Abstract
Regulation of the sterol-synthesizing mevalonate pathway occurs in part through feedback-regulated endoplasmic reticulum degradation of 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-R). In yeast, the Hmg2p isozyme of HMG-R is regulated in this manner. We have tested the involvement of ubiquitination in the regulated degradation of Hmg2p, by using both genetic and direct biochemical approaches. Hmg2p degradation required the UBC7 gene, and Hmg2p protein was directly ubiquitinated. Hmg2p ubiquitination was dependent on UBC7 and was specific for the degraded yeast Hmg2p isozyme. Furthermore, Hmg2p ubiquitination was regulated by the mevalonate pathway in a manner consistent with regulation of Hmg2p stability. Thus, regulated ubiquitination appeared to be the mechanism by which Hmg2p stability is controlled in yeast. Finally, our data indicated that the feedback signal controlling Hmg2p ubiquitination and degradation was derived from farnesyl diphosphate, and thus implied conservation of an HMG-R degradation signal between yeast and mammals.
3-hydroxy-3-methylglutaryl-CoA reductase (HMG-R) is a key enzyme of the mevalonate pathway from which sterols such as cholesterol are synthesized. HMG-R is an integral membrane protein of the endoplasmic reticulum (ER), and is one of a growing group of proteins that is subject to ER degradation. In both mammals and yeast (1, 2), ER degradation of HMG-R is regulated by the mevalonate pathway: slowing pathway flux causes slowed degradation, whereas increasing pathway flux causes increased degradation. Neither the mechanism of ER protein degradation nor the manner in which the mevalonate pathway selectively controls HMG-R degradation is understood (3–9).
In yeast, the HMG-R isozyme Hmg2p is subject to regulated ER degradation (2). Our recent genetic studies indicated that the 26S proteasome is required for this process (10–12). Accordingly, we have examined the role of ubiquitin (Ub) in the regulated degradation of Hmg2p, because ubiquitination targets many proteins for proteasomal destruction (10, 13, 14). The studies below demonstrated a direct role for ubiquitination in the regulated degradation of Hmg2p in yeast. The data also indicated that features of the signaling mechanism that couples the mevalonate pathway to Hmg2p degradation are broadly conserved in eukaryotes.
METHODS
Yeast and DNA Methods.
Culture and DNA transformation of yeast strains, and all DNA manipulations were performed as described (2, 11, 16).
Epitope Tagging of the Hmg2p Protein and Construction of myc-Hmg2p-Expressing Yeast Strains.
A double-stranded oligonucleotide encoding the myc epitope tag (NDQKLISEEDLFA) was inserted into the BstBI site, between codons 618 and 619, of the HMG2 coding region in pRH144–2 (2). This insertion resulted in plasmid pRH423, which expressed myc-Hmg2p from the strong glyceraldehyde-3-phosphate dehydrogenase promoter. The two synthetic oligonucleotides were: 5′-CG AAT GAG CAA AAG CTA ATT TCA GAA GAG GAC TTA TTC G-3′ (sense strand) and 5′-CGC GAA TAA GTC CTC TTC TGA AAT TAG CTT TTG CTC ATT-3′ (antisense strand). The myc tag had no effect on regulation, expression, or distribution of the Hmg2p protein. Plasmid pRH423 was integrated into yeast strains at the HMG2 locus to allow the study of Hmg2p degradation (by myc immunoblotting) without interference from endogenous HMG-R protein. Strains MHY501 (wild type), MHY495 (ubc6::HIS3), and MHY507 (ubc7::LEU2) served as an otherwise isogenic set of strains with null mutations in the indicated UBC gene (provided by M. Hochstrasser, University of Chicago; ref. 15). The resulting myc-Hmg2p expressing strains were RHY506 (wild type), RHY507 (ubc6::HIS3), and RHY511(ubc7::LEU2). The ubc7::LEU2 is an insertional disruption of the coding region and is abbreviated ubc7Δ in the figures.
Stationary Chase Degradation Assay in ubcΔ Strains.
The stationary chase assay was performed as described (11, 16). As yeast grow into stationary phase, protein synthesis ceases, and net degradation of Hmg2p is observed as a loss of immunoreactivity. Samples of early log phase (OD600 approximately 0.25) experimental cultures were diluted so that after 15 hr the diluted cultures were once again in early log phase, and the original cultures were in early stationary phase. Lysates of each culture then were prepared and immunoblotted as described.
Cycloheximide Chase Assay of Hmg2p Degradation.
The stability of the entire pool of Hmg2p was assayed by addition of cycloheximide to log phase cultures and subsequent immunoblotting, as described (2, 11, 16).
Methionine Chase Assay of Hmg2p-Green Fluorescent Protein (GFP) Protein Degradation.
To study the degradation of the entire pool of Hmg2p-GFP in vivo, cultures of RHY513 were deprived of methionine to stop protein synthesis, and subsequently examined for GFP fluorescence (see Fig. 5A). Early log phase (OD600 approximately 0.2) cells were harvested and resuspended in fresh medium lacking methionine, in the presence or absence of drug, and incubated at 30°C.
Figure 5.
ZA stimulated degradation of Hmg2p-GFP in vivo. (A) Hmg2p-GFP degradation of Hmg2p-GFP (in strain RHY513) was examined in the absence (no drug) and presence (plus ZA) of ZA by methionine chase. In each panel, 20–25 cells are photographed at the indicated times with identical optical settings. (B) Effect of ZA on steady-state levels of Hmg2p-GFP. Early log phase cells expressing Hmg2p-GFP (RHY513) were grown for 3 hr in the presence of ZA, ZA plus HMG-CoA synthase inhibitor L-659,699 (Fig. 3), or no drug, and then analyzed by FACS with 10,000 cells per histogram. Horizontal axis is log of relative fluorescence.
Coimmunoprecipitation [Immunoprecipitation (IP)/Immunoblotting (IB)] Assay of Ubiquitination.
Strains RHY415, RHY416, and RHY420 were used in the experiment in Fig. 2A. The genotype of each strain (before the addition of plasmids) was (a,lys2–801,ade2–101,ura3–52,his3Δ200,met,HMG2,hmg1::LYS2, trp1Δ). The strain used in Fig. 2B that expressed Hmg2p was also RHY416. RHY415 and RHY416 each harbored YEp105 (2 μ, TRP1) that expressed the myc-Ub protein on induction with Cu+2 (17) (provided by M. Hochstrasser). RHY415 (used in the low lane) expressed only the endogenous HMG2 gene from the genomic locus, and RHY416 (used in the high lane) harbors pRH144-2 (integrating, URA3), that expressed the HMG2 coding region from the strong constitutive glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter (2). Strain RHY420 expressed myc-Hmg2p from the GAPDH promoter (from pRH423) and did not harbor any tagged Ub plasmid. It served as a control (myc-Hmg2p) for the IP/IB protocol and provided a size standard for nonubiquitinated Hmg2p. The Hmg1p-expressing strain used in Fig. 2B was otherwise isogenic to RHY416 but harbored an integrated copy of pRH105–25 (2), which expressed Hmg1p at similar levels to those of Hmg2p in RHY416. The strains used in Fig. 2C were RHY506 and RHY511 (used in Figs. 1 and 5) but transformed with YEp112 (from K. Kuchler, University and Biocenter of Vienna), that expressed hemagglutinin (HA)-tagged Ub (18). All lysis and IPs were performed on 4 OD600 units of log-phase cells as described (2), except Mops (pH 6.8) replaced Tris as the buffer in the lysis solution, and 5 mM N-ethylmaleimide also was added to the lysis and IP buffers. In all assays, the expression of the tagged Ub was up-regulated by a 1-hr preincubation of each strain with 150 μM Cu+2 before the lysis and IP steps.
Figure 2.
Hmg2p was ubiquitinated. (A) (Left) Strains expressing myc-Ub along with high levels (HIGH) or low levels (low) of Hmg2p were assayed for ubiquitination by IP of Hmg2p followed by immunoblotting for coprecipitated myc-tagged-Ub (myc-Ub). (Right) myc-Hmg2p was the result of subjecting an isogenic strain that expressed myc-Hmg2p but not myc-Ub to the identical assay. Arrows represent (single or double- sided) the mobility of Hmg2p (Mr 115). Line segments represent the mobility of a 190-kDa protein standard. (B) Comparison of myc-Ub coprecipitation in isogenic strains expressing comparable levels of degraded Hmg2p (lane 2) or stable Hmg1p (lane 1). IP lanes are anti-myc immunoblots of the immunoprecipitated proteins. The lanes are heavily exposed to indicate the extreme difference obtained with the distinct isozymes. Lysate indicated cell lysates that were immunoblotted for myc immunoreactivity to assess myc-Ub induction. (C) Effect of ubc7::LEU2 on Hmg2p ubiquitination. Wild-type (RHY506) and ubc7::LEU2 (RHY511) strains were transformed with YEp112, a plasmid analogous to YEp105 that expresses HA-Ub. Lysates were immunoprecipitated then immunoblotted either with monoclonal 12CA5 to detect HA-Ub (Left, 4/5 total) or 9E10 to detect myc-Hmg2p (Right, 1/5 total). The lysates displayed identical induction of the HA-tagged Ub (not shown)
Figure 1.
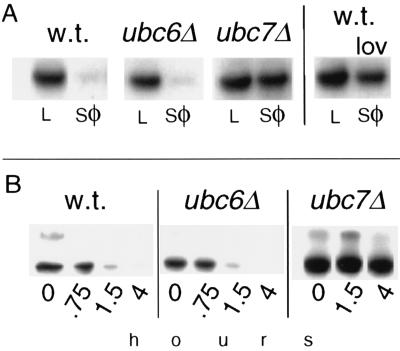
Hmg2p degradation required the UBC7 gene. (A) Stability of myc-Hmg2p in wild-type, ubc6::HIS3 (ubc6Δ), or ubc7::LEU2 (ubc7Δ) strains as measured by stationary chase. Equal numbers of cells in log phase (L) or early stationary phase (sφ) were analyzed by SDS/PAGE and anti-myc IB. A sample of the wild-type strain also was grown into stationary phase in the presence of lovastatin (25 μg/ml) to show the regulation of myc-Hmg2p degradation caused by slowing of the mevalonate pathway (Right). (B) Degradation of myc-Hmg2p by cycloheximide chase. Log-phase cultures of the indicated strains were treated with cycloheximide (50 μg/ml final). At the indicated times, equal numbers of cells were lysed and analyzed by IB.
Anti-Epitope Tag IB.
SDS/PAGE or dot IB was performed as described (2, 11), by using 9E10 anti-myc hybridoma (ATCC CRL 1729) cell culture supernatant [1:5 dilution in Tris-buffered saline with Triton-X-100 and milk (TBSTM)] or 12CA5 anti-HA tag mAbs (1/3,000 in TBSTM; Babco, Richmond, CA) as 1o antibodies, and horseradish peroxidase-goat anti-mouse IgG (1/15,000) was used as a 2o antibody. All immunoblots in all figures were detected with ECL reagent (Amersham).
Optical Detection of Hmg2p-GFP.
The strain expressing Hmg2p-GFP (RHY513; Ade−, Met−) had essential HMG-R activity supplied by 6 myc-Hmg2p, and Hmg2p-GFP expressed from plasmid pRH469 (11). Cells were examined by fluorescence microscopy or FACS as described (11, 16).
RESULTS AND DISCUSSION
Our first test of a role for ubiquitination in Hmg2p degradation involved examining the effects of mutations in several genes encoding Ub conjugating enzymes (UBCs). The UBC-encoded enzymes are required for the specific conjugation of Ub to target proteins. We tested the UBC6 and UBC7 genes, which previously had been implicated in the degradation of several ER proteins (19, 23), for effects on Hmg2p degradation. A ubc7::LEU2 null mutation strongly inhibited Hmg2p degradation, whereas the corresponding ubc6::HIS3 mutant was without significant effect (Fig. 1). Two assays of Hmg2p stability were used: the loss of Hmg2p that occurred when cells were grown into early stationary phase (Fig. 1A) (stationary chase; refs. 11 and 16), and the time-dependent loss of Hmg2p that occurred after inhibition of protein synthesis by cycloheximide (Fig. 1B) (cycloheximide chase; refs. 2, 11, and 16). In the stationary chase experiment, the stabilization caused by the ubc7::LEU2 null mutation was as strong as the regulatory stabilization caused by growing the cells in lovastatin, a mevalonate pathway inhibitor (ref. 2; Fig. 1A, Right). The half-life of Hmg2p in each strain was estimated from the cycloheximide chase data: 1.1 hr for wild type, 1.2 hr for ubc6Δ, and >4 hr for ubc7Δ. The higher molecular weight band is caused by aggregation after lysis of the cells.
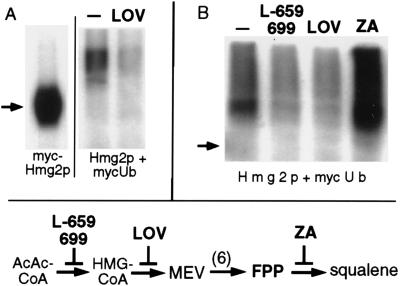
The requirement for UBC7 in Hmg2p degradation indicated that Hmg2p might be directly ubiquitinated. To test this possibility, we used strains expressing an epitope-tagged form of Ub to facilitate detection of ubiquitinated Hmg2p (17, 20–22). Covalent attachment of myc-tagged Ub to Hmg2p was assessed by IP of Hmg2p with polyclonal antibodies directed against HMG-R, followed by IB of the immunoprecipitated protein with a mAb (9E10) directed against the myc tag. An identical strain expressing only myc-Hmg2p served as a control for the IP/IB procedure. When strains expressing Hmg2p and myc-Ub were assayed, coprecipitated myc-Ub immunoreactivity migrated as a collection of molecular weights higher than myc-tagged Hmg2p itself (Fig. 2A, arrow). The intensity of the coprecipitated myc-Ub was dependent on the amount of Hmg2p expressed: in an otherwise identical strain with low (approximately 30-fold less) levels of Hmg2p (genomic vs. the powerful glyceraldehyde-3-phosphate dehydrogenase promoter) the amount of coprecipitated myc-Ub was far less (low vs. high lanes in Fig. 2A). Coprecipitated myc immunoreactivity in the low lane could be detected by increasing immunoblot exposure times 20- to 30-fold (data not shown). Expression of myc-Ub in these two strains was identical, as assessed from immunoblotting lysates of each strain with the anti-myc antibody.
Hmg2p is rapidly degraded in yeast, whereas the closely related Hmg1p isozyme is extremely stable (2). We therefore tested the Hmg1p protein for ubiquitination in the IP/IB assay by comparing otherwise identical strains with similar levels of either Hmg2p or Hmg1p. The polyclonal antibody used in the comparison is equally effective at precipitating either isozyme (2). The stable Hmg1p protein showed no evidence of ubiquitination by this assay (Fig. 2B, IP), even at the very high chemiluminescent exposure times shown. Immunoblotting of cellular lysates of either strain indicated that the expression and utilization of the myc-tagged Ub was identical between the two strains (Fig. 2B, lysates). Thus, the isozyme specificity of HMG-R ubiquitination paralleled the isozyme specificity of degradation.
Next, the ubc7::LEU2 null mutation that strongly stabilized Hmg2p was tested for a direct effect on Hmg2p ubiquitination (Fig. 2C). Ub tagged with the HA epitope (HA-Ub) was coexpressed with myc-Hmg2p in strains with the wild-type or null alleles of UBC7. In this way, both the amount of immunoprecipitated myc-Hmg2p and coprecipitated HA-Ub was assayed in parallel on the same samples by immunoblotting with the appropriate monoclonal. In samples with the same amount of immunoprecipitated myc-Hmg2p (Fig. 2C, anti-myc), the amount of coprecipitated HA-Ub was approximately 9-fold less in the ubc7::LEU2 null strain (Fig. 2C, anti-HA). This result indicated that UBC7 played a significant and direct role in Hmg2p ubiquitination. Some HA-Ub appeared to be coprecipitated even in the mutant, possibly because of the participation of other UBCs. Nevertheless, the major role of UBC7 in Hmg2p degradation paralleled its role in Hmg2p ubiquitination.
Hmg2p stability is regulated by the mevalonate pathway (2) (Fig. 3, diagram). Drugs such as lovastatin (an inhibitor of Hmg2p) or L-659,699 (an inhibitor of HMG-CoA synthase) slow Hmg2p degradation, presumably by decreasing the availability of a mevalonate-derived signal that controls degradation. This signal has been proposed to be derived from a pathway molecule upstream of squalene, because zaragozic acid (ZA), an inhibitor of squalene synthase, does not slow Hmg2p degradation (2). We wanted to determine if these drugs had effects on Hmg2p ubiquitination consistent with their effects on Hmg2p stability. For example, if the stabilizing effect of lovastatin occurred through altering ubiquitination, then the drug would be expected to decrease Hmg2p ubiquitination. Hmg2p ubiquitination was assayed after a 30-min preincubation with a dose of lovastatin (25 μg/ml) previously demonstrated to slow degradation. Lovastatin treatment caused an approximate 3- or 4-fold attenuation of Hmg2p ubiquitination (Fig. 3 A and B). We then compared the effects of lovastatin, L-659,699, and ZA in the same experiment. Ubiquitination of Hmg2p was decreased to a similar extent by lovastatin or L-659,699, drugs that each slow Hmg2p degradation (2). In contrast, ZA caused an increase in Hmg2p ubiquitination. In all treatments the global labeling of proteins by myc-Ub, as assessed by immunoblotting cellular lysates, was unaffected by any drug (data not shown). The data indicated that Hmg2p ubiquitination was responsive to alterations of the mevalonate pathway. Furthermore, drugs previously shown to slow degradation of Hmg2p caused concomitant decrease of Hmg2p ubiquitination.
Figure 3.
Hmg2p ubiquitination was regulated by the mevalonate pathway. (A) Cultures of RHY416 coexpressing Hmg2p and myc-Ub were preincubated with or without lovastatin (25 μg/ml) for 30 min, followed by the IP/IB assay for coprecipitated myc-Ub. An isogenic strain expressing only myc-Hmg2p was run in parallel (myc-Hmg2p) as an assay control and a size standard. (B) The procedure was used to compare several drugs that affect Hmg2p degradation. Both L-659,699 and lovastatin (LOV) stabilize Hmg2p (2), whereas ZA hastens degradation (ref. 2 and see below). The actions of each agent are depicted in mevalonate pathway diagram at bottom: L-659,699 blocks HMG-CoA synthase, LOV blocks HMG-CoA reductase, and ZA blocks squalene synthase.
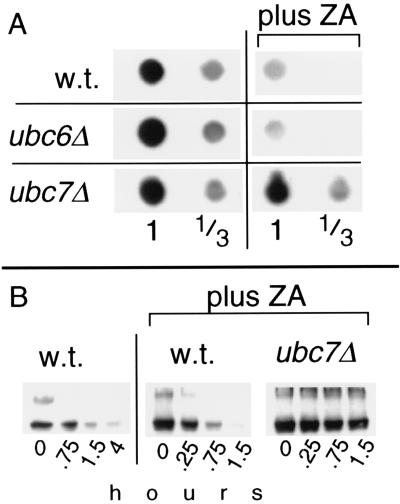
The increase in ubiquitination caused by ZA was further studied. This effect of ZA provided a further test of the relevance of Hmg2p ubiquitination in the regulation of Hmg2p stability. If the ubiquitination of Hmg2p was important in determining the degradation rate, then ZA would be expected to increase the degradation rate of Hmg2p and other substrates that are similarly regulated. Although we previously had shown that ZA did not slow degradation, the assays were not optimized to detect a hastening of degradation (2). Accordingly, we examined more carefully the effect of ZA on the steady-state levels and degradation rate of myc-Hmg2p (Fig. 4). Three hours after addition of the drug, the steady-state level of myc-Hmg2p, as determined by dot IB of the myc tag, dropped approximately 4-fold as visually estimated by comparison with 3-fold dilutions of control lysates on the same blot (Fig. 4A, Top). This effect of ZA was totally inhibited by a ubc7::LEU2 null mutation, but was unaffected by the ubc6::HIS3 null. Furthermore, a strain with null mutations in both UBC4 and UBC5 showed normal response to ZA (data not shown). The UBC7-dependent drop in Hmg2p steady-state level was consistent with an enhancement of Hmg2p degradation by ZA. We directly tested the effect of ZA on the rate of Hmg2p degradation in a cycloheximide chase. Hmg2p degradation was hastened by the presence of ZA (Fig. 5B, Left vs. Center). The estimated half-life of Hmg2p in ZA-treated cultures was 0.35 hr (21 min.) vs. 1.1 hr for the untreated controls. The degradation-accelerating effect of ZA was strongly inhibited by the ubc7::LEU2 mutation (Hmg2p half-life ≫2 hr), but was unaffected by the ubc6::HIS3 null mutation (data not shown). These experiments demonstrated ZA hastened the degradation of Hmg2p, as predicted from the effects of the drug on Hmg2p ubiquitination. Taken together, the results of these and earlier studies with mevalonate pathway inhibitors suggested that Hmg2p stability is physiologically regulated at the level of ubiquitination.
Figure 4.
ZA stimulated Hmg2p degradation. (A) Effect of ZA on the steady-state level of myc-Hmg2p. Strains with the indicated mutations (also used in Fig. 1) were grown into early log phase, treated with no drug or 25 μg/ml ZA, and allowed to incubate at 30°C for an additional 3 hr (final OD600 < 0.5) after which equal numbers of cells were lysed and dot-immunoblotted for myc-Hmg2p. Lysate samples were diluted 1:3 and blotted at the same volume as the undiluted samples to assist quantitation. (B) ZA-stimulated degradation of Hmg2p by cycloheximide chase in wild-type or ubc7::LEU2 strains. The wild-type, no ZA series (far left four lanes labeled w.t.) is the same one as shown in Fig. 1, because both experiments were run simultaneously.
We extended our studies with ZA to the degradation of the Hmg2p-GFP reporter protein (11, 16). Hmg2p-GFP has only the transmembrane portion of Hmg2p. This portion of Hmg2p has been shown to be sufficient for regulation of Hmg2p stability by the mevalonate pathway (2, 16). Accordingly, examination of ZA effects on the stability of Hmg2p-GFP provided a further test of the relevance of its actions to the known features of Hmg2p regulation (2). The fluorescent Hmg2p-GFP fusion allows optical detection of degradation in vivo, by FACS or fluorescence microscopy (11, 16). Addition of a small dose of ZA increased the degradation rate of the Hmg2p-GFP reporter (Fig. 5A). This effect of ZA on degradation was directly observed by fluorescence microscopy after cessation of protein synthesis by methionine removal (Fig. 5A). The degradation-enhancing effect of ZA also was observed as a lowering of the steady-state levels of the reporter protein in log phase cells. Three hours after the addition of ZA to living cells, there was a significant change in steady-state Hmg2p-GFP fluorescence as measured by FACS analysis (Fig. 5B, plus ZA). ZA treatment caused an average 4-fold shift in the fluorescence histogram of the treated population, consistent with the observed increase in the degradation rate of the fluorescent reporter shown in Fig. 5A. Thus, the actions of ZA were generalized to the Hmg2p-GFP reporter, and like other regulatory perturbations, only required the Hmg2p transmembrane region.
ZA most likely hastened degradation through blockade of squalene synthase and the resultant buildup of a mevalonate pathway product that performs a signaling function. Alternatively, the effect of ZA could have occurred by direct action of the drug on the apparatus that controls Hmg2p degradation. We examined these alternatives by testing the effects of L-659,699, which blocks the upstream enzyme HMG-CoA synthase (Fig. 3, diagram), on the ability of ZA to alter Hmg2p-GFP levels. The effect of ZA on the steady-state FACS histogram was entirely blocked by L-659,699 (Fig. 5B). L-659,699 similarly reversed the enhanced ubiquitination of Hmg2p caused by ZA (data not shown). These results indicated that the effect of the squalene synthase inhibitor ZA on Hmg2p degradation required the production of a mevalonate pathway product that otherwise would not be allowed to build up (Fig. 3, diagram). Because the substrate of squalene synthase is farnesyl diphosphate (FPP), this molecule is a reasonable source, or possibly a candidate, for the molecule that stimulates Hmg2p degradation.
In this work we have shown that the degradation of Hmg2p is mediated by ubiquitination. The UBC7 gene played a central role in the ubiquitination and degradation of the Hmg2p protein, in addition to its previously reported role in ER degradation of other membrane and luminal proteins (19, 23). Furthermore, Hmg2p ubiquitination was regulated by the mevalonate pathway in a manner consistent with pathway regulation of Hmg2p stability. Drugs that slow degradation of Hmg2p decreased ubiqitination of Hmg2p. In contrast, ZA, shown herein to increase ubiquitination of Hmg2p, increased the degradation rate of Hmg2p or Hmg2p-GFP. The effect of ZA on Hmg2p degradation required the UBC7 gene and a functioning mevalonate pathway. Thus, a reasonable model would be that the pathway product FPP, the substrate of squalene synthase, was a source of the regulatory signal that controlled UBC7-dependent Hmg2p ubiquitination and degradation. Similar degradation-enhancing effects of ZA on mammalian HMG-R have been reported both in cultured cells and in vivo in rat liver (9, 24). Furthermore, the molecule farnesol, derived from FPP, has been posited to be a signal for HMG-R degradation in mammals (25, 26). Thus, FPP-derived molecules may control HMG-R degradation in all eukaryotes.
It is unclear if the entire train of events from generation of the mevalonate-derived signal to the final degradation of HMG-R by the proteasome is conserved between mammals and yeast. Recent studies indicate that mammalian HMG-R degradation may similarly involve the proteasome (27). However, in that work no involvement of Ub could be demonstrated. In contrast, the ER degradation of both mutant and normal cystic fibrosis transmembrane conductance regulator (CFTR) in mammalian cells is mediated by ubiquitination (5), indicating that ubiquitination can mediate ER protein degradation in mammals. Thus, it may be that some molecular details of HMG-R degradation differ between mammals and yeast. Nevertheless, it is clear that yeast Hmg2p is destroyed by a degradation pathway widely used in eukaryotes (3–7), that both mammalian and yeast HMG-R appear to require the proteasome in vivo, and that FPP is likely to play a conserved role as a source of a degradation signal for HMG-R. A detailed mechanistic picture of HMG-R regulated degradation will allow understanding of both the medical and evolutionary questions that arise in the study of this regulatory axis.
Acknowledgments
We thank Mark Hochstrasser for strains, plasmids, and numerous intelligent conversations, Karl Kuchler for plasmids, and Steven Hedrick for the use of the FACS Star flow cytometer, and members of the Hampton lab for continuous high-level interaction and enthusiasm. R.Y.H. wishes to further thank Southern station KEAD for excellent reception and great listening. This work was supported by start-up funds from the University of California at San Diego Department of Biology, and grants from the California chapter (96–299) and the national chapter (96013020) of the American Heart Association, and the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (Grant DK51996).
ABBREVIATIONS
- HMG
3-hydroxy-3-methylglutaryl
- ER
endoplasmic reticulum
- HMG-R
HMG-CoA reductase
- FPP
farnesyl diphosphate
- GFP
green fluorescent protein
- HA
hemagglutinin
- IP
immunoprecipitation
- IB
immunoblotting
- UB
ubiquitin
- UBC
UB conjugating enzyme
- HA-Ub
Ub tagged with the HA epitope
- ZA
zaragozic acid
References
- 1.Chun K T, Bar N S, Simoni R D. J Biol Chem. 1990;265:22004–22010. [PubMed] [Google Scholar]
- 2.Hampton R Y, Rine J. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino J S, Lippincott S J. Curr Opin Cell Biol. 1991;3:592–600. doi: 10.1016/0955-0674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- 4.Wojcikiewicz R J, Furuichi T, Nakade S, Mikoshiba K, Nahorski S R. J Biol Chem. 1994;269:7963–7969. [PubMed] [Google Scholar]
- 5.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 6.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 7.Wiertz E J H J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 8.Roitelman J, Simoni R D. J Biol Chem. 1992;267:25264–25273. [PubMed] [Google Scholar]
- 9.Keller R K, Zhao Z, Chambers C, Ness G C. Arch Biochem Biophys. 1996;328:324–330. doi: 10.1006/abbi.1996.0180. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 11.Hampton R Y, Gardner R, Rine J. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMartino G N, Moomaw C R, Zagnitko O P, Proske R J, Chu P M, Afendis S J, Swaffield J C, Slaughter C A. J Biol Chem. 1994;269:20878–20884. [PubMed] [Google Scholar]
- 13.Finley D, Chau V. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 14.Jentsch S. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 16.Hampton R Y, Koning A, Wright R, Rine J. Proc Natl Acad Sci USA. 1996;93:828–833. doi: 10.1073/pnas.93.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellison M J, Hochstrasser M. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 18.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biederer T, Volkwein C, Sommer T. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 20.Kolling R, Hollenberg C P. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicke L, Riezman H. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 22.Egner R, Kuchler K. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- 23.Hiller M M, Finger A, Schweiger M, Wolf D H. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 24.Peffley D M, Gayen A K. Arch Biochem Biophys. 1997;337:251–260. doi: 10.1006/abbi.1996.9796. [DOI] [PubMed] [Google Scholar]
- 25.Meigs T E, Roseman D S, Simoni R D. J Biol Chem. 1996;271:7916–7922. doi: 10.1074/jbc.271.14.7916. [DOI] [PubMed] [Google Scholar]
- 26.Correll C C, Ng L, Edwards P A. J Biol Chem. 1994;269:17390–17393. [PubMed] [Google Scholar]
- 27.McGee T P, Cheng H H, Kumagai H, Omura S, Simoni R D. J Biol Chem. 1996;271:25630–25638. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]