Abstract
The zebrafish u-boot (ubo) gene encodes the transcription factor Prdm1, which is essential for the specification of the primary slow-twitch muscle fibres that derive from adaxial cells. Here, we show that Prdm1 functions by acting as a transcriptional repressor and that slow-twitch-specific muscle gene expression is activated by Prdm1-mediated repression of the transcriptional repressor Sox6. Genes encoding fast-specific isoforms of sarcomeric proteins are ectopically expressed in the adaxial cells of ubotp39 mutant embryos. By using chromatin immunoprecipitation, we show that these are direct targets of Prdm1. Thus, Prdm1 promotes slow-twitch fibre differentiation by acting as a global repressor of fast-fibre-specific genes, as well as by abrogating the repression of slow-fibre-specific genes.
Keywords: skeletal muscle, fibre type, Blimp1/Prdm1, Sox6, zebrafish, u-boot
Introduction
During vertebrate development, cells become committed to the myogenic fate through the activation of myogenic regulatory factors in the paraxial mesoderm. Subsequently, the committed cells or myoblasts differentiate into muscle fibres with distinct contractile speeds, the so-called slow- and fast-twitch fibres. Terminal differentiation of these different fibre types requires the expression of specific isoforms of sarcomeric proteins, such as the myosin light and heavy chains (MyLC and MyHC, respectively) and troponins. In the zebrafish embryo, progenitors of the slow- and fast-twitch fibres can be identified on the basis of their morphology and positioning within the segmental plate before somitogenesis (Devoto et al, 1996). Paraxial mesodermal cells that lie in direct contact with the notochord—designated adaxial cells—differentiate into slow myoblasts that migrate out through the developing myotome to form a superficial layer of slow-twitch fibres. These slow fibres are mononuclear, span the entire length of each somite and express the homeodomain protein Prox1, as well as the slow myosin heavy chain 1 (smyhc1) and the slow-specific Troponin C (stnnC; Devoto et al, 1996; Roy et al, 2001; Xu et al, 2006). The fast-twitch fibres derive from more laterally located paraxial mesodermal cells that start to differentiate in the wake of the migrating slow myoblasts (Blagden et al, 1997). In contrast to the slow myoblasts, fast-twitch myoblasts undergo fusion to generate multinucleated fibres (Moore et al, 2007; Srinivas et al, 2007) and express fast MyLC and MyHC isoforms, as well as troponin T3a (tnnt3a) and troponin I2 (tnni2) (Xu et al, 2000; Hsiao et al, 2003).
The specification of adaxial cells to follow the slow-twitch fibre differentiation programme depends crucially on inductive signals from the notochord and floorplate mediated by members of the Hedgehog protein family (Currie & Ingham, 1996; Blagden et al, 1997; Du et al, 1997; Lewis et al, 1999; Barresi et al, 2000; Wolff et al, 2003). Reception of the Hedgehog signals by adaxial cells results in the activation of transcription of the u-boot (ubo) gene, the function of which is both necessary and sufficient to drive slow-twitch differentiation in myoblasts (Roy et al, 2001; Baxendale et al, 2004). In ubotp39 mutants, presumptive slow-twitch fibres lose sMyHC and Prox1 expression, and seem to differentiate into fast-twitch fibres (Roy et al, 2001). The ubo gene encodes the B-lymphocyte-induced maturation protein Blimp1 or Prdm1, a PR-domain-containing protein, which, in mammals, is involved in the terminal differentiation process of B lymphocytes, the response to viral infection and primordial germ cell specification (Keller & Maniatis, 1991; Turner et al, 1994; Ohinata et al, 2005; Kallies & Nutt, 2007). In these contexts, Prdm1 has been shown to mediate transcriptional repression, acting as a scaffold that recruits co-repressors and chromatin-modifying enzymes to specific target genes (Yu et al, 2000; Gyory et al, 2004). Here, we investigate the nature of Prdm1 function and the regulatory networks underlying fibre type specification, and identify several direct targets of Prdm1.
Results And Discussion
Adaxial cells transform from slow into fast in ubotp39 mutants
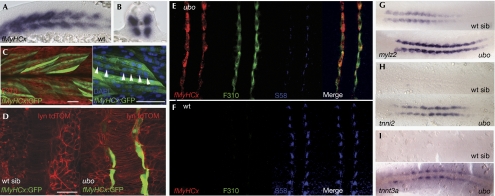
In wild-type embryos, adaxial cells are characterized by their expression of a slow isoform of the MyHC—detected by the S58 antibody—but are devoid of staining with F310—a fast MyLC isoform-specific antibody. Differentiation of adaxial cells into slow-twitch muscle starts several hours earlier than that of the fast-twitch fibres. Previous studies of ubo mutants have shown that loss of Prdm1 expression causes adaxial cells to transform from slow- to fast-twitch character (Roy et al, 2001). Consistent with this, we found that adaxial cells were labelled with F310 in ubotp39 homozygotes well before the normal onset of fast-muscle differentiation. To confirm that this represents the precocious differentiation of adaxial cells into fast-twitch fibres, we constructed a reporter gene, using sequences upstream from the previously uncharacterized fMyHC gene, which drives green fluorescent protein (GFP) expression strictly in fast-muscle cells (Fig 1A–C). When injected into ubotp39 mutant embryos, this fMyHCx:GFP transgene was ectopically expressed in adaxial cells (Fig 1D), indicative of their transformation from slow to fast character (Fig 1E,F). By using in situ hybridization (ISH), we found that tnnt3a and tnni2, encoding fast-specific isoforms of troponin, were also ectopically expressed in the adaxial cells of ubotp39 mutants, whereas expression of the gene encoding MyLC (mylz2) was significantly elevated above the levels found in wild-type adaxial cells (Fig 1G–I). Thus, Prdm1 acts to repress fast-specific genes, as well as to promote expression of slow-specific genes in adaxial cells.
Figure 1.
Fast-muscle-fibre-specific genes are ectopically expressed in the slow domain of ubotp39 mutant embryos. In situ hybridization showing fast-muscle-specific expression of the fast myosin heavy chain gene (fMyHCx) at the 18-somite stage in (A) lateral and (B) transverse views. (C) An fMyHCx:GFP construct expresses GFP specifically in the fast-muscle fibres in the trunk. Transient expression of fMyHCx:GFP in the fast fibres in a 30 hpf (hours post fertilization) embryo, as shown by the colocalization with the fast-fibre marker F310 antigen. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) to highlight the polynuclear arrangement (arrowheads) of the fast fibres expressing fMyHCx:GFP. (D) The construct fMyHCx:GFP is detected in the adaxial cells of ubotp39 mutants in transient assays. The fMyHCx:GFP construct is not expressed at the 12-somite stage in wild-type embryos, but is ectopically expressed in the adaxial cells of a 12-somite-stage ubotp39 embryo. Lyn tdTomato (red) was used to highlight cellular membranes and the outline of the notochord. (E,F) Ectopic expression of fMyHCx messenger RNA (red) and F310 (green) is detected in the adaxial cells of a 12-somite-stage ubotp39 embryo (100% of embryos assayed; n>30). In wild-type siblings (wt sib), adaxial cells are only labelled with the slow MyHC S58 (blue) antibody. Genes that normally are expressed exclusively in the fast-muscle domain are ectopically expressed in the adaxial cells of the ubotp39 mutant: (G) mylz2, (H) tnni2 and (I) tnnt3a. Scale bars, 25 μm. GFP, green fluorescent protein; mylz2, myosin light chain 2; tnni2, troponin I2; tnnt3a, troponin T3a; ubo, u-boot; wt, wild type.
Prdm1 acts as a repressor to promote slow twitch fibre type
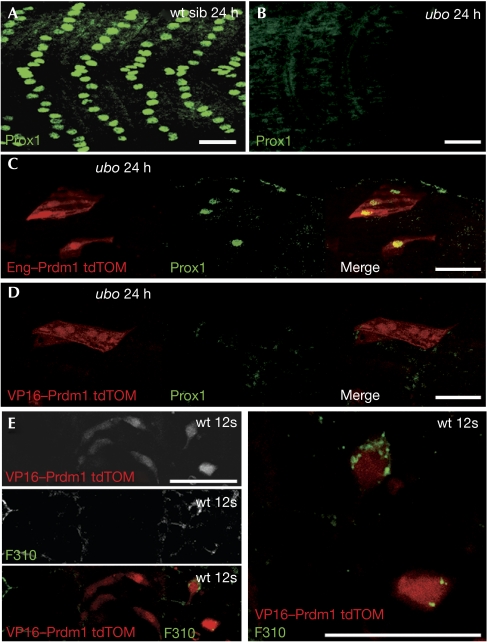
To investigate whether Prdm1 acts as an activator or repressor of transcription during slow-twitch muscle development in zebrafish, constructs in which the Prdm1 DNA-binding domain is fused to either the Engrailed (Eng) repressor or the VP16 activator domain (Kessler, 1997) were tested for their ability to substitute for the wild-type protein. Sequences encoding the fusion constructs were cloned downstream from a heat-shock-inducible promoter that simultaneously drives expression of the fluorescent protein tdTomato. Transient expression of the Eng–Prdm1 protein induced by heat shock was sufficient to rescue the expression of Prox1 in ubotp39 mutant embryos (Fig 2C). By contrast, transient expression of the Vp16–Prdm1 fusion protein was unable to rescue Prox1 expression or suppress myoblast fusion (Fig 2D,E), but was sufficient to activate fast-myosin expression. Taken together, these data indicate that Prdm1 acts as a repressor rather than an activator to promote slow-muscle differentiation, and show that activation of prox1 and smyhc1 expression is an indirect effect of Prdm1 activity. As the PR domain is absent from the Eng–Prdm1 protein fusion, it also follows that this domain is not essential for Prdm1 function, at least in this context. Consistent with this, previous studies have shown the PR domain to be dispensable for Prdm1 function in the context of β-interferon repression (Gyory et al, 2004).
Figure 2.
Slow-fibre differentiation requires the repressive function of Prdm1. (A) Prox1 expression in slow-muscle fibres of wild-type (wt) embryos at 24 h. (B) ubotp39 mutant embryos lack Prox1-expressing slow fibres at 24 h. (C) Eng–Prdm1 expression, marked by tdTomato (tdTOM), rescues mononucleate fibre differentiation and Prox1 expression in ubotp39 homozygotes (49 out of 94 Eng–Prdm1-expressing fibres were Prox1 positive; n=8). (D) VP16–Prdm1 expression, marked by tdTOM, was found exclusively in multinucleate fibres lacking Prox1 staining in ubotp39 homozygotes (none of the 134 Vp16–Prdm1-expressing fibres was Prox1 positive; n=11). (E) Precocious labelling with the fast-specific F310 antibody in VP16–Prdm1-expressing muscle precursors at the 12-somite stage (12s; 12 out of 30 F310-positive cells; n=4). Scale bars, 25 μm. Eng, Engrailed; Prox1, prospero-related homeobox gene 1; sib, sibling; ubo, u-boot.
Prdm1 regulates Sox6—a repressor of slow fibre identity
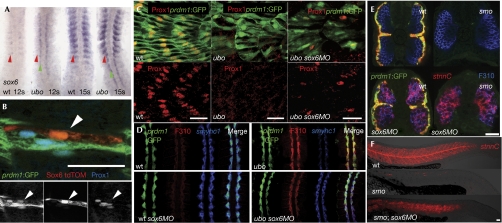
In mice, the transcription factor Sox6 acts as a repressor of fetal slow-twitch fibre differentiation (Hagiwara et al, 2005, 2007). We analysed expression of the zebrafish sox6 gene and found that it was expressed in the fast-muscle progenitor domain of the somites but excluded from adaxial cells (Fig 3A). In ubotp39 mutant embryos, by contrast, sox6 is ectopically expressed in adaxial cells, indicating that Prdm1 represses sox6 expression in slow-muscle progenitors. Forcing ectopic expression of sox6 in the adaxial cells of wild-type embryos caused an inhibition of Prox1 expression (Fig 3B). Conversely, morpholino-mediated knockdown of sox6 in ubotp39 embryos partly rescued Prox1 expression and restored expression of smyhc1 to normal levels in adaxial cells (Fig 3C). Although neither Prox1 nor smyhc1 was expressed ectopically in fast fibres in response to sox6 knockdown, robust expression of stnnC was induced in the fast muscle of both wild-type and smoothened (smo) mutant (that lack all Hedgehog signalling activity) embryos injected with the sox6 morpholino (Fig 3E,F). This disparity might reflect a differential sensitivity of these slow-twitch-specific genes to sox6 activity, revealed by incomplete knockdown by the morpholino. Taken together, these data indicate that Sox6 acts as a repressor of slow-twitch-specific gene expression and suggest that Prdm1 activates such expression by repressing transcription of sox6.
Figure 3.
Slow-specific gene expression is upregulated in sox6 morphant embryos. (A) sox6 is expressed in the fast-muscle precursors (red arrowheads) in wild-type (wt) embryos and ectopically expressed in the adaxial cells (green arrowheads) in ubotp39 embryos at the 12-somite (12s) and 15-somite (15s) stage. (B) Heat-shock-induced expression of Sox6 (marked with tdTomato (tdTOM)) inhibits Prox1 expression at 24 hpf (hours post fertilization) in adaxially derived cells marked by prdm1:GFP (arrowhead; 100% of the adaxially derived cells lacked Prox1 when ectopically expressing Sox6 (from the analysis of 14 fibres in 4 embryos)). (C) Lateral views of the tail at 24 hpf showing partial rescue of Prox1 expression in ubotp39 embryos injected with sox6MO (ubotp39: 0 rescued fibres in 20 embryos; ubotp39; sox6MO: mean of 58 rescued fibres per embryo, n=5, s.d.=29). (D) At 12s, ubotp39 mutants have reduced expression of smyhc1 and ectopic expression of fast-MyLC (recognized by the F310 antibody) in the adaxial cells marked with prdm1. sox6MO causes rescue of the smyhc1 expression in ubotp39 mutants but does not prevent ectopic fast-MyLC expression. (E) Transverse views of 24 hpf wt and smoothened (smo) embryos carrying the prdm1:GFP reporter gene (upper panel) and siblings of the same genotypes that have been injected with the sox6 morpholino (sox6MO; lower panels), showing that loss of sox6 causes Hedgehog-independent ectopic expression of stnnC (red) in fast muscle (identified by F310 antigen (blue)). (F) Lateral views of stnnC expression (red) in wt, smo and smo;sox6MO embryos at 24 hpf (9 out of 9 smo;sox6MO embryos, and 15 out of 15 wt embryos showed strong ectopic stnnC expression). Scale bars, 0.25 μm. GFP, green fluorescent protein; Prox1, prospero-related homeobox gene 1; smyhc1, slow myosin heavy chain 1; sox6MO, sox6 morpholino; stnnC, slow-specific Troponin C; ubo, u-boot.
Fast-twitch-specific genes are direct targets of Prdm1
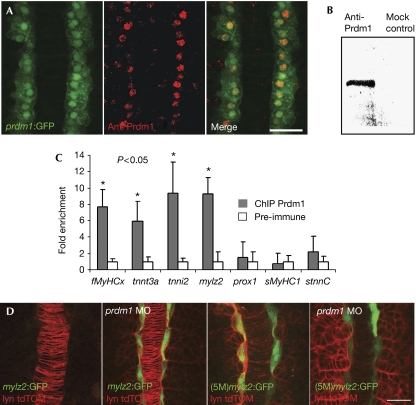
The coordinated repression of multiple genes encoding fast-twitch isoforms of sarcomeric proteins could be accomplished by Prdm1-mediated repression of a fast-specific global transcriptional activator; alternatively, Prdm1 might itself act directly to repress transcription of these genes. To distinguish between these two scenarios, we used chromatin immunoprecipitation (ChIP) to test for binding of the protein to upstream regulatory regions of the putative targets. A polyclonal antibody was raised against 186 amino acids from the Prdm1 PR domain; as expected, this labelled adaxial cell nuclei during early- and mid-myogenesis (Fig 4A), and bound to Prdm1 specifically in immunoprecipitation assays (Fig 4B). DNA from chromatin immunoprecipitated with this antibody was amplified by using primer pairs specific for sequences proximal to the transcription initiation sites of the mylz2, fMyHCx, tnnt3a and tnni2 genes. These sequences were all found to be enriched in the Prdm1-precipitated chromatin; by contrast, none of the slow-muscle-specific genes smyhc1, stnnC or Prox1 was enriched (Fig 4C). These data indicate that Prdm1 selectively binds to putative regulatory regions of fast-fibre-specific genes in vivo, suggesting that Prdm1 acts as a direct repressor of their transcription.
Figure 4.
Prdm1 acts as a direct repressor of fast-fibre-specific gene expression in adaxial cells. (A) prdm1:GFP expression is specific to adaxial cells in the somite area at the eight- to ten-somite stage. A Prdm1-specific polyclonal antibody detects Prdm1 protein in adaxial cells, consistent with the prdm1:GFP expression pattern. Scale bar, 0.25 μm. (B) Prdm1 is specifically immunoprecipitated using the Prdm1 antibody. (C) Amplification of the fast-fibre-specific genes fMyHCx, tnnt3a, tnni2 and mylz2 using Taqman real-time PCR showed significant enrichment compared with the pre-immune controls. Three slow-muscle-specific genes, prox1, sMyHC1 and stnnC, were tested as negative controls and were not enriched in the chromatin immunoprecipitated (ChIP) chromatin. (D) The mylz2:GFP promoter fusion is not expressed in wild-type embryos at the 12-somite stage, but is expressed in the adaxial cells in prdm1 morphants at the 12-somite stage. Mutation of the Prdm1-binding sites (5M) resulted in ectopic adaxial GFP expression in both control and prdm1 morphant embryos. Scale bar, 0.25 μm. fMyHC, fast myosin heavy chain; GFP, green fluorescent protein; MO, morpholino; mylz2, myosin light chain 2; prox1, prospero-related homeobox gene 1; sMyHC1, slow myosin heavy chain 1; stnnC, slow-specific Troponin C; tnni2, troponin I2; tnnt3a, troponin T3a.
The mylz2 promoter has functional Prdm1 binding sites
Expression of a GFP reporter gene containing 2.3 kb of the mylz2 promoter sequence is specifically repressed in adaxial cells by Prdm1 activity (Fig 4D). Although the consensus binding site for Prdm1 has not been determined in zebrafish, we identified five putative Prdm1-binding sites in this fragment, containing the GAAAG core of the sequence (A/C)AG(T/C)GAAAG(T/C)(T/G) that has been defined as mediating Prdm1-dependent gene regulation in mammals (Kuo & Calame, 2004). The introduction of point mutations in each of these five potential Prdm1-binding sites in this construct led to ectopic adaxial GFP expression in wild-type embryos, similar to that seen with the wild-type construct in ubo morphants (Fig 4D). This finding is consistent with Prdm1 acting directly to repress the mylz2 gene in adaxial cells at the 12-somite stage.
Identification of Prdm1 target genes by ChIP on chip
To confirm and extend the findings of our candidate gene analysis, we used a recently constructed zebrafish promoter array, consisting of 60-mer probes for more than 11,000 genes within the zebrafish genome (Wardle et al, 2006), to probe the DNA isolated by ChIP of myoblast extracts (supplementary information online, accession code GSE10883 at GEO). By setting the gene array threshold for enrichment to the level of mylz2, which was at the level of significant P-value 0.0078 and P[Xbar] 0.0075, we identified 381 putative target genes (supplementary information online). Gene ontology analysis showed various genes enriched in the ChIP-on-chip sample: 11% were documented transcription factors, 24% were new genes or genes without known function, 15% were genes encoding proteins with enzyme activity, such as kinases and phosphatases, whereas others had gene ontology terms linking them to the immune and haematopoietic systems, cell-cycle regulation or apoptosis. Significantly, we found several genes encoding fast-fibre-specific isoforms of sarcomeric proteins, including those encoding fast MyHC and troponins described above, whereas no genes encoding slow-specific sarcomeric proteins or Prox1 were identified (supplementary information online). Surprisingly, sox6 was not among the transcription factor-encoding genes identified in this analysis. However, we note that representation of regulatory regions on the gene array is restricted to sequences 9 kb upstream from the 5′ end of the complementary DNAs used in its design (Wardle et al, 2006). We have identified additional sox6 sequences 30 kb upstream from the transcription start site used in the array (J.v.H., S.E. & P.W.I., unpublished data); whether sox6 is a direct target of Prdm1 remains to be determined.
Conclusion
Our data underline the pivotal role of Prdm1 in switching between alternative muscle fibre type programmes in the zebrafish embryo. We have shown that it accomplishes its function in two ways: first, by repressing the transcription of a repressor of slow-specific gene transcription, sox6, in a manner analogous to its repression of Pax5 in B cells (Lin et al, 2002) and, second, by acting directly as a global repressor of fast-specific differentiation genes. Although Prdm1 is expressed in the myotome of the mouse (Chang et al, 2002; Vincent et al, 2005), at present it is unclear whether it has an analogous role in fibre type specification in amniotes. Our finding that Sox6 suppresses slow-twitch fibre specification in zebrafish, however, establishes that at least some aspects of the regulatory network underlying fibre type specification (Hagiwara et al, 2005, 2007) have been conserved in evolution.
Methods
Fish strains, cloning of gene promoters and injection of embryos. Zebrafish mutants ubotp39 and smob641, and the transgenic line Tg(actal:GFP)zf13 have been described previously (van Eeden et al, 1996; Higashijima et al, 1997; Barresi et al, 2000; Chen et al, 2001; Roy et al, 2001; Baxendale et al, 2004). The Tg(prdm1:gfp)i111 and Tg(prdm1:gfp)i106 are described by Elworthy et al (2008). The mylz2:GFP promoter construct was generated by using 2,239 bp of the mylz2 upstream region (Ju et al, 2003; Moore et al, 2007) to generate a stable line Tg(mylz2:GFP)i135. The mylz2:GFP plasmid was also used as a template for in vitro mutagenesis of the five sites containing the Prdm1 GAAAG core sequence using the QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) following the manufacturer's instructions to generate the Tg((5M)mylz2:GFP)i130 transgenic line. A 6.8 kb fragment upstream from the fMyHCx ATG was isolated by PCR from bacterial artificial chromosome zCR392328 by using a left primer containing an AscI restriction site (TGGCGCGCCTGCATGGTGTTTGACA) and a right primer containing NcoI (ACCCATGGTGGCGGCTTACCGT). The promoter DNA was in all cases subcloned into a GFP vector with flanking I-sceI sites. One-cell-stage embryos were microinjected with 4–8 nl of plasmid at a concentration of 40 ng/nl. Embryos were kept at 28.5°C and analysed for GFP at the 12-somite stage. Ubo morpholino-mediated knockdown of Prdm1 activity was carried out as described previously (Baxendale et al, 2004). The sox6 translation targeted morpholino (GTGGCTTGCTTGGAAGACATGATTC) was injected into one-cell-stage embryos at 0.9 mM. All fish were raised, staged and maintained as described previously (Kimmel et al, 1995; Westerfield, 2000).
Prox1 rescue assay. Eng–Prdm1 and VP16–Prdm1 fusion constructs (gifts from Dr Johaness Bischof; for details, see the supplementary information online) were used in attempts to rescue Prox1 expression. Both constructs and the complete sox6 cds (EU532205) were subcloned into a pSGH2 vector containing a bidirectional heat-shock promoter (Bajoghli et al, 2004) that drives expression of both the fusion protein and the fluorescent protein tdTomato. One-cell-stage embryos were microinjected with plasmid at a concentration of 40 ng/μl. At the three- to four-somite stage, the injected embryos were heat shocked by incubation at 39°C for 2 h. Injected embryos were fixed in 4% paraformaldehyde at 24 hpf (hours post fertilization) and analysed by using confocal microscopy.
Antibodies, immunohistochemistry and ChIP. A Prdm1 polyclonal antibody was raised against a fragment of the protein corresponding to amino acids 161–346 expressed as a His-tag fusion protein using the pET19b vector (Novagen, Darmstadt, Germany). Immunoprecipitation using the Prdm1 antibody was carried out according to Link et al (2006) using crude protein extracts from zebrafish embryos at the ten-somite stage. The precipitated proteins were analysed on SDS–polyacrylamide gel electrophoresis gel after Coomassie staining.
A Prox1 antibody was raised against recombinant zebrafish Prox1 purified from Escherichia coli (A.M. Taylor & P.W.I., unpublished data). Whole-mount immunohistochemical analysis using F310 fast MyLC (1:50, DSHB), S58 slow MyHC (1:10, DSHB), Prox1 (1:5,000) and Prdm1 (1:15,000) antibodies was performed on embryos fixed in 4% paraformaldehyde (for protocols, see the supplementary information online). For ChIP analysis, α-actin:GFP embryos were injected with dominant negative Protein Kinase A (dnPKA) at the one- to two-cell stage and were kept in embryonic medium until the 12- to 14-somite stage. The chorions were removed using pronase and cells were fixed for 15 min in 1.85% formaldehyde. For the ChIP-on-chip assay (supplementary information online), the embryos were dissociated using collagenase and GFP-positive cells were isolated. In addition to the ChIP on chip, three replicates of 300 embryos were used in the ChIP assay, which was performed as described previously (Wardle et al, 2006) using the Prdm1 antibody or rabbit preserum. Precipitated chromatin was analysed using Custom TaqManR Assays (Applied Biosystems, Foster City, CA, USA), specific to regions within the first 1 kb of upstream sequences from the first codon of mylz2 (NM_131188), fMyHCx (EU115994), tnnt3a (NM_131565), tnni2 (NM_205742), stnnC (AF281003), prox1 (NM_131405) and sMyHC1 (NM_001020507; for oligonucleotides and probes, see the supplementary information online). For whole-mount ISH, antisense digoxigenin (DIG) probes for mylz2, tnnt3a, tnni2, fMyHCx, sox6 and stnnC (subcloned from IMAGE_6899234, zgc:86932; Thisse et al, 2001; Xu et al, 2006) were generated and the ISH was performed as described previously (Thisse & Thisse, 1998). Fluorescent ISH used anti-dig POD (Roche, Basel, Switzerland) at 1:10,000 with tyramide signal amplification (TSA) Cyanine 3 (Perkin Elmer, Waltham, MA, USA).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Supplement Data 1
Acknowledgments
We thank J. Bischof for the Eng–Prdm1 and Vp16–Prdm1 fusion constructs, and A.M. Taylor for generating the His–Prdm1 fusion protein and the Prox1 antibody. This work was funded by a UK MRC Programme Grant (G0100151) to P.W.I., a Wellcome Trust Programme grant (077592) to J.C.S. and the EU FP6 ‘Cells into Organs' Network of Excellence. J.v.H. is a Wenner-Gren Foundation fellow.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bajoghli B, Aghaallaei N, Heimbucher T, Czerny T (2004) An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev Biol 271: 416–430 [DOI] [PubMed] [Google Scholar]
- Barresi MJ, Stickney HL, Devoto SH (2000) The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development 127: 2189–2199 [DOI] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S (2004) The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet 36: 88–93 [DOI] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM (1997) Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev 11: 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DH, Cattoretti G, Calame KL (2002) The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev 117: 305–309 [DOI] [PubMed] [Google Scholar]
- Chen W, Burgess S, Hopkins N (2001) Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development 128: 2385–2396 [DOI] [PubMed] [Google Scholar]
- Currie PD, Ingham PW (1996) Induction of a specific muscle cell type by a hedgehog-like protein in zebrafish. Nature 382: 452–455 [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M (1996) Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122: 3371–3380 [DOI] [PubMed] [Google Scholar]
- Du SJ, Devoto SH, Westerfield M, Moon RT (1997) Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-β gene families. J Cell Biol 139: 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S, Hargrave M, Knight R, Mebus K, Ingham PW (2008) Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135: 2115–2126 [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejér G, Seto E, Wright KL (2004) PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5: 299–308 [DOI] [PubMed] [Google Scholar]
- Hagiwara N, Ma B, Ly A (2005) Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Dev Dyn 234: 301–311 [DOI] [PubMed] [Google Scholar]
- Hagiwara N, Yeh M, Liu A (2007) Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn 236: 2062–2076 [DOI] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G (1997) High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol 192: 289–299 [DOI] [PubMed] [Google Scholar]
- Hsiao CD, Tsai WY, Horng LS, Tsai HJ (2003) Molecular structure and developmental expression of three muscle-type troponin T genes in zebrafish. Dev Dyn 227: 266–279 [DOI] [PubMed] [Google Scholar]
- Ju B, Chong SW, He J, Wang X, Xu Y, Wan H, Tong Y, Yan T, Korzh V, Gong Z (2003) Recapitulation of fast skeletal muscle development in zebrafish by transgenic expression of GFP under the mylz2 promoter. Dev Dyn 227: 14–26 [DOI] [PubMed] [Google Scholar]
- Kallies A, Nutt SL (2007) Terminal differentiation of lymphocytes depends on Blimp-1. Curr Opin Immunol 19: 156–162 [DOI] [PubMed] [Google Scholar]
- Keller AD, Maniatis T (1991) Identification and characterization of a novel repressor of β-interferon gene expression. Genes Dev 5: 868–879 [DOI] [PubMed] [Google Scholar]
- Kessler DS (1997) Siamois is required for formation of Spemann's organizer. Proc Natl Acad Sci USA 94: 13017–13022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Kuo TC, Calame KL (2004) B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol 173: 5556–5563 [DOI] [PubMed] [Google Scholar]
- Lewis KE, Currie PD, Roy S, Schauerte H, Haffter P, Ingham PW (1999) Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signalling. Dev Biol 216: 469–480 [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K (2002) Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol 22: 4771–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V, Shevchenko A, Heisenberg CP (2006) Proteomics of early zebrafish embryos. BMC Dev Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW (2007) A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 134: 3145–3153 [DOI] [PubMed] [Google Scholar]
- Ohinata Y et al. (2005) Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213 [DOI] [PubMed] [Google Scholar]
- Roy S, Wolff C, Ingham PW (2001) The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev 15: 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas BP, Woo J, Leong WY, Roy S (2007) A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat Genet 39: 781–786 [DOI] [PubMed] [Google Scholar]
- Thisse B et al. (2001) Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission (http://zfin.org)
- Thisse C, Thisse B (1998) High resolution whole-mount in situ hybridization. Zebrafish Sci Monitor 5: 8–9 [Google Scholar]
- Turner CA Jr, Mack DH, Davis MM (1994) Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77: 297–306 [DOI] [PubMed] [Google Scholar]
- van Eeden FJ et al. (1996) Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123: 153–164 [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ (2005) The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132: 1315–1325 [DOI] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC (2006) Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol 7: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (2000) The Zebrafish Book. Eugene, OR, USA: University of Oregon Press [Google Scholar]
- Wolff C, Roy S, Ingham PW (2003) Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol 13: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Xu J et al. (2006) Genomewide expression profiling in the zebrafish embryo identifies target genes regulated by Hedgehog signaling during vertebrate development. Genetics 174: 735–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, He J, Wang X, Lim TM, Gong Z (2000) Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev Dyn 219: 201–215 [DOI] [PubMed] [Google Scholar]
- Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K (2000) Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol 20: 2592–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplement Data 1