenic properties of these lesions.
MATERIALS AND METHODS
General Materials and Instrumentation
Radiolabeling was done using [γ-32P]ATP purchased from Amersham Life Sciences with T4 polynucleotide kinase which was obtained from New England BioLabs. A Milli-Q PF system was used to purify distilled deionized water that was used to make all the buffers. All buffers were passed through a 0.45 μm filter before use. Storage phosphor autoradiography was performed on a Typhoon 9400 phosphorimager system. Data analysis was performed using ImageQuaNT software (version 5.2a), and the rate constants were determined using GraFit 5.0 software. All other chemicals used for these experiments were purchased from Fisher Scientific, VWR, or Sigma. Fpg, hOGG1 and Ntg1 were purified as previously described (45, 46).
Oligonucleotides
The FapyG and FapyA containing oligonucleotides were synthesized as previously described (17–19). The oligonucleotides containing OG and the complementary strands were synthesized at the University of Utah oligonucleotide synthesis core facility. These samples were then purified via HPLC on a Beckman Gold Nouveau system with a Dionex 100 ion exchange column. Sequences of the oligonucleotides used and the relevant duplexes studied are shown in Figure 2.
Figure 2.

Sequence of the duplexes used in this study. Duplex 1 is a 36-base-pair duplex with a centrally located FapyG or OG lesion. Duplex 2 is a 30-base-pair duplex with a centrally located FapyA or OG lesion.
Substrate DNA preparation
For all experiments, 2.5 pmol of the X-containing strand was radiolabeled on the 5′ end using [γ-32P]-ATP by T4 kinase at 37 °C. Excess [γ-32P]-ATP was removed using a Pharmacia Microspin G-50 spin column according to the manufacturer’s protocol. For the glycosylase activity assays, additional nonradioactive X-containing DNA was added to the labeled strand to yield a solution containing 5% labeled DNA. This was then annealed to the complement (added at 20% excess) by heating at 55 °C for 5 min for FapyG and FapyA (to prevent degradation) and 90 °C for OG in annealing buffer (20 mM Tris-HCl (pH 7.6), 10 mM EDTA, and 150 mM NaCl), followed by cooling overnight. To determine the effect of different annealing temperatures on the extent of duplex formation, the OG:A duplex annealed under both conditions was analyzed via native gel (no urea) electrophoresis followed by quantification with storage phosphor autoradiography. This analysis indicated only a slightly lower amount of duplex formed at the lower duplex annealing temperature (98% vs 100%).
Single-Turnover Kinetics
Single-turnover experiments, where [Enz] > [DNA], were performed using the base pair duplexes in Figure 2 to evaluate the glycosylase activity of the enzymes. In each case, the total reaction volume was 60 μL with a final duplex DNA concentration of 20 nM. The duplex was incubated with either 200 nM active Fpg or 100 nM active hOGG1/Ntg1 in an assay buffer (20 mM Tris-HCl, pH 7.6, 10 mM EDTA, 0.1 mg/mL BSA, and 30 mM NaCl, (70 mM NaCl for hOGG1)) at 37 °C. Aliquots were removed at various times (15 s to 60 min) and quenched by the addition of 5 μL of formamide denaturing dye (80% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue in TBE buffer) (for the glycosylase/lyase activity) and placed on dry ice. To measure only the base excision (glycosylase) activity, aliquots were quenched by the addition of 5 μL of 0.5 N NaOH, heated to 55 °C (90 °C for OG) for 2 min and then frozen with dry ice, until ready for gel loading. A control experiment with the OG:A duplex annealed at 55 °C gave identical results.
For reactions under single-turnover conditions in which the glycosylase reaction was too fast to measure manually, a Rapid Quench Flow instrument (RQF-3) from Kintek was used in a manner similar to that previously reported (46, 47). The relevant enzyme was mixed with the DNA duplex (20 nM final). Aliquots were removed at various time points between 0.1 s and 5 min and quenched by the addition of 0.5 M NaOH. The samples were then heated for 2 min at 55 °C for Fapy lesions and 90 °C for OG, and then the formamide denaturing dye was added. The samples were then run on a 15% denaturing polyacrylamide gel in 1X TBE at 1600 V for 2 h. The separation of the product and the substrate was visualized using autoradiography by exposure to a storage phosphor screen overnight. In all cases, the data are reported as the average of at least three separate experiments, and the error is reported as the standard deviation of the sample set.
RESULTS
General Considerations of Glycosylase Assays
The glycosylase activity of the three enzymes was analyzed using the DNA duplexes shown in Figure 2, utilizing methods similar to those reported previously (45–47). In each case, the lesion-containing strand was end-labeled with [γ-32P]-ATP, and the activity was assessed by the extent of strand scission at the lesion site. All three glycosylases have lyase activities that lead to strand scission. The base excision activity can be monitored using the enzyme to provide strand scission or by quenching with NaOH to cleave abasic sites. The glycosylase and lyase activities are tightly coupled in Fpg such that both types of experiments provide identical amounts of strand scission. However, with hOGG1 and Ntg1, a greater extent of strand scission is observed with the NaOH quench, indicating a decoupling of the glycosylase and β-lyase activities, with the former being more efficient. A representative storage phosphor autoradiogram of a PAGE analysis with hOGG1 is shown in Figure 3A and 3B.
Figure 3.

Representative assays for glycosylase/lyase and glycosylase actvity of hOGG1. (A) Storage phosphor autoradiogram of the removal of FapyG opposite A by hOGG1 with a dye quench to determine the glycosylase/lyase activity. Reactions were performed at 37 °C, with a DNA concentration of 20 nM and a hOGG1 concentration of 100 nM (active site concentration). The first lane is a control without the enzyme, and the triangle indicates an increase in incubation time of up to 60 min. “S” and “P” refer to the bands arising from the substrate and product duplex, respectively. (B) Storage phosphor autoradiogram of the removal of FapyG opposite A by hOGG1 with a NaOH quench to determine the glycosylase activity. Reactions were performed at 37 °C, with a DNA concentration of 20 nM and a hOGG1 concentration of 100 nM (active site concentration). The first lane is a control without the enzyme, and the triangle indicates an increase in incubation time of up to 60 min. (C) Representative plot of the removal of FapyG from FapyG:A base-pair-containing duplex 1 by hOGG1 with a dye (red circles) or a NaOH quench (green inverted triangles). The graph was derived from the quantitation of the storage phosphor autoradiograms shown in panels A and B. The data were fit to a single exponential to determine kgl or kg. Rate constants from several experiments are averaged to provide the values in Table 1.
Previous work showed that following the rate of strand scission at OG mediated by Fpg, hOGG1 and Ntg1 under single-turnover conditions ([E] > [DNA]) provides data that follows first-order kinetics and may be fitted to a single exponential (46). Under these conditions the observed rate constant associated with this process (kobs) is equal to kgl (Scheme 1). The rate constant kgl includes the base excision (glycosylase) and the strand scission (β-lyase) reactions. The rate constants kg and kgl are determined from reaction using NaOH or formamide dye quenching, respectively. Notably, Fapy lesions are more sensitive to NaOH treatment at 90 °C than OG. As a result, the quenching steps for all the Fapy lesions were carried out at 55 °C to reduce degradation. Under these conditions, in the absence of a glycosylase enzyme, minimal cleavage at the FapyG lesion site was observed. However, 20–30% of the FapyA substrate strand was cleaved under these conditions. Consequently, the extent of cleavage at the FapyG or FapyA site under the reaction conditions in the absence of the enzyme was subtracted from the amount of cleavage produced in the presence of enzyme to correct for the nonenzyme catalyzed degradation of the substrate.
Scheme 1.
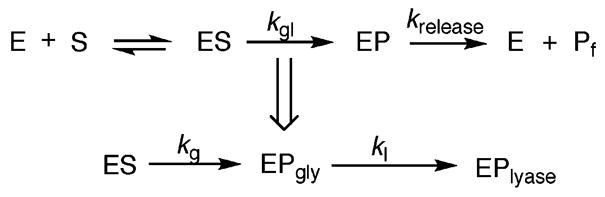
Minimal Kinetic Scheme Used in Analysis of Glycosylase Activity of Fpg, hOGG1 and Ntg1a
a In the case of Fpg, the lyase step is fast so it is not possible to measure the lyase rate. In contrast, glycosylase and lyase steps are uncoupled in hOGG1 and Ntg1. The kg denotes the rate constant for the glycosylase step determined by quenching the reaction with a NaOH/heat treatment. The strand cleavage step is determined by quenching the reaction with a formamide dye treatment and is denoted by kgl. In the case of Fpg, kgl = kg, while with hOGG1 and Ntg1 kgl = kl
Removal of FapyG by Fpg, hOGG1 and Ntg1
Steady-state experiments show that polymerases incorporate dAMP opposite FapyG more readily than opposite G (20). Moreover, mutagenesis studies in COS-7 cells and E. coli indicate that FapyG causes G → T transversions, thus also implicating the formation of FapyG:A base pairs (21, 24). These investigations indicated that base pairs consisting of FapyG opposite C and A are potential substrates for BER enzymes. The efficiency of base-excision of these lesions by the three glycosylases (Fpg, hOGG1 and Ntg1) was determined from single-turnover experiments with duplex 1 containing either FapyG or OG opposite C or A (Table 1). When the lesions are paired with C in this duplex, Fpg catalyzes the removal of FapyG with an ~4-fold faster rate than OG. Quenching with NaOH or relying upon Fpg to provide strand scission gave similar observed rates, consistent with the tight coupling of the enzyme’s glycosylase and lyase activities. With the FapyG:A-containing substrate, the rate constant for base removal (kg = 17 ± 1 min−1) represented a 30-fold increase over the rate constant observed for OG removal from the corresponding OG:A substrate (kg = 0.6 ± 0.1 min−1). A comparison of the relevant rate constants indicates a 5-fold preference of Fpg for removal of FapyG from the FapyG: C- over the FapyG:A-containing duplex. This is compared to a 35-fold greater rate of removal of OG by Fpg from the OG:C over OG:A substrate.
Table 1.
Rate Constants Determined at 37 °C under Single-Turnover Conditions for Fpg, hOGG1 and Ntg1 with FapyG and OG in Duplex 1
| Fpg
|
hOGG1
|
Ntg1
|
|||
|---|---|---|---|---|---|
| substrate | kg (min−1) | kgl (min−1) | kg (min−1) | kgl (min−1) | kg (min−1) |
| FapyG:C | 86 ± 10 | 0.23 ± 0.01 | 57 ± 6 | 0.24 ± 0.04 | 0.9 ± 0.4 |
| FapyG:A | 17 ± 1 | <0.02 | 1.2 ± 0.1 | 0.29 ± 0.05 | 2.4 ± 0.3 |
| OG:C | 21 ± 1 | 0.12 ± 0.06 | 57 ± 7 | MCa | MC |
| OG:A | 0.6 ± 0.1 | <0.01 | <0.02 | MC | MC |
MC: minimal cleavage (<5% after background correction at all time points).
A significant difference between Fpg and hOGG1 or Ntg1 is that the glycosylase (base excision) step is faster than the lyase (strand cleavage) step in the eukaryotic enzymes. This is evident in the differences in the rate constants kg and kgl (Table 1). With hOGG1, the rate constant for FapyG removal from FapyG:C bps (kg = 57 ± 6 min−1) is more than 200-fold greater than the rate constant observed for the combined reaction (kgl = 0.23 ± 0.01 min−1). The difference in the two rate constants indicates that kgl is the rate constant for the β-lyase step. With the hOGG1 enzyme, the rate constant for the FapyG glycosylase activity from FapyG:C is within error of that for OG glycosylase activity from the corresponding OG:C-containing duplex (kg = 57 ± 7 min−1). In contrast, hOGG1 removes FapyG from FapyG:A bps (kg = 1.2 ± 0.1 min−1) with a rate constant that is 60-fold greater than for OG from OG:A base pairs (kg < 0.02 min−1). By comparing the rate constants, FapyG removal by hOGG1 from the FapyG:C substrate is 47-fold more efficient than from the corresponding FapyG:A substrate. The magnitude of the effect of the opposite base for FapyG removal is reduced compared to the activity of hOGG1 on the corresponding OG-containing duplexes, where there is 3000-fold preference of hOGG1 for OG:C over OG:A base pairs.
As reported previously (46), Ntg1 does not catalyze removal of OG opposite either C or A. However, Ntg1 removes FapyG in both base-pairing contexts. As observed with hOGG1, the rate of the glycosylase step with Ntg1 is much faster than the β-lyase step. The rate constant for the glycosylase reaction with FapyG:A (kg = 2.4 ± 0.3 min−1) is almost 8-fold larger than the rate constant for the β-lyase step (kgl = 0.29 ± 0.05 min−1). Interestingly, the enzyme-catalyzed strand scission (kgl) shows no preference for the base opposite the lesion (Table 1). However, in monitoring the glycosylase step, Ntg1 catalyzes removal of the FapyG lesion from the promutagenic FapyG:A base pair 3-fold faster than from the corresponding FapyG:C base pair duplex (kg = 0.9 ± 0.4 min−1).
Removal of FapyA by Fpg, hOGG1 and Ntg1
In vitro experiments show that KF exo− incorporates dAMP and dGMP opposite FapyA more readily than it does opposite A, suggesting that base pairs containing FapyA opposite A, G and T are relevant biological substrates for BER glycosylases (23). The efficiency of base-excision of the FapyA lesion in the potential biological contexts by the OG glycosylases (Fpg, hOGG1 and Ntg1) was determined from single-turnover experiments in the sequence context of duplex 2 (Table 2). In addition, the FapyA excision activity was also evaluated in an FapyA:C base pair context to further elaborate the sensitivity to the opposite base and the potential role of base pair stability. To provide a benchmark for the comparison of the data with FapyG, the rate constants for the corresponding duplex 2 substrates containing OG were also determined.3 Like FapyG, FapyA is a much better substrate for Fpg than OG. However, in contrast to the processing of FapyG, Fpg shows very little discrimination with respect to the identity of the opposing base when excising FapyA. For example, the rate constant for FapyA removal from the FapyA:A substrate (kg = 99 ± 7 min−1) is similar to that measured with the FapyA:T substrate (kg = 103 ± 7 min−1). This feature of the removal of FapyA by Fpg makes the difference in processing of FapyA relative to OG quite different depending on the base pair context being compared. For example, the rate constant for the FapyA glycosylase activity of Fpg with the FapyA:G-containing substrate is more than 4-fold faster than its OG glycosylase activity with OG:G-containing substrate (Table 2). However, relative to the activity of Fpg with OG:A, removal of FapyA when paired with A is 150-fold more efficient.
Table 2.
Rate Constants Determined at 37 °C under Single-Turnover Conditions for Fpg, hOGG1 and Ntg1 with FapyA and OG in Duplex 2
| Fpg
|
hOGG1
|
Ntg1
|
|||
|---|---|---|---|---|---|
| substrate | kg (min−1) | kgl (min−1) | kg (min−1) | kgl (min−1) | kg (min−1) |
| FapyA:G | 64 ± 6 | MCb | < 0.02 | 0.36 ± 0.03 | 69 ± 6 |
| FapyA:A | 99 ± 7 | MC | 0.34 ± 0.04a | 0.4 ± 0.1 | 99 ± 2 |
| FapyA:T | 103 ± 7 | MC | 0.21 ± 0.03a | 0.41 ± 0.02 | 141 ± 8 |
| FapyA:C | 84 ± 2 | 0.05 ± 0.01 | 1.9 ± 0.3 | 0.4 ± 0.1 | 77 ± 5 |
| OG:G | 14 ± 1 | < 0.02 | 0.030 ± 0.002 | 0.04 ± 0.01 | 0.10 ± 0.03 |
| OG:A | 0.57 ± 0.01 | MC | < 0.02 | MC | MC |
| OG:T | 14 ± 1 | 0.08 ± 0.02 | 57 ± 2 | MC | MC |
| OG:C | 14 ± 1 | 0.06 ± 0.01 | 50 ± 7 | MC | MC |
Reaction only 50% complete after 60 min.
MC: Minimal cleavage (<5% after background correction at all time points).
Some activity of removal of FapyA by hOGG1 was observed in all four base pair contexts tested, although the reaction with the FapyA:G duplex was minimal (kg < 0.02 min−1) (Table 2). Notably, the reaction was only observed to go to completion with the FapyA:C duplex. The exact reason for the lack of complete reactions with the FapyA:A and FapyA:T substrates is not clear, but may be a consequence of the slow rate of the reaction and instability of the duplex under the reaction conditions. The rate constant for the glycosylase reaction with the FapyA:C substrate (kg = 1.9 ± 0.3 min−1) is considerably slower than the corresponding rate for removal of OG in the corresponding duplex (kg = 50 ± 7 min−1). As observed in duplex 1, the lyase activity for OG removal by hOGG1 is considerably slower than the glycosylase step. For example, with the OG:T-containing duplex 2, the rate constant for the glycosylase step (kg = 57 ± 2 min−1) is ~600-fold faster than the lyase step (kgl = 0.08 ± 0.02 min−1).
A distinctive feature of the glycosylase activity of Ntg1 is that the activity toward FapyA is greater than toward FapyG. With the FapyA:A-containing duplex, the rate constant for the glycosylase reaction (kg = 99 ± 2 min−1) is nearly 45-fold faster than with the FapyG:A substrate (kg = 2.4 ± 0.3 min−1). Like hOGG1, Ntg1 also catalyzes deglycosylation much more readily than the strand scission reaction. With the FapyA:A substrate, the glycosylase step (kg = 99 ± 2 min−1) is nearly 300-fold faster than the lyase step (kgl = 0.4 ± 0.1 min−1). Similarly, with another biologically relevant base pair substrate, FapyA:G, the glycosylase step (kg = 69 ± 6 min−1) is nearly 200-fold faster than the lyase step (kgl = 0.36 ± 0.03 min−1). In addition, like Fpg, excision of FapyA by Ntg1 is insensitive to the identity of the opposite base.
DISCUSSION
Studies on DNA containing randomly generated lesions established that FapyG and FapyA are substrates for various OG glycosylases (34, 35). However, due to limitations in studying randomly generated lesions, the absolute efficiency of repair of these lesions was unknown. Moreover, the magnitude of the effects of the base opposite the lesions could not be established. Using defined oligonucleotide duplex substrates containing formamidopyrimidine lesions and quantitative kinetics, such features have now been revealed. An important aspect of the analysis of the activity of the glycosylases in this study was the evaluation of the glycosylase reaction separately from the strand scission (lyase) reaction. With Fpg these two reactions are tightly coupled; however, with hOGG1 and Ntg1 the glycosylase step is much faster than the lyase step. In fact, the extent of the activity of these enzymes may be underestimated or differences in substrate preferences may not be revealed if the enzyme is used to provide strand scission. This can be illustrated with hOGG1 in which the relative preference for C over A opposite the lesion is larger when comparing the rate constants for the glycosylase reaction (kg) than the lyase reaction (kgl). Similarly, the preference of Ntg1 for removal of FapyG in the promutagenic FapyG:A bp over the FapyG:C bp is only revealed when analyzing the glycosylase reaction.
Importantly, this approach revealed the robust activity of Fpg and hOGG1 for removal of FapyG lesions. Indeed, the measured rate constants show that Fpg excises FapyG lesions much more efficiently than OG. Similarly, FapyG was removed as efficiently as OG by the human BER glycosylase hOGG1. The fact that both lesions are substrates for Fpg and hOGG1 may be rationalized by the structural similarity between FapyG and OG. Indeed, though the structures of hOGG1 and Fpg are completely different from each other, the strategy for OG recognition by the two enzymes is similar (9). Both enzymes recognize OG by extracting the lesion base from the helix for placement within a lesion specific pocket where contacts are made to NH7 of OG which distinguishes it from G (48, 49). X-ray structural studies of Lactococcus lactis Fpg (LlFpg) bound to lesion-containing DNA have shown that a carbocyclic analogue of FapyG is specifically recognized within the OG base-binding site by residues that are strictly conserved among Fpg enzymes from different species (48, 50). However, the manner of recognition of FapyG within the LlFpg active site was found to be significantly different than that observed for OG recognition by Bacillus stearothermophilus Fpg (BsFpg). The flexibility to adopt alternate recognition complexes may be a feature that enables Fpg to recognize a wide variety of substrates (26). The ability of Fpg to remove FapyA and FapyG with similar efficiencies also is consistent with the idea that the Fpg base recognition site is open and plastic. In contrast, X-ray structural studies of hOGG1 bound to DNA substrates have revealed a rigid, preformed site into which OG fits in snugly (9, 49). There are many contacts with the Watson–Crick face of OG, as well as residues that interact on opposite sides of the planar OG heterocycle to sandwich the base within the active site. Calculations have also indicated the importance of favorable dipole–dipole interactions in hOGG1 recognition of OG (51). The minimal activity of hOGG1 toward removal of FapyA lesions is consistent with a restricted substrate scope that is limited to lesions that are very similar to OG. The “A”-like features of FapyA may not allow for proper engagement in the OG-specific pocket of hOGG1 to allow for efficient excision (9, 51). The fact that FapyA is only removed when paired with C is also consistent with the preference of hOGG1 for lesions derived from G that would be found paired with C.
The identity of the base opposite has a marked influence of the observed rate of lesion excision by Fpg and hOGG1 (Table 1). Single-turnover experiments revealed that both enzymes more readily remove FapyG or OG when paired with C over A. Similar results were previously observed in the steady-state kinetics studies with Fpg with FapyG containing substrates (43). This selectivity for FapyG and OG in the correct Watson–Crick context provides a means of protecting the genome from mutagenesis. Selective removal of FapyG or OG opposite C helps in restoring the original base pair, while slower removal of the lesions opposite A prevents G →T transversion mutations. In the structural studies, hOGG1 and Fpg provide specific contacts to the intrahelical C left behind after expulsion of OG (49, 52). However, the extent of C-specific interactions is more extensive with hOGG1. This is consistent with a more stringent selection against the presence of A opposite OG by hOGG1. This is illustrated by the relative rate constants (Table 1) that demonstrate a preference of hOGG1 for C over A for OG removal that is quite large (~3000-fold). Notably, the relative preference of hOGG1 for C over A opposite FapyG is significantly reduced (47-fold). This is due primarily to the greater ability of hOGG1 to remove FapyG from FapyG:A base pairs than OG from OG:A base pairs. The relative preference of Fpg for C over A when the G lesion is FapyG is also reduced compared to OG; however the magnitude of the difference is not quite as large as was observed with hOGG1.
Though often thought of as an OG glycosylase, Ntg1 shows minimal excision activity with OG (46). However, Ntg1 catalyzes removal of both formamidopyrimidine lesions. In fact the observed rates show that the activity toward the FapyA lesion is quite efficient. The activity of Ntg1 toward FapyG is approximately 100-fold slower than FapyA; however the rate constants measured are similar to those of other glycosylases with standard substrates (46, 47, 53). Ntg1 is similar in terms of sequence and substrate specificity to the E. coli BER glycosylase endonuclease III and its human homologue, NTH1 (26). Endonuclease III has been previously shown to be capable of removal of FapyA and FapyG, with more activity toward FapyA (54). The fact that Ntg1 cleaves FapyG and FapyA is consistent with these lesions resembling pyrimidines. Though no structural data on Ntg1 is available, it is possible that the lack of activity toward OG is due to the inability of a large planar purine heterocycle to fit into the lesion-binding site. Another unusual feature of the glycosylase activity of Fpg and Ntg1 toward FapyA is insensitivity to the identity of the opposite base. For example, FapyA is removed as efficiently when paired with A, G or C as it is when base paired with T. Removal of a lesion from a promutagenic base pair leaves the incorrect nucleotide thus leading to permanent mutations. The fact that FapyA is weakly mutagenic may suggest that repair of this lesion may not be as crucial, and therefore highly discriminatory recognition of this lesion may not have been developed. Indeed, the activity of Fpg and Ntg1 toward removal of FapyA may be a consequence of the structural similarity of this lesion to other substrates (e.g., OG for Fpg and thymine glycol for Ntg1).
A factor that influences the recognition of a lesion and the opposite base is the stability of the lesion-containing base pair. If the lesion base pair is more easily disrupted, this may compensate for the absence of favorable interactions with the proper opposite base. Reduced selectivity for the base opposite a lesion has previously been observed in studies on Fpg excision of the helix-destabilizing hydantoin lesions, guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) (45–47). Duplexes containing the FapyG lesion do not appear to be less stable than those containing OG (20), however, the local stability may be reduced due to the structural flexibility of the ring-opened FapyG relative to the planar OG. This property may also result in reduced stability at the damaged base pair upon interrogation by the glycosylase enzyme. Recent structural studies of BsFpg trapped via disulfide cross-linking to normal nonlesion DNA suggest that this glycosylase actively tests the robustness of base pairs as part of the damage search process (51). Notably, FapyA lesions are more duplex destabilizing than FapyG (23), and this may be a factor resulting in the lack of opposite base sensitivity with both Fpg and Ntg1.
An important aspect of the prevention of mutations associated with OG is the removal of misincorporated adenines from OG:A base pairs by the adenine glycosylase MutY. This enzyme intercepts this promutagenic base pair and allows for recreation of a proper substrate for Fpg. MutY has been shown to remove adenine from FapyG:A base pairs (43). The rate of adenine removal mediated by MutY from FapyG:A bps is not as fast as from OG:A bps, but faster than when G is paired with A. Surprisingly, however, recent in vivo studies in E. coli have shown that FapyG is not particularly mutagenic, and moreover this mutagenesis is not enhanced in mutM– mutY– E. coli. This suggests that the activity of MutY toward FapyG:A base pairs may not be as important for preventing mutagenesis as the activity toward OG:A. The origin of the low mutagenesis of FapyG was proposed to be due to the processing by the DNA polymerase. This proposal was based on detailed analysis of the kinetics of KF exo+ processing of FapyG-containing DNA templates (21). The observed inefficient extension of Fapy-G:A base pairs coupled with the inherently lower insertion frequency would provide an opportunity for the exonuclease activity of the DNA polymerase to correct its mistakes. It is also possible that removal of FapyG may occur via alternative BER glycosylases or other repair pathways (e.g., NER). Redundant mechanisms for preventing mutations from a given lesion may make observing the effects of a specific repair enzyme difficult. Moreover, such redundancies are likely important to mitigate the properties of mutagenic and toxic lesions, like FapyG.
However, the striking observation that FapyG is more mutagenic in simian kidney (COS-7) cells than in E. coli suggests that the processing of FapyG by polymerases and repair enzymes may be distinctly different in eukaryotic cells. Moreover, the presence of FapyG was also more mutagenic than OG in simian kidney cells. This is particularly interesting in light of the more relaxed specificity of hOGG1 for the proper opposite base when the lesion is FapyG. In fact, this may contribute to the higher mutation frequency of FapyG over OG lesions in COS-7 cells. Analysis of the details of the activity of other human glycosylases toward removal of FapyG and the effects of the base opposite may be quite illuminating. In particular, the mammalian NEIL1 glycosylase has been shown to be able to remove Fapy lesions (37). This glycosylase appears to remove mainly damaged pyrimidines (55) and therefore one might find that this enzyme also has activity toward FapyG in promutagenic contexts. Thus, detailed analysis of the kinetics of FapyG with other BER glycosylases in defined base pairing and sequence contexts may provide insight into their role as potential modifiers of mutagenesis caused by FapyG lesions. Moreover, such studies would aid in the design of cellular experiments to evaluate how such effects translate to a cellular environment.
Acknowledgments
We thank Hillary Workman, Andrea Voth and Dr. Margaret McGowan for initial work with the FapyA- and OA-containing lesion substrates.
Footnotes
This work was supported by the National Cancer Institute of the National Institutes of Health CA-90689 (S.S.D.) and CA-074954 (M.M.G.).
Abbreviations: BsFpg, Bacillus stearothermophilus Fpg; BER, base excision repair; bp, base pair; BSA, bovine serum albumin; DTT, dithiothreitol; EDTA, ethylenediamine-tetraacetic acid; exo, exonuclease; FapyA, 4,6-diamino-5-formamidopyrimidine; FapyG, 2,6-di-amino-4-hydroxy-5-formamidopyrimidine; Fpg, formamidopyrimidine glycosylase or MutM; hOGG1, human OG glycosylase; IPTG, isopropyl β-D-thiogalactoside; KF, Klenow fragment of DNA polymerase I; NER, nucleotide excision repair; nt, nucleotide; OA, 8-oxo-7,8-dihydroadenine; OG, 8-oxo-7,8-dihydroguanine; PAGE, polyacrylamide gel electrophoresis; ROS, reactive oxygen species; TBE, tris-borate-EDTA; Tris, tris(hydroxymethyl)aminomethane; yOGG2 (Ntg1), yeast OG glycosylase.
The nucleotides referred to throughout this manuscript are 2′-deoxynucleotides, although not explicitly designated in terms of the abbreviations used. For simplicity’s sake, the same abbreviation is used for the base and the nucleotide. The context makes it clear which is being referred to.
Though FapyA is structurally similar to OA, this was not used as a benchmark since OA was found to be a poor substrate for these glycosylases (Voth, A, David, S. S., unpublished results).
References
- 1.Delaney S, Delaney JC, Essigmann JM. Chemical-Biological Footprinting: Probing Properties of DNA Lesions formed by Peroxynitrite. Chem Res Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neeley WL, Essigmann JM. Mechanisms of Formation, Genotoxicity, and Mutation of Guanine Oxidation Products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 3.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Klaunig JE, Kamendulis LM. The Role of Oxidative Stress in Carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–67. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 5.Cheadle JP, Sampson JR. MUTYH-associated polyposis-From defect in base excision repair to clinical genetic testing. DNA Repair. 2006;6:274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Beckman KB, Ames BN. Oxidative Decay of DNA. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 7.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. Hydroxyl Radicals and DNA Base Damage. Mutation Res. 1999;424:9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 9.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candeias LP, Steenken S. Reaction of HO. with guanine derivatives in aqueous solution:formation of two different redox-active OH-adduct radicals and their unimolecular transformation reactions. Chem Eur J. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg MM. In vitro and in vivo effects of oxidative damage to deoxyguanosine. Biochem Soc Trans. 2004;32:46–50. doi: 10.1042/bst0320046. [DOI] [PubMed] [Google Scholar]
- 12.Xue L, Greenberg MM. Facile quantification of Lesions Derived from 2-deoxyguanosine in DNA. J Am Chem Soc. 2007;129:7010–7011. doi: 10.1021/ja072174n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation-generated free radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 14.Pouget J-P, Douki T, Richard M-J, Cadet J. DNA Damage Induced in Cells by Gamma and UVA Radiation as Measured by HPLC/GCMS and HPLC-EC and Comet Assay. Chem Res Toxicol. 2000;2000:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 15.Jaruga P, Speina E, Gaskowski D, Tudek B, Olinski R. Endogenous oxidative DNA base modifications analysed with repair enzymes and GC/MS technique. Nucleic Acids Res. 2000;28(6):e16. doi: 10.1093/nar/28.6.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodepudi V, Shibutani S, Johnson F. Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8-dihydro-8-oxoadenosine and their incorporation into DNA. Chem Res Toxicol. 1992;5:608–617. doi: 10.1021/tx00029a004. [DOI] [PubMed] [Google Scholar]
- 17.Haraguchi K, Greenberg MM. Synthesis of oligonucleotides containing Fapy·dG (N6-(2-Deoxy-α,α-D-erythro-pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine. J Am Chem Soc. 2001;123:8636–8637. doi: 10.1021/ja0160952. [DOI] [PubMed] [Google Scholar]
- 18.Haraguchi K, Delaney MO, Wiederholdt CJ, Sambandam A, Hantosi Z, Greenberg MM. Synthesis and Characterization of Oligodeoxynucleotides Containing Formamidopyrimidine Lesions and Nonhydrolyzable Analogues. J Am Chem Soc. 2002;124:3263–3269. doi: 10.1021/ja012135q. [DOI] [PubMed] [Google Scholar]
- 19.Jiang YL, Wiederholdt CJ, Patro JN, Haraguchi K, Greenberg MM. Synthesis of Oligonucleotides Containing Fapy·dG(N6-(α′-Deoxy-α,β-D-erythropentofuranosyl)-2,6-di-amino-4-, hydroxy-5-formamidopyrimidine) Using a 5′-Dimethox-ytrityl Dinucleotide Phosphoramidite. J Org Chem. 2005;70:141–149. doi: 10.1021/jo048253o. [DOI] [PubMed] [Google Scholar]
- 20.Wiederholdt CJ, Greenberg MM. Fapy-dG Instructs Klenow Exo- to Misincorporate Deoxyadenosine. J Am Chem Soc. 2002;124:7278. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 21.Patro JN, Widederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the Replication of the Ring Opened Formamidopyrimidine, Fapy·dG in E. coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 22.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 23.Delaney MO, Wiederholt CJ, Greenberg MM. Fapy dA includes nucleotide misincorporation translesionally by a DNA polymerase. Angew Chem, Int Ed. 2002;41:771–773. doi: 10.1002/1521-3773(20020301)41:5<771::aid-anie771>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 24.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Maasaki M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages:comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxopurines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaels ML, Miller JH. The GO system Protects Organisms from the Mutagenic Effect of the Spontaneous Lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved In Base-Excision Repair. Chem Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 27.Fromme JC, Verdine GL. Base Excision Repair, Adv. Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 28.Barnes DE, Lindahl T. Repair and Genetic Consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 29.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3683. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-Oxoguanine (8-Hydroxyguanine) DNA glycosylase and its Substrate Specificity. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eide L, Bjoras M, Pirovano M, Alseth I, Berdal KG, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katafuchi A, Nakanu T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential Specificity of Human and Escherichia coli Endonuclease III and VIII Homologues for Oxidative Base Lesions. J Biol Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 33.Rosequist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair. 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 34.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate Specificity of the Escherichia coli FPG Protein (Formamidopyrimidine-DNA glycosylase): Excision of Purine Lesions in DNA Produced by Ionizing Radiation or Photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 35.Karakaya A, Jaruga P, Bohr VA, Grollman AP, Dizdaroglu M. Kinetics of excision of purine lesions from DNA by Escherichia coli FPG protein. Nucleic Acids Res. 1997;25:474–479. doi: 10.1093/nar/25.3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karahalil B, Girard PM, Boiteux S, Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 1998;26:1228–1232. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaruga P, Birincioglu M, Rosequist TA, Dizdaroglu M. Mouse NEIL1 Protein is Specific for Excision of 2,6-Diamino-4-hydroxy-5-formamidopyrimidine and 4,6-Diamino-5-formamidopyrimidine from Oxidatively Damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 38.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senturker S, van der Kemp PA, You HJ, Doetsch PW, Dizdaroglu M, Boiteux S. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Der Kemp PA, Thomas D, Barbey R, De Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc Natl Acad Sci USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjørås M. The Saccharomyces cerevisiae homologues of Endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol Cell Biol. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and Characterization of a human DNA glycosylase for repair of modified oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiederholdt CJ, Delaney MO, Pope MA, David SS, Greenberg MM. Repair of DNA Containing FapydG and its C-Nucleoside Analogue by Formamidopyrimidine DNA Glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- 44.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 45.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of Hydantoin Products of 8-Oxoguanine Oxidation by the Escherichia coli DNA Repair Enzyme, Fpg. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 46.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Recognition and Removal of Oxidized Guanines in Duplex DNA by the Base Excision Repair Enzymes hOGG1, yOGG1 and yOGG2. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 47.Krishnamurthy N, Muller JG, Burrows CJ, David SS. Unusual Structural Features of Hydantoin Lesions Translate into Efficient Recognition by Escherichia coli Fpg. Biochemistry. 2007;46:9355–9365. doi: 10.1021/bi602459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 49.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 50.Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural Basis for the Recognition of the FapydG Lesion (2,6-Diamino-4-hydroxy-5-formamidopyrimidine) by Formamidopyrimidine-DNA Glycosylase. J Biol Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 52.Fromme JC, Verdine GL. Structural Insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Stuct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 53.Porello SL, Leyes AE, David SS. Single-turnover and Pre-Steady-State Kinetics of the Reaction of the Adenine Glycosylase MutY with Mismatch-Containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 54.Widederholt CJ, Patro JN, Jiang YL, Haraguchi K, Greenberg MM. Excision of formamidopyrimidine lesions by endonuclease III and VII is not a major repair pathway in Escherichia coli. Nucleic Acids Res. 2005;33:331–3338. doi: 10.1093/nar/gki655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]


