Abstract
Brd4 is a member of the BET (bromodomains and extraterminal) subfamily of bromodomain proteins that includes chromatin-modifying proteins and transcriptional regulators. Brd4 has a role in cell cycle progression, making it indispensable in mouse embryos and cultured cells. The N-terminal domain of Brd4 participates in a fusion oncogene. Brd4 associates with acetylated histones in chromatin, and this association persists during mitosis implicating Brd4 in epigenetic memory. Brd4 sequence, particularly the bromodomains and ET domain, is conserved in the zebrafish and Xenopus laevis proteins reported here. Brd4 is expressed and localized on mitotic chromosomes in early zebrafish embryos before and after the midblastula transition (MBT), indicating that the Brd4-chromosome association is a conserved property that is maintained even prior to zygotic transcription. The association of Brd4 with acetylated histones may also be conserved in early embryos as we found that histones H3 and H4 are already acetylated during pre-MBT stages.
Keywords: Bromodomain, midblastula transition, Brd4, zebrafish, Xenopus, acetylated histones, chromosomes
Introduction
The bromodomain is a conserved motif of approximately 110 amino acids that contains four alpha-helices (Haynes et al., 1992; Zeng and Zhou, 2002). It is found in many proteins involved in transcriptional regulation, and in chromatin-associated proteins such as histone acetylases and chromatin remodeling factors (Dhalluin et al., 1999; Winston and Allis, 1999; Aparicio, 2000). The bromodomain binds acetylated lysine residues in histones, and shows specificity for certain modified residues (Jeanmougin et al., 1997; Zeng and Zhou, 2002; Kasten et al., 2004).
Brd4 is a member of the BET subfamily of proteins which contain two copies of the bromodomain and a functionally undefined ET domain (Jeanmougin et al., 1997; Wu and Chiang, 2007). Members of the BET family include Drosophila Fsh (Digan et al., 1986; Haynes et al., 1989), yeast BDF1 (Chua and Roeder, 1995), and mammalian BRD2, BRD3, and BRDT (Jones et al., 1997; Shang et al., 2007). Brd4 has been shown to regulate cell growth in cultured cells (Dey et al., 2000; Maruyama et al., 2002). Supporting the role of Brd4 in growth regulation, brd4 null mice display early embryonic lethality, and heterozygote mutant mice show growth defects associated with reduced cell proliferation (Houzelstein et al., 2002).
A previous study demonstrated that Brd4 dynamically interacts with acetylated histones in interphase nuclei (Dey et al., 2003). Another characteristic feature of Brd4 is its localization on condensed chromosomes during mitosis as its binding to acetylated histones persists (Dey et al., 2000; Dey et al., 2003). Since acetylated histones are generally found in transcriptionally active regions of the genome, Brd4 may recognize acetylated histone codes during interphase and mitosis, thereby maintaining the information in the codes across cell division (Jeanmougin et al., 1997; Strahl and Allis, 2000). A recent study found that Brd4 also associates with meiotic chromosomes (Nagashima et al., 2007). The interaction of Brd4 with mitotic chromosomes is disturbed upon treatment with antimicrotubule drugs such as nocodazole (Maruyama et al., 2002). When nocodazole was removed, Brd4 was reloaded onto the chromosomes, and the cells proceeded to complete division. Therefore, Brd4 also plays an integral part in mitotic progression.
Recently, Brd4 was found to interact with the P-TEFb complex that phosphorylates polymerase II during transcription. Brd4 plays a critical role in stimulating the activity of P-TEFb and in transcription elongation (Jang et al., 2005; Yang et al., 2005). Furthermore, Brd4 interacts with papilloma virus E2 protein, and this association is important for transcriptional regulation and mitotic segregation of viral genomes (You et al., 2004; Wu et al., 2006).
Because of its continued association with mitotic chromosomes, Brd4 may have a role in epigenetic memory. The determination state of many cells in vivo is stable for prolonged periods of time during which the cell may undergo multiple rounds of division. This stability requires information transfer through cell division. As most DNA-binding proteins are released in mitosis, those that remain associated with mitotic chromosomes may transmit information across mitosis. Because Brd4 has been implicated in epigenetic memory, it is interesting to study its behavior in the early embryo of lower vertebrates where zygotic transcription is initiated only after completion of multiple rounds of cell division at the midblastula transition (MBT). For this reason we have isolated the zebrafish and Xenopus brd4 genes, and studied the localization of Brd4 protein in the zebrafish embryo during early embryogenesis. We found that zebrafish Brd4 is co-localized with acetylated histones on mitotic chromosomes even before zygotic transcription is initiated.
Results
Identification of zebrafish and Xenopus Brd4
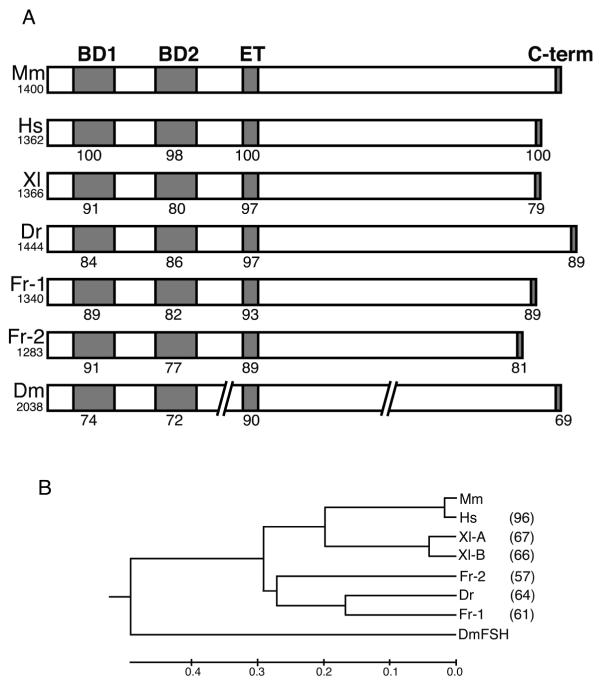
To obtain the brd4 genes from zebrafish and Xenopus we identified EST clones that represent this gene, generated full-length ORFs by RT-PCR, and re-sequenced the genes from both species (see Experimental Procedures). As is common in X. laevis, two closely similar pseudoalleles of the brd4 gene exist in this species. We compare the Xenopus and zebrafish proteins with those from human and mouse as well as two isoforms of Fugu brd4 which we identified in the Fugu genome database. The domain structure of Brd4 is highly conserved, as is the sequence in the two bromodomains and the ET domain (Fig. 1A; Supplemental Fig. 1). Each bromodomain (BD1 and BD2) is more highly conserved between species than the sequence between BD1 and BD2 within the same protein (Supplemental Table 1), as expected from the conservation of domain structure in the BET family from yeast to flies to mammals (Jeanmougin et al., 1997). An additional conserved 11 amino acid region occurs between BD1 and BD2 which includes a nuclear localization signal. In addition, striking homology is found in several patches in the C-terminal domain (Supplemental Fig.1). Among them, the highly conserved sequence at the extreme C-terminus (Fig. 1A) has been shown to interact with the papilloma virus E2 and P-TEFb (You et al., 2004; Bisgrove et al., 2007). Amino acid sequences outside of the conserved domains diverge, particularly in the C-terminal portion of the protein. As a result, the overall sequence identity between species is only 57-67 %, except between mouse and human (Fig. 1B). The zebrafish brd4 gene maps to chromosome 3 near the telomere.
Fig. 1.
(A) Schematic structure of Brd4 proteins, containing two bromodomains, an ET domain, and a C-terminal domain. Amino acid identity with mouse Brd4 is indicated for each domain. Total amino acid length is shown under the name of the species. Mm, mouse; Hs, human; Xl, Xenopus; Dr, zebrafish; Fr, fugu; Dm, Drosophila. The Drosophila sequence is interrupted to indicate its substantially longer length. (B) Phylogenetic tree of Brd4 proteins. Numbers at the right indicate amino acid identity compared to the mouse. No value is entered for Drosophila because of the substantial length differences.
brd4 is widely expressed in zebrafish and Xenopus embryos
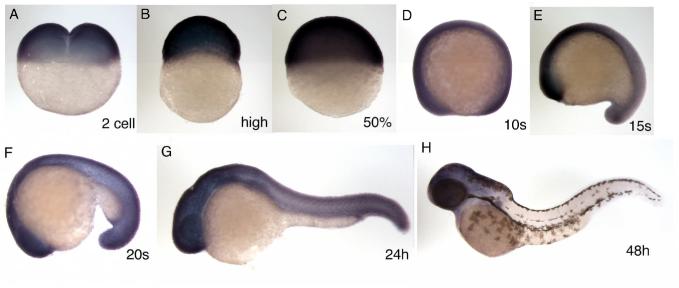
Zebrafish brd4 mRNA was found in the fertilized egg before MBT (Fig. 2A, 2 cell), and was expressed ubiquitously at relatively high levels from gastrulation to mid-somitogenesis stages (Fig. 2C-E). At about the 20 somite stage, brd4 expression started to decrease in the notochord, but remained high in the head and CNS (Fig. 2F). This pattern persisted until about 24 hours post fertilization (hpf) (Fig. 2G). At 48 hpf, brd4 mRNA levels were dramatically reduced in the posterior part of the body, and the signal became restricted to the head area (Fig. 2H). The expression pattern of the Xenopus brd4 gene is similar to that of zebrafish brd4, showing a substantial zygotic component, ubiquitous expression in the early embryo, and later concentration to the head (Supplemental Fig. 2).
Fig. 2.
Expression pattern of the zebrafish brd4 gene. The stage examined by in situ hybridization, carried out as described by (Toyama and Dawid, 1997), is shown in each panel. A, 2 cell stage; B, high stage; C, 50% epiboly; D, 10 somite; E, 15 somite; F, 20 somite; G, 24 hpf; H, 48 hpf. All lateral view, anterior is left (D-H).
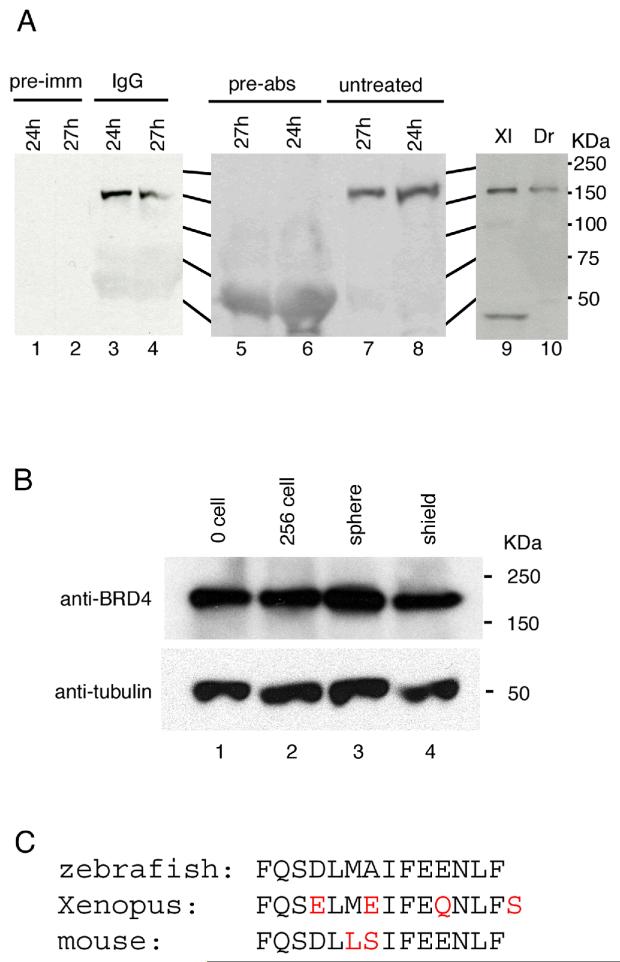
To study the expression of Brd4 protein, we generated a rabbit polyclonal antibody against a peptide containing the C-terminal 14 amino acids of zebrafish Brd4 (Fig. 3C). The antibody recognized an approximately 150 kDa single polypeptide in whole cell extracts of 24 hpf zebrafish embryos (Fig. 3A). To test its specificity, the antibody was pre-incubated with the peptide antigen, resulting in the disappearance of the 150 kDa protein from the Western blot (Fig. 3A). The zebrafish anti-Brd4 antibody detected a band of approximately predicted size in Xenopus extracts (Fig. 3A). Using this antibody we examined the expression of Brd4 protein during early zebrafish embryogenesis; the protein was detected at all stages tested, including the fertilized egg stage (Fig. 3B). Thus, Brd4 protein is present in the egg as is brd4 mRNA. The amount of Brd4 protein appeared relatively constant throughout early embryogenesis (Fig. 3B).
Fig. 3.
Brd4 protein analysis. (A) Antibody characterization. Polyclonal antibody against the Brd4 C-terminal 14 residues detected an approximately150 kD protein in zebrafish embryonic extracts (A, lanes 3, 4, 7, 8, and 10). Pre-immune serum (pre-imm, lanes 1 and 2) did not detect this protein. Antibody pre-incubated with antigen peptide (pre-abs, lane 5 and 6) failed to detect the 150 kD protein. Anti-Brd4 antibody cross-reacts with a protein of predicted size in extracts from Xenopus embryos (lane 9). (B) Brd4 protein during zebrafish embryogenesis. Extracts equivalent to two embryos were loaded in each lane. Lower panel shows α-tubulin as a loading control. Stages are indicated for each lane. (C) The C-terminal amino acid sequence of zebrafish, Xenopus, and mouse is shown with differences in red.
Brd4 protein is localized to the mitotic chromosome
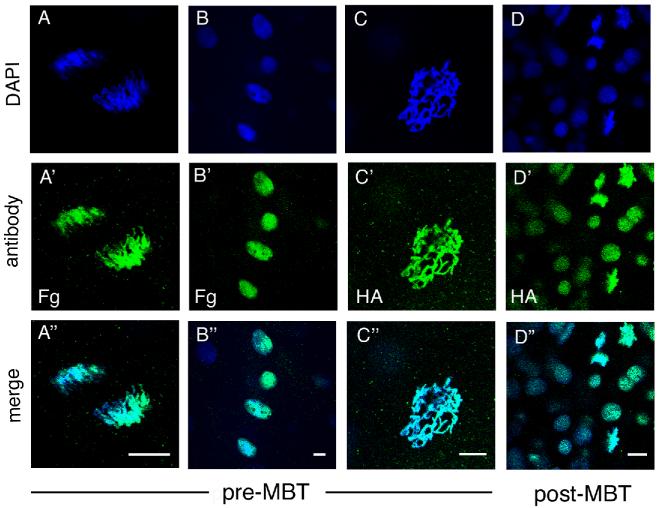
Previous work has shown that Brd4 remains associated with mitotic and meiotic chromosomes in mammalian cells (Dey et al., 2003; You et al., 2004; Nishiyama et al., 2006; Nagashima et al., 2007); this unusual behavior implicates Brd4 in epigenetic memory. Since no zygotic transcription occurs before MBT in the zebrafish embryo, it was of interest to examine the behavior of Brd4 during pre-MBT as compared to post-MBT stages of embryogenesis. In vitro synthesized FLAG- or HA-tagged zebrafish brd4 RNA was injected at the 1-2 cell stage, and Brd4 was visualized with anti-FLAG or anti-HA antibodies in embryos fixed at different stages of development. Confocal microscopy visualized the epitope-tagged Brd4 in nuclei of interphase cells or associated with mitotic chromosomes, as indicated by the coincidence of antibody staining with DAPI staining for DNA (Fig. 4A-D). We found that zebrafish Brd4 was localized in interphase nuclei and was associated with mitotic chromosomes in both pre- and post-MBT stages (Fig. 4A’-D’).
Fig. 4.
Localization of epitope-tagged Brd4 protein. RNA encoding HA- or FLAG-tagged zebrafish brd4 was injected into 1-2 cell stage embryos and visualized by antibody staining. Each column represents a separate embryo at the stage indicated. (A-A” and B-B”): FLAG-Brd4, (C-C” and D-D”): HA-Brd4. (A-C) Pre-MBT (512 cell stage) embryos with mitotic chromosomes (A, anaphase and C, prophase), and interphase nuclei (B) shown. (D) Post-MBT (sphere to dome stage) embryo presents both mitotic chromosomes and interphase nuclei in one embryo. Scale bar: 10 μm.
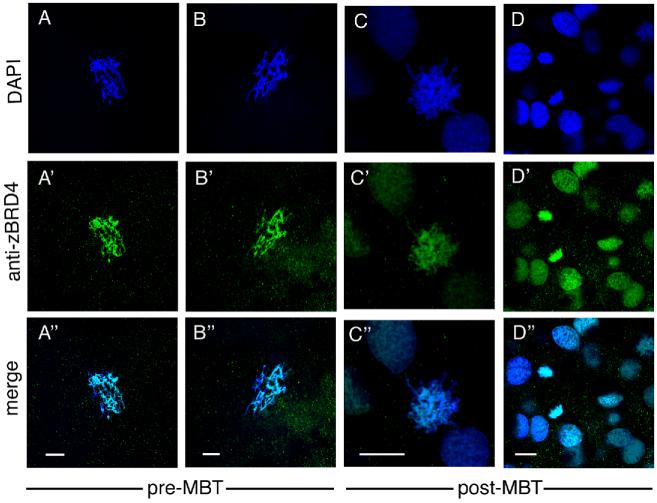
We asked whether endogenous zebrafish Brd4 protein behaves similarly to injected epitope-tagged protein in the early embryo. As illustrated in Figure 5, these experiments confirmed that endogenous Brd4 protein is localized in the nuclei of interphase cells and found associated with mitotic chromosomes before and after MBT in the zebrafish embryo.
Fig. 5.
Localization of endogenous Brd4 protein visualized by staining with anti-zebrafish Brd4 antibody. (A-A” and B-B”): pre-MBT (256-512 cell stage) embryos, (C-C” and D-D”): post-MBT (sphere to dome stage) zebrafish embryos. A-C, prophase. Scale bar: 10 μm.
In all protein staining experiments examining an overexpressed tagged version of zebrafish Brd4 or endogenous Brd4 we analyzed at least 3 embryos per stage and studied more than 20 interphase nuclei and over 10 mitotic chromosome sets per embryo. Since cells divide synchronously in pre-MBT embryos, we observed interphase nuclei and mitotic chromosomes in different embryos. In all cases, we obtained consistent results in that Brd4 was always localized to mitotic chromosomes. Therefore, Brd4 remains associated with mitotic chromosomes regardless of the transcriptional status of the embryo. The localization of Brd4 to mitotic chromosomes in the pre-MBT zebrafish embryo is not a common behavior of chromatin-binding proteins in this organism. We examined the localization of RNA polymerase II in the early zebrafish embryo and observed that this protein does not associate with mitotic chromosomes, both before and after MBT (Supplemental Fig. 3). Therefore, the ability of Brd4 to bind mitotic chromosome appears to be a characteristic feature of this protein.
Brd4 and acetylated histones co-localize on zebrafish chromosomes
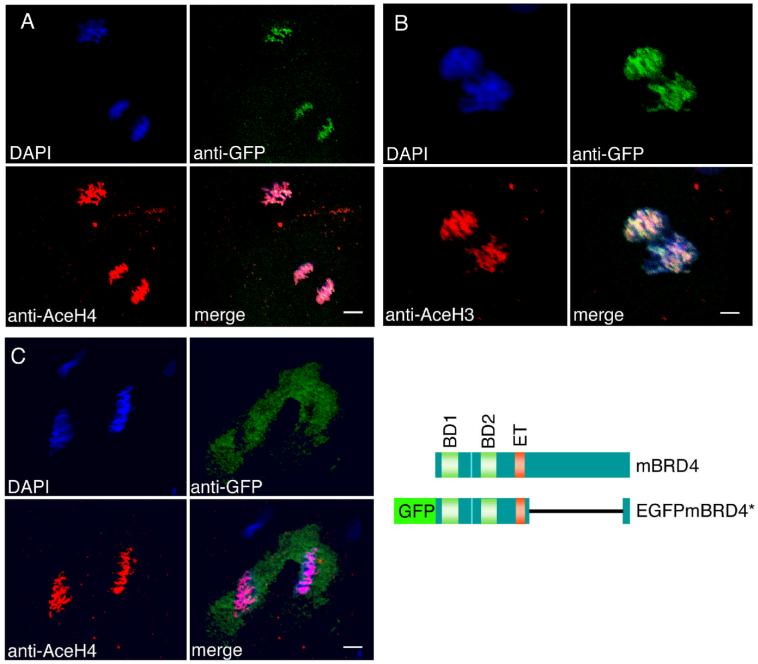
In the mouse, Brd4 binds to acetylated histones in vivo and in vitro (Dey et al., 2003; Nishiyama et al., 2006). To test whether the interaction of Brd4 with mitotic chromosomes in the zebrafish embryo is correlated with histone acetylation, we injected RNA encoding a fusion protein of EGFP and a portion of mouse Brd4 into 1-2 cell embryos. The portion of mouse Brd4 included in this construct retains the bromo- and ET domains and retains its ability to interact with acetylated histones (K.O., unpublished results). Injected embryos were fixed and immunostained with anti-GFP and anti-acetyl-histone H4 or H3 antibody. In both pre-MBT and post-MBT embryos, the fusion protein was associated with mitotic chromosomes and co-localized with acetylated histone H4 (Fig. 6A) and H3 (Fig. 6B). EGFP itself, used as control, did not co-localize with acetylated histones and was not associated with mitotic chromosomes (Fig. 6C).
Fig. 6.
Brd4 protein is co-localized with acetylated histones in zebrafish embryos. (A, B) EGFP-mBrd4* RNA lacking aa 700-1317 was injected into 1-2 cell embryos; EGFP RNA was injected as control in (C). The embryos were fixed before MBT at 256 cell (A, C) and 128-256 cell stage (B), and immunostained with anti-acetyl-histone antibodies (red) and anti-GFP antibodies (green). (A) Anti-tetra-acetyl-histone H4; (B) anti-di-acetyl-histone H3; (C) anti-tetra-acetyl-histone H4. The structures of wild type and fusion brd4 constructs are shown. Scale bar: A and C, 8 μm; B, 4 μm.
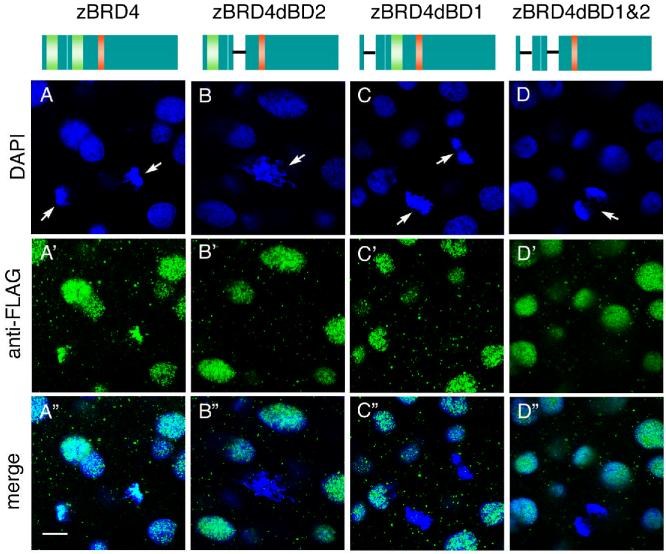
Studies with mammalian cells suggest that the bromodomains mediate the association of acetylated histones with Brd4 (Dey et al., 2003; Nishiyama et al., 2006). To examine whether the association of Brd4 with the chromosomes likewise requires the bromodomains in the zebrafish embryo, we used deletion mutants of zebrafish Brd4 that lacked bromodomain 1 (dBD1), bromodomain 2 (dBD2), or both (dBD1 and 2). RNAs encoding FLAG-tagged versions of the wild type and mutant proteins were injected into 1-2 cell embryos, and the localization of the tagged proteins was examined by staining with anti-FLAG antibody. We studied more than 20 mitotic chromosome sets and at least 60 interphase nuclei in three to five embryos per construct. All mitotic chromosomes found in embryos injected with either of the three mutant constructs (dBD1, dBD2 or dBD1 and 2) were Brd4 negative. In contrast, FLAG-tagged wild type Brd4 was found on 86% of mitotic chromosome sets examined. Therefore, interaction of Brd4 with mitotic chromosomes requires the presence of both bromodomains. All of the proteins examined localized to the interphase nuclei (Fig. 7). Protein localization was unchanged between pre- and post-MBT stages (Fig. 7 and data not shown).
Fig. 7.
Bromodomain-deleted zebrafish Brd4 proteins do not associate with mitotic chromosomes. Constructs are shown schematically at the top: Brd4, FLAG-tagged construct containing both bromodomains; constructs with deletions of bromodomain 1 (dBD1), bromodomain 2 (dBD2), or both bromodomains (dBD1 and 2). RNAs were injected into 1-2 cell stage zebrafish embryos, and stained at the sphere stage with anti-FLAG antibody (green). Arrows indicate mitotic chromosomes. Scale bar: μm.
Discussion
We report here the isolation of the brd4 genes from zebrafish and Xenopus. Brd4 proteins have been well studied in mammals, but have not been previously characterized in lower vertebrates. The BET subfamily, of which Brd4 is a member, also includes BRD2, BRD3, and BRDT in mammals. In searching the EST and genomic databases we found brd2 and 3 in the zebrafish in addition to brd4. Two zebrafish brd2 and two brd3 paralogs were identified, although we could find only a single brd4 gene. It is well known that zebrafish may contain duplicated genes as compared to the mammalian homologs due to an additional genome duplication during evolution (Aparicio, 2000; Van de Peer et al., 2001).
The founding member of the bromodomain gene family, Drosophila fsh, encodes two splice variants, Fsh (S) and Fsh (L) (Digan et al., 1986; Haynes et al., 1989). Fsh (S) lacks the C-terminal region after the ET domain. Overexpression of Fsh (S), but not of Fsh (L), can induce ectopic expression of HOX genes (Chang et al., 2007), suggesting functional differences between the two isofos. It has also been reported that mouse and human Brd4 may exist as short splice variants containing a similar C-terminal truncation, but functional differences between the mammalian splice variants remain to be defined. We have not found any evidence for the existence of brd4 splice variants in zebrafish.
Brd4 interacts with acetylated histones, and these interactions may mediate that association of Brd4 with the chromosomes (Dey et al., 2003; Nishiyama et al., 2006). The association of Brd4 with the chromatin is notable for the fact that this protein remains bound to mitotic chromosomes, leading to the suggestion that Brd4 has a role in epigenetic memory. While most chromatin-associate proteins dissociate during mitosis, the forkhead transcription factor FoxI1 likewise remains bound to the condensed mitotic chromosomes (Yan et al., 2006). ChIP analysis revealed that FoxI1 binds to the consensus sequence characteristic of all Fox proteins (Yan et al., 2006). The known functions of FoxI1 include a requirement in otic placode and jaw development in zebrafish (Nissen et al., 2003; Solomon et al., 2003), but it remains unknown how these functions may relate to the persistent chromosome association of the protein.
Brd4 has a role in cell cycle progression and in transcriptional elongation (Maruyama et al., 2002; Jang et al., 2005; Yang et al., 2005; Bisgrove et al., 2007). Early embryonic development in the zebrafish separates these two processes to the extent that rapid cell division and suppression of transcriptional activity characterize the first three hours of development (Kane and Kimmel, 1993; Yang et al., 2005). Many organisms including Drosophila, Xenopus, and zebrafish, undergo a series of very short and synchronous cell cycles in the period immediately after fertilization. These cell cycles consist of alternating S- and M-phases without G1 and G2 phases and no functional cell cycle checkpoints. The period of rapid division comes to and end with the MBT, which in zebrafish occurs around the tenth cell division. At this time the cell cycle loses its synchrony and lengthens to include G1 and G2 phases, and zygotic transcription initiates (Newport and Kirschner, 1982b; Newport and Kirschner, 1982a; Newport and Dasso, 1989; Clute and Masui, 1995; Clute and Masui, 1997; Ikegami et al., 1997). These changes appear to be linked, as inhibition of zygotic transcription at MBT abolished the appearance of the G phase, while induction of a G1 phase prior to MBT caused premature activation of zygotic transcription (Newport and Kirschner, 1982b; Zamir et al., 1997).
Given these large changes in cell physiology we asked whether Brd4 is present throughout early embryogenesis and whether there are any changes in its association with the chromosomes. We find that both brd4 mRNA and Brd4 protein are stored in the egg and continue to be present throughout embryogenesis. This behavior is shared with many other macromolecules and even cell organelles whose storage in the egg may be the basis for the embryo’s ability to undergo very rapid cleavage divisions and enter morphogenesis within half a day after fertilization. More interesting than just the presence of Brd4 in the egg is the fact that this protein associates with mitotic chromosomes both before and after MBT (Figs. 4, 5), a behavior not shared by other DNA-binding proteins such as RNA polymerase II (Supplemental Fig. 3). Association of Brd4 with mitotic chromosomes has led to the hypothesis that Brd4 has a role in epigenetic memory (Dey et al., 2003). Brd4 binds specifically to certain acetylated histones, and this binding is required for its chromatin association (Dey et al., 2003). Little is known about the status of histone modifications during pre-MBT development. Despite the lack of zygotic transcription, we observed that lysine residues of histone H3 and H4 were acetylated, some as early as the 4 cell stage (data not shown). Very early acetylation was detected on histone H4 Lys8 and Lys12, followed by Lys5. Interestingly, these are the acetyl-lysine residues that mediate preferential Brd4 binding in vitro (Dey et al., 2003). Thus early histone acetylation may mediate Brd4-chromosome association in the embryo. It is possible that Brd4 bound to acetylated histones marks those genes that are slated to be transcribed at the MBT. On the other hand, the early Brd4-chromosome interactions may simply function to support the precision of mitotic progression, a possibility suggested by the fact that Brd4 has a role in protecting cells against mitotic stress (Kanno et al., 2004; Nishiyama et al., 2006).
Although the bulk of zygotic transcription does not start until MBT, there are some notable exceptions. Zebrafish bozozok and Churchill mRNA were found to be transcribed in pre-MBT embryos (Leung et al., 2003; Londin et al., 2007), and a recent global gene expression analysis reported a significant number of additional transcripts that are synthesized before MBT (Mathavan et al., 2005). Selective pre-MBT transcription has also been reported in Xenopus (Yang et al., 2002). It is possible that the association of Brd4 with chromosomes in cleavage stage embryos is involved in these early transcription events. While pre-MBT transcription does occur, it is the exception rather than the rule, and thus the question arises why acetylated histones that are associated with transcriptionally active chromatin and Brd4 should be localized on chromosomes before the onset of most zygotic transcription. No definitive answer is available to this question, but we suggest that this apparently premature assembly of components is another manifestation of the need to develop quickly. Just as storing many macromolecules and organelles in the egg allows the zygote to get off to a fast start, so the assembly of transcriptionally competent domains before the start of transcription will allow the embryo to make RNA quickly once it has reached MBT.
Experimental Procedures
Isolation of zebrafish and Xenopus brd4
In both species, a search of the EST databases led to sequences of parts of the brd4 gene. PCR-based techniques were used to isolate the remaining portions of each gene which were then sequenced. The sequences were submitted under Accession Numbers: EU236152, zebrafish; EU236153, Xenopus A; EU236154, Xenopus B. The Fugu brd4 genes were identified in the genome sequence through Blast searches in the Fugu genome database (http://genome.jgi-psf.org/Takru4/Takru4.home.html). The sequence of one of the genes required a correction of the splice sites, see legend to Supplemental Fig. 1. The chromosomal position of the zebrafish brd4 gene was analyzed by using the Sanger center web site (http://vega.sanger.ac.uk/Danio_rerio/index.html). Sequence comparisons were done with Clustal W methods to identify conserved domains.
Expression constructs and RNA synthesis
The EGFP fusion constructs of full length murine brd4 (mBrd4) cDNA were previously described (Maruyama et al., 2002). The EGFP-brd4 coding sequences were subcloned into the pCS2 vector. The entire zebrafish brd4 coding region was cloned into the pCS2 vector and HA- or FLAG-epitopes were fused to the N-terminus. Zebrafish bromodomain deletion constructs (lacking bromodomain 1 (dBD1), bromodomain 2 (dBD2), or both (dBD1 and 2) were generated from a FLAG-tagged construct; dBD1 lacks amino acid 1 to 152; dBD2 lacks amino acids 356-473; dBD1 and 2 lacks 1-152 and 356-473. Deletions were introduced by using appropriate PCR primers, and the nucleotide sequences of the regions generated by PCR were confirmed by sequencing. RNA was synthesized using mMESSAGE mMACHINE (Ambion/Applied Biosystem) and injected into 1-2 cell stage zebrafish embryos.
Immunostaining
Embryos were fixed with 4% paraformaldehyde at 4°C overnight, manually dechorionated and stored in methanol at -20°C. Embryos were rehydrated in a methanol/PBST0.1 (PBS, 2% BSA, 0.1% Triton X100) series, and incubated with blocking solution (10% lamb serum in PBST0.1) at room temperature for at least one hour prior to incubation with the primary antibody. Anti-di-acetyl histone H3 antibody (Upstate 07-108) and Anti-tetra-acetyl histone H4 antibody (Upstate, 06-598) were used at 1:200 and 1:100 dilution, respectively. Anti-GFP antibody was obtained from SantaCruz (sc-9996) and used at a 1:250 dilution. Secondary antibodies (Alexa Fluor 488 and 568; Molecular Probe/Invitrogen) were used at a dilution of 1:1000. Staining was observed with a Leica SP2 or Zeiss LSM510 confocal microscope. In some cases, contrast was adjusted in Photoshop for the entire image.
Generation of zebrafish Brd4 antibody
Rabbit polyclonal antibody against the C-terminal 14 amino acids (FQSDLMAIFEENLF) of zebrafish Brd4 protein was generated by Spring Valley Laboratories, Inc. After fixation, embryos were stored in PBST0.1 at 4°C (no methanol treatment). Samples were treated with blocking solution at room temperature for at least one hour prior to incubation with 1:1000 diluted zebrafish anti-Brd4 IgG fraction as primary antibody.
Western blotting
Embryonic extract was prepared by homogenizing embryos in NP40 extraction buffer (10 mM Tris-HCl, pH 7.5, 137 mM NaCl, 5 mM EDTA, 0.5% NP40, protease inhibitor (Roche)). Anti-zebrafish Brd4 was used at a dilution of 1:20,000-50,000. Anti-mouse α-tubulin was obtained from Calbiochem (Cat. number CP06) and used at 1:4000.
Supplementary Material
Amino acid sequence comparison of Brd4 proteins among different species. Conserved amino acids are highlighted in gray. Conserved domains are color-coded in the zebrafish sequence: bromodomain 1, blue; bromodomain 2, green; ET domain, orange; C terminal domain, red. Mm, mouse; Hs, human; Xl, Xenopus; Dr, zebrafish; Fr, fugu; Dm, Drosophila.
The sequence of one of the genes (Fr-1) required a correction of splice sites. Based on the predicted exon-intron boundaries (http://genome.jgi-psf.org/Takru4/Takru4.home.html), the amino acid sequence of the first half of bromodomain 2 differed from that of other species (splice accepter-donor sites on genomic sequence of Fr-1: 757457-757402, 757229-757176). We examined nucleotide sequences of this corresponding region manually and found alternative splice junctions which resulted in an amino acid sequence similar to those in other species (splice accepter-donor sites on genomic sequence of Fr-1: 757266-757188). Since bromodomain 2 is highly conserved among different species, we decided to use the revised sequence for comparison.
Expression pattern of the Xenopus brd4 B gene. A, gastrula at stage 10.5, animal pole view; B, st 14; C, st 17; D, st 23; E, st 37. (B-E) lateral view, anterior is left.
RNA polymerase II does not associate with mitotic chromosomes in pre-MBT or post-MBT embryos. Zebrafish embryos were fixed at the 256 cell and sphere stages. Mouse monoclonal anti-RNA polymerase II antibody (8WG16) was obtained from Covance. (A-A” and B-B”) pre-MBT (256 cell), (C-C”) post-MBT (sphere). (A-C) DAPI staining, (A’-C’) anti-RNA polymerase II, (A”-C”) merged image. (A-A”) Interphase nuclei, (B-B”) mitotic chromosomes. Scale bar: 10 μm.
Amino acid sequence comparison of BD1 to BD2 in mouse Brd4 and zebrafish Brd4. The bromodomain sequences are more conserved among species than between BD1 and BD2 within one Brd4 protein.
Acknowledgments
We thank Mark Rath for excellent technical assistance.
This research was supported by the Intramural Research Program of the NICHD, National Institutes of Health.
Contributor Information
Reiko Toyama, Laboratory of Molecular Genetics, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, Bethesda, MD, USA.
Martha L. Rebbert, Laboratory of Molecular Genetics, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, Bethesda, MD, USA
Igor B. Dawid, Laboratory of Molecular Genetics, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, Bethesda, MD, USA
References
- Aparicio S. Vertebrate evolution: recent perspectives from fish. Trends Genet. 2000;16:54–6. doi: 10.1016/s0168-9525(99)01934-4. [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–5. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YL, King B, Lin SC, Kennison JA, Huang DH. A double-bromodomain protein, FSH-S, activates the homeotic gene ultrabithorax through a critical promoter-proximal region. Mol Cell Biol. 2007;27:5486–98. doi: 10.1128/MCB.00692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P, Roeder GS. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol. 1995;15:3685–96. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P, Masui Y. Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Dev Biol. 1995;171:273–85. doi: 10.1006/dbio.1995.1280. [DOI] [PubMed] [Google Scholar]
- Clute P, Masui Y. Microtubule dependence of chromosome cycles in Xenopus laevis blastomeres under the influence of a DNA synthesis inhibitor, aphidicolin. Dev Biol. 1997;185:1–13. doi: 10.1006/dbio.1997.8540. [DOI] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–63. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–49. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–6. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Digan ME, Haynes SR, Mozer BA, Dawid IB, Forquignon F, Gans M. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev Biol. 1986;114:161–9. doi: 10.1016/0012-1606(86)90392-1. [DOI] [PubMed] [Google Scholar]
- Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SR, Mozer BA, Bhatia-Dey N, Dawid IB. The Drosophila fsh locus, a maternal effect homeotic gene, encodes apparent membrane proteins. Dev Biol. 1989;134:246–57. doi: 10.1016/0012-1606(89)90094-8. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami R, Rivera-Bennetts AK, Brooker DL, Yager TD. Effect of inhibitors of DNA replication on early zebrafish embryos: evidence for coordinate activation of multiple intrinsic cell-cycle checkpoints at the mid-blastula transition. Zygote. 1997;5:153–75. doi: 10.1017/s0967199400003828. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–34. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–3. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- Jones MH, Numata M, Shimane M. Identification and characterization of BRDT: A testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics. 1997;45:529–34. doi: 10.1006/geno.1997.5000. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. Embo J. 2004;23:1348–59. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Soll I, Arnold SJ, Kemler R, Driever W. Direct binding of Lef1 to sites in the boz promoter may mediate pre-midblastula-transition activation of boz expression. Dev Dyn. 2003;228:424–32. doi: 10.1002/dvdy.10408. [DOI] [PubMed] [Google Scholar]
- Londin ER, Mentzer L, Gates KP, Sirotkin HI. Expression and regulation of the zinc finger transcription factor Churchill during zebrafish development. Gene Expr Patterns. 2007;7:645–50. doi: 10.1016/j.modgep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22:6509–20. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–76. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima T, Maruyama T, Furuya M, Kajitani T, Uchida H, Masuda H, Ono M, Arase T, Ozato K, Yoshimura Y. Histone acetylation and subcellular localization of chromosomal protein BRD4 during mouse oocyte meiosis and mitosis. Mol Hum Reprod. 2007;13:141–8. doi: 10.1093/molehr/gal115. [DOI] [PubMed] [Google Scholar]
- Newport J, Dasso M. On the coupling between DNA replication and mitosis. J Cell Sci Suppl. 1989;12:149–60. doi: 10.1242/jcs.1989.supplement_12.13. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–86. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30:687–96. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Dey A, Miyazaki J, Ozato K. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol Biol Cell. 2006;17:814–23. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess SM. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–54. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–15. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Kudoh T, Dawid IB, Fritz A. Zebrafish foxi1 mediates otic placode formation and jaw development. Development. 2003;130:929–40. doi: 10.1242/dev.00308. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Toyama R, Dawid IB. lim6, a novel LIM homeobox gene in the zebrafish: comparison of its expression pattern with lim1. Dev Dyn. 1997;209:406–17. doi: 10.1002/(SICI)1097-0177(199708)209:4<406::AID-AJA8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Taylor JS, Braasch I, Meyer A. The ghost of selection past: rates of evolution and functional divergence of anciently duplicated genes. J Mol Evol. 2001;53:436–46. doi: 10.1007/s002390010233. [DOI] [PubMed] [Google Scholar]
- Winston F, Allis CD. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6:601–4. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–96. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26:155–68. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–52. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–60. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol Cell Biol. 1997;17:529–36. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–8. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence comparison of Brd4 proteins among different species. Conserved amino acids are highlighted in gray. Conserved domains are color-coded in the zebrafish sequence: bromodomain 1, blue; bromodomain 2, green; ET domain, orange; C terminal domain, red. Mm, mouse; Hs, human; Xl, Xenopus; Dr, zebrafish; Fr, fugu; Dm, Drosophila.
The sequence of one of the genes (Fr-1) required a correction of splice sites. Based on the predicted exon-intron boundaries (http://genome.jgi-psf.org/Takru4/Takru4.home.html), the amino acid sequence of the first half of bromodomain 2 differed from that of other species (splice accepter-donor sites on genomic sequence of Fr-1: 757457-757402, 757229-757176). We examined nucleotide sequences of this corresponding region manually and found alternative splice junctions which resulted in an amino acid sequence similar to those in other species (splice accepter-donor sites on genomic sequence of Fr-1: 757266-757188). Since bromodomain 2 is highly conserved among different species, we decided to use the revised sequence for comparison.
Expression pattern of the Xenopus brd4 B gene. A, gastrula at stage 10.5, animal pole view; B, st 14; C, st 17; D, st 23; E, st 37. (B-E) lateral view, anterior is left.
RNA polymerase II does not associate with mitotic chromosomes in pre-MBT or post-MBT embryos. Zebrafish embryos were fixed at the 256 cell and sphere stages. Mouse monoclonal anti-RNA polymerase II antibody (8WG16) was obtained from Covance. (A-A” and B-B”) pre-MBT (256 cell), (C-C”) post-MBT (sphere). (A-C) DAPI staining, (A’-C’) anti-RNA polymerase II, (A”-C”) merged image. (A-A”) Interphase nuclei, (B-B”) mitotic chromosomes. Scale bar: 10 μm.
Amino acid sequence comparison of BD1 to BD2 in mouse Brd4 and zebrafish Brd4. The bromodomain sequences are more conserved among species than between BD1 and BD2 within one Brd4 protein.