Abstract
In the highly concentrated environment of the cell, polypeptide chains are prone to aggregation during synthesis (as nascent chains await the emergence of the remainder of their folding domain), translocation, assembly, and exposure to stresses that cause previously folded proteins to unfold. A large and diverse group of proteins, known as chaperones, transiently associate with such folding intermediates to prevent aggregation, but in many cases the specific functions of individual chaperones are still not clear. In vivo, Hsp90 (heat shock protein 90) plays a role in the maturation of components of signal transduction pathways but also exhibits chaperone activity with diverse proteins in vitro, suggesting a more general function. We used a unique temperature-sensitive mutant of Hsp90 in Saccharomyces cerevisiae, which rapidly and completely loses activity on shift to high temperatures, to examine the breadth of Hsp90 functions in vivo. The data suggest that Hsp90 is not required for the de novo folding of most proteins, but it is required for a specific subset of proteins that have greater difficulty reaching their native conformations. Under conditions of stress, Hsp90 does not generally protect proteins from thermal inactivation but does enhance the rate at which a heat-damaged protein is reactivated. Thus, although Hsp90 is one of the most abundant chaperones in the cell, its in vivo functions are highly restricted.
All cellular processes rely on the ability of linear polypeptide sequences to acquire and maintain appropriate three-dimensional conformations. In vitro, experiments with unfolded proteins have established that the primary amino acid sequences of many small proteins contain all the information necessary for the formation of native three-dimensional conformations (1). Larger, multidomain proteins have much greater difficulty reaching their native structures and are prone to aggregation (2–4). In the living cell, problems in achieving proper structure are compounded by the very high concentrations of aggregation-prone intermediates generated during synthesis, at points of translocation across membranes, and during times of stress. Mechanisms for shifting the balance away from aggregation and toward proper folding are essential. Molecular chaperones provide an important contribution by binding to nonnative polypeptides, stabilizing folding intermediates, preventing the occurrence of inappropriate inter- and intramolecular interactions, and resolving kinetically trapped intermediates (2–4). All cells possess a large complement of molecular chaperones to carry out these functions but, in many cases, the importance of particular chaperones in general protein folding is unclear.
In bacterial cells two abundant chaperone complexes are essential for the proper and efficient folding of nascent polypeptides, the DnaK (Hsp70)/DnaJ/GrpE complex and the GroEL/GroES (Hsp60/Hsp10) chaperonin complex. In addition to acting during de novo protein synthesis, both chaperone complexes also help prevent the permanent inactivation of proteins by heat (3, 4). In vitro studies indicate cooperativity between these two chaperone complexes, as incompletely folded polypeptides can be transferred from one chaperone to the other (5, 6). Moreover, the two chaperone complexes interact with similar subsets of proteins at different stages of folding. DnaK binds hydrophobic regions of polypeptides lacking secondary structure, such as those found in translating and translocating polypeptides. GroEL binds to partially folded or kinetically trapped intermediates and promotes their folding within its central cavity (2–4). Thus, in bacterial cells the DnaK/DnaJ/GrpE and GroEL/ES complexes form a network of chaperone activities that mediate the proper folding of most cellular proteins.
A similar chaperone network has not been identified in the eukaryotic cytosol. Homologs of DnaK and DnaJ are abundant in the cytosol of all eukaryotic cells. However, in most cell types the GroEL homolog of the eukaryotic cytosol, TriC, is not sufficiently abundant to play a major role in the folding of most proteins. Indeed, the chaperone activities of TriC appear highly restricted; its primary function is apparently to promote the maturation of cytoskeletal proteins (3, 4). It is not known what other chaperones contribute to the bulk folding activities of the eukaryotic cytosol.
Recently, the hypothesis that the Hsp90 (heat shock protein 90) chaperone complex might play this role has received much discussion at meetings and in the literature (4, 7). Hsp90 is highly conserved and very abundant. It constitutes 1–2% of all protein in the eukaryotic cytosol and its levels increase when cells are exposed to stress (8). Whereas the Escherichia coli homolog of Hsp90, HtpG, is dispensable (9), Hsp90 is essential for viability in both of the eukaryotic organisms in which it has been tested, Saccharomyces cerevisiae (8) and Drosophila melanogaster (10).
In vitro, at high concentrations, Hsp90 exhibits limited chaperone activity with a wide range of substrates (11–18). It does not possess refolding activity. Rather, Hsp90 interacts with nonnative proteins that have already attained a high degree of secondary structure, preventing their aggregation and maintaining them in folding-competent conformations that can complete folding upon the addition of other chaperones (7, 14, 19). In vitro experiments with reticulocyte lysates and genetic experiments in vivo indicate that Hsp90 forms a dynamic chaperone complex with several accessory proteins: hop (p60, Sti1), hip (p48), p23, DnaJ, p50 (cdc37), and the immunophilins FKBP52 (hsp56, p59), FKBP51, and Cyp-40 (Cpr6, Cpr7) (7, 14, 20). Most of these proteins are also abundant, and all but hip and hop have some chaperoning activity in vitro (7, 19–21). The complexity of this chaperone system makes a complete analysis of its functions difficult in a purified system.
As yet, there is little evidence indicating a general chaperone function for the Hsp90 complex in vivo. Schneider and co-workers (15) recently demonstrated that the Hsp90 inhibitor geldanamycin inhibits the refolding of heat-inactivated firefly luciferase in vertebrate cells. Otherwise, all of the known chaperone functions of the Hsp90 complex in vivo are restricted to the maturation of certain signal-transducing proteins, such as steroid hormone receptors and nonreceptor protein tyrosine kinases (22–25). Moreover, unlike GroEL and Hsp70, a large excess of Hsp90 to substrate is required to achieve the more general chaperone activities of Hsp90 in vitro (11–18). The abundance of Hsp90 in the cell makes such a general chaperone function plausible, but experiments in yeast demonstrate that Hsp90 can be reduced to a small fraction of its normal concentration without reducing growth rates at normal temperatures (8). Thus, it is still an open question what role Hsp90 plays in general protein folding in vivo.
A classic method for determining the function of a protein in vivo is to employ a temperature-sensitive (ts) mutant. After a shift to nonpermissive temperature, the mutant protein unfolds and loses activity, causing the processes in which it functions to be lost. This approach is problematic with heat-induced chaperones (Hsps) because higher levels of Hsp function are required for growth at higher temperatures. Thus, mutations that impair or alter the chaperone’s function can produce a ts growth phenotype, not because the chaperone loses function at high temperatures, but because its activity is insufficient to satisfy the cell’s requirements at high temperatures. Indeed, when we characterized the biochemical activities of eight S. cerevisiae Hsp90 ts mutants, all but one proved to have reduced function at normal temperatures and to retain substantial function at nonpermissive temperatures. That is, their ts phenotype is not due to unfolding at nonpermissive temperatures.
One mutant, however, behaves as a classic ts mutant: hsp90G170D has wild-type activity at 25°C and rapidly loses function upon a shift to high temperature (24). A case in point is provided by the effects of this mutation on the glucocorticoid receptor (GR). GR depends upon continuous cycles of interaction with Hsp90 to maintain its activity (25). When yeast cells expressing GR are shifted to high temperatures, GR activity is immediately lost in hsp90G170D cells (24). In wild-type cells, as well as in all seven of our other Hsp90 ts mutants, GR activity actually increases.
The hsp90G170D mutant carries a glycine to aspartic acid substitution at amino acid 170. Its instability at high temperatures is explained by the recent solution of the crystal structure of this domain. Residue 170 maps to the hydrophobic core of this N-terminal domain (26, 27), which should produce structural instability. The other mutants in this domain map to the ATP binding pocket or to the surface. Here, we take advantage of this unique mutant to directly test the breadth of Hsp90 chaperone activities in vivo. By shifting hsp90G170D cells to nonpermissive temperatures we assess the role of Hsp90 in de novo protein folding, protecting proteins from thermal inactivation, and reactivating proteins after thermal inactivation.
MATERIALS AND METHODS
Plasmid and Strain Construction.
The p60v-src expression plasmid Y316v-src was obtained from D. Morgan (28), and the β-galactosidase expression plasmid pLGSD5 (+ATG) was obtained from D. Gottschling (29). Plasmids encoding the bacterial luciferase fusion proteins from either Vibrio harveyi MAV [(plx709-fab9; (30)] or V. harveyi CTP5 [(pLX408facbc9; (31))] were obtained from A. Szalay. BamHI–SacI fragments containing these genes were amplified by the PCR (32). These fragments were then subcloned into pRS316 Gal1-10, in which the EcoRI-BamHI fragment of Gal1-10 is inserted into the corresponding sites of the vector pRS316, to create the galactose-inducible bacterial luciferase expression plasmids Gal ts-lux (bacterial luciferase-V.h.MAV) and Gal tr-lux (bacterial luciferase-V.h.CTP5). The firefly luciferase expression plasmid pGAL-FFL[SEL] was constructed by subcloning a HindIII–KpnI fragment containing the SEL allele of firefly luciferase (33) into pRS316 Gal1-10. Hsp82 expression plasmids piHGpd/P82 and piHGpd/G170D were constructed by subcloning the ClaI–NotI fragment from pTGpd/P82 and pTGpd/G170D (24) into pRS303 (34).
The haploid S. cerevisiae strains iP82a (wild type) and iG170Da (hsp90G170D) used in this study are derivatives of W303 (ade2-1 can1-100 his3-12,16 leu2-3,112 trp1-1 ura3-1; R. Rothstein, Columbia University). In these strains, both endogenous Hsp90 genes, HSP82 and HSC82, are deleted and wild-type or hsp90G170D sequences, regulated by the strong constitutive promoter GPD, are integrated into the genome. To create these strains, plasmids piHGpd/P82 and piHGpd/G170D were linearized with NheI and then used to transform ΔPCLDa (24). Transformants were grown on 5-fluoroorotic acid medium to select for those that had lost the wild-type HSC82 expression plasmid from ΔPCLDa. These strains, iP82a and iG170Da, were then transformed with expression plasmids for various test substrates: for p60v-src, plasmid Y316v-src; for firefly luciferase, plasmid pGAL-FFL[SEL]; for β-galactosidase, plasmid pLGSD5 (+ATG); for bacterial luciferase-V.h.MAV, plasmid Gal ts-lux, and for bacterial luciferase-V.h.CTP, plasmid Gal tr-lux.
Media.
Cells were cultured in SD, SR, or SGal (20 g of glucose, raffinose, or galactose, respectively, 5 g of ammonium sulfate, and 1.7 g of yeast nitrogen base without amino acids per liter, supplemented with essential amino acids and nucleotides) or YNBD [20 g of glucose, 1.7 g of yeast nitrogen base without amino acids, and 200 ml of phthalic acid (0.25 M, pH 5.5) per liter, supplemented with essential amino acids].
Assessment of Test Protein Activities.
To induce expression of the test proteins, cells were first grown overnight to 2 × 106 cells per ml in SR medium at 25°C. The raffinose medium allowed rapid induction of genes controlled by the GAL1 promoter when cells were shifted to medium containing galactose. Cells harvested by centrifugation were transferred to SGal medium that had been pre-equilibrated at 25°C, 34°C, 35°C, or 36°C. Cells were maintained in galactose medium for the minimal time required to obtain expression sufficient for reliable assays of individual test substrates: for p60v-src, 6 hr; for bacterial luciferase-V.h.MAV, 4 hr; and for firefly luciferase, β-galactosidase, and bacterial luciferase-V.h.CTP, 3 hr.
For heat inactivation experiments, cells were grown overnight to 2 × 106 cells per ml in galactose medium at 25°C to induce reporter enzyme expression. Glucose (2% final concentration) was added and cultures were incubated at 25°C for 30 min to inhibit further synthesis and allow newly synthesized proteins to mature. Cells were then transferred to prewarmed flasks at 25°C, 34°C, 37°C, 38°C, 39°C, 40°C, 41°C, or 42°C for 15 min to inactivate the proteins. For recovery experiments, cells were treated in the same manner, but cycloheximide (0.01 mg/ml) was added after heat inactivation, and cells were transferred to 34°C to recover for 0.5, 1, 2, 3, 4, and 5 hr.
To measure firefly luciferase activity, 100 μl of 0.5 mM luciferin (Sigma) was added to 100 μl of cells and light emission was immediately measured in an Opticomp 1 luminometer (MGM Instruments, Hamden, CT). β-Galactosidase activity was assessed as described (24). Briefly, 2 × 107 cells per sample were collected by centrifugation, washed with water, and frozen at −80°C. Thawed cells were resuspended in 150 μl of lysis buffer [0.1 M potassium phosphate, pH 7.8/20% (vol/vol) glycerol/1 mM dithiothreitol/2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride/2 μg of leupeptin per ml/2 μg of aprotinin per ml/2 μg of pepstatin A per ml]. Then 150 μl of glass beads (425–600 μm) were added and cells were lysed by agitation for 4 min at 4°C in a multitube Vortexer (VWR Scientific). β-Galactosidase activity was measured with the Glacto-light Kit (Tropix, Bedford, MA) in an Opticomp 1 luminometer and normalized to protein concentration (determined by the Bradford assay; Bio-Rad). Prior to measurement of bacterial luciferase-V.h.MAV activity, cells (50 ml) were collected by centrifugation and resuspended in 5 ml of raffinose medium containing 0.01 mg/ml cycloheximide to prevent further luciferase synthesis. Activity of bacterial luciferase was measured as light emission in an Opticomp 1 luminometer after the addition of 5 μl of n-decyl aldehyde (Sigma) to 0.5 ml of cells.
Protein Solubility Assay.
Cells were cultured in SD medium to a density of 2 × 106 cells per ml and transferred to YNBD medium supplemented with methionine, uracil, tryptophan, and adenine for 2 hr. Cells, 2 × 107 cells per sample, were collected by centrifugation and incubated for 15 min at 25°C, 34°C, and 36°C in the same medium without methionine. [35S]Methionine (Amersham) was added, 10 μCi per sample (1 μCi = 37 kBq), and incubation was continued for an additional 45 min without temperature change. Cells were washed two times with unlabeled medium, then incubated in unlabeled medium for 1 hr at the same temperature. Labeled cells were washed twice in cold medium. Cells were then resuspended in 300 μl of ice-cold lysis buffer [50 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/10 mM KCl/0.1 mM EDTA/1 mM benzamidine/0.1 mM dithiothreitol/2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride/10 μg cycloheximide per ml/2 μg of leupeptin per ml/2 μg of aprotinin per ml/2 μg of pepstatin A per ml]. Glass beads (300 μl) were added and cells were lysed by agitation in a multitube Vortexer for 4 min at 4°C. Beads and debris were removed by centrifugation at 1,500 × g for 5 min at 4°C. The beads were washed twice with 200 μl of ice-cold lysis buffer, which was pooled with the lysate.
For analysis of the total lysate, 200 μl of the pooled lysate was removed, 1/6 vol of concentrated sample buffer [390 mM Tris⋅HCl, pH 6.8/12% SDS (wt/vol)/60% glycerol (vol/vol)/30% 2-mercaptoethanol (vol/vol)/0.01 mg/ml bromophenol blue] was added, and the sample was immediately boiled for 5 min. Another 200 μl of the lysate was spun in a Beckman Optimal TL ultracentrifuge at 340,000 × g for 10 min. Sample buffer was added to the supernatant, and the sample was boiled for 5 min. The pellet was resuspended in 200 μl of lysis buffer, sample buffer was added, and the sample was boiled for 5 min.
Equal amounts of each sample were separated by SDS/10% PAGE and transferred to Immobilon-P membranes (Millipore). Proteins were stained with Coomassie blue. The membranes were then dried and exposed directly to Hyperfilm MP (Amersham) and developed in a Kodak RP X-Omat Processor.
Western Blot Analysis.
Cells (2 × 107 cells per sample) were collected by centrifugation, washed with water, and resuspended in 200 μl of ethanol containing 2 mM phenylmethylsulfonyl fluoride. Cells were lysed with glass beads for 4 min at 4°C. Proteins were precipitated at −35°C, dried in a SpeedVac Concentrator (Savant), resuspended in 200 μl of sample buffer 965 mM Tris⋅HCl, pH 6.8/2% SDS/10% glycerol/5% 2-mercaptoethanol/bromophenol blue), and boiled for 5 min.
Proteins were separated by SDS/10% PAGE, transferred to Immobilon-P membranes, and stained with Coomassie blue. Membranes were incubated in 10% nonfat dehydrated milk in PBS (180 g of NaCl per liter, 23 g of Na2HPO4 per liter, 4 g of KCl per liter, 0.02% thimerosal) for l hr, in primary antibody for 1 hr, in affinity-purified rabbit anti-mouse IgG antibody (Cappel) (when applicable) for 30 min, and with staphylococcal protein A-conjugated horseradish peroxidase (Boehringer Mannheim) for 1 hr. Immune complexes were visualized with the ECL reagent (Amersham). Antibody 4G10 (Upstate Biotechnology, Lake Placid, NY) was used to detect phosphotyrosine residues, antibody LA074 (Quality Biotech, Camden, NJ) to detect p60v-src, rabbit anti-luciferase (Promega) to detect firefly luciferase, antibody B-LUX (gift of A. Szalay, University of Alberta) to detect bacterial luciferase, and antibody 62b (gift of J. Warner, Albert Einstein College of Medicine) to detect yeast ribosomal protein L3.
RESULTS
Role of Hsp90 in de Novo Protein Folding.
To examine the role of Hsp90 in de novo protein folding, we placed the coding sequences of several test substrates under the control of the galactose-inducible promoter GAL1 (35). Cells were grown in raffinose medium and shifted to galactose medium that had been equilibrated at various temperatures. This allowed synthesis of the test substrates in cells with various levels of Hsp90 function.
We have previously shown that the oncogenic tyrosine kinase p60v-src, a protein whose maturation is highly dependent on Hsp90 (36), is inactive when synthesized in hsp90G170D cells at high temperatures (24). To titrate the activity of Hsp90G170D more precisely, we analyzed p60v-src activity when the protein was synthesized at 34°C, 35°C, and 36°C. Activity was assessed by the ability of p60v-src to promiscuously phosphorylate a broad array of yeast proteins (Fig. 1A). (Endogenous levels of tyrosine kinase activity are very low in yeast; see lane C, Fig. 1A, cells not expressing p60v-src.)
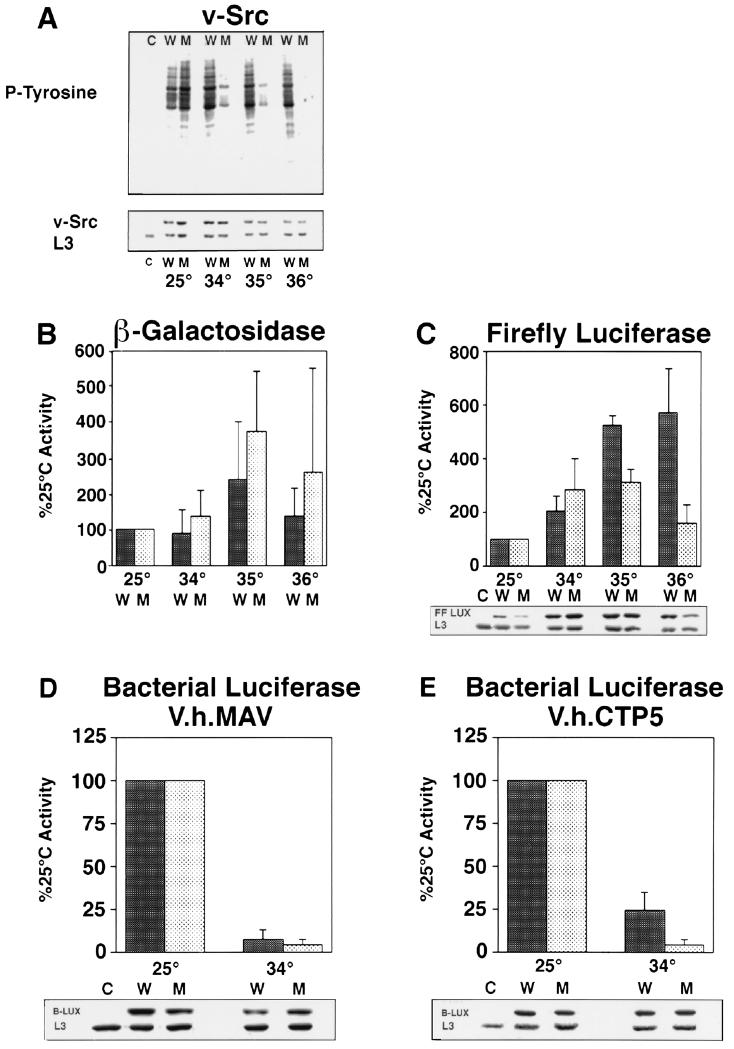
Figure 1.
Role of Hsp90 in de novo protein folding of test substrates. Activity and accumulation of newly synthesized proteins were assayed in wild-type (W) or hsp90G170D mutant (M) cells as follows: p60v-src (A), β-galactosidase (B), firefly luciferase (C), bacterial luciferase-V.h.MAV (D), and bacterial luciferase-V.h.CTP5 (E). Cells grown to midlogarithmic phase at 25°C in glucose were shifted to galactose at the indicated temperatures to simultaneously induce the expression of test substrates and inactivate Hsp90G170D. At 25°C, activities in wild-type and hsp90G170D cells were similar, but they varied somewhat in individual cultures grown from different colonies of each strain. Values at higher temperatures (means and standard deviations) are therefore normalized to the activity of each culture at 25°C. For each substrate, at least three independent experiments were performed, each including three independent transformants of each strain. Boxed data below activity measurements show the accumulation of substrate proteins as determined by immunological reaction with specific antibodies (Western blotting). Equal loading was confirmed by probing blots with antibody specific for the yeast ribosomal protein L3. C indicates wild-type cells lacking a substrate expression plasmid. Loss of Hsp90 function does not affect the maturation or accumulation of β-galactosidase, and it affects the accumulation of firefly luciferase only at temperatures close to the denaturation temperature of the protein, but it strongly affects the maturation of a bacterial luciferase protein that has difficulty folding even in wild-type cells.
Wild-type cells expressing p60v-src exhibited high levels of tyrosine kinase activity at all temperatures. hsp90G170D cells exhibited high levels of activity at 25°C. At 34°C, p60v-src activity was greatly reduced in hsp90G170D cells, and at 36°C, no activity was detected.
The accumulation of p60v-src was slightly compromised in hsp90G170D cells at higher temperatures. However, this effect was small relative to the decrease in p60v-src activity. The loss of p60v-src function was not a secondary consequence of cell death: hsp90G170D cells remained as viable as wild-type cells during the course of this experiment (data not shown). Similar results were obtained with another Hsp90 substrate, GR, the glucocorticoid receptor (ref. 24 and Y. Kimura and S.L., unpublished data). GR lost transcriptional activity (but continued to accumulate) over the same temperature range as p60v-src. Using these criteria, we conclude that in the hsp90G170D mutant, Hsp90 function is greatly reduced at 34°C and absent at 36°C.
This information was employed to examine the requirement for Hsp90 in the de novo folding of other proteins. With two of our test substrates, β-galactosidase and firefly luciferase, a chaperone function for Hsp90 has been demonstrated previously in vitro (13–17). When these proteins are unfolded in denaturant and diluted into aqueous buffers in a large excess of Hsp90, they are maintained in a folding-competent state; maturation into enzymatically active proteins can then be completed upon the addition of other chaperones (14, 17). When β-galactosidase was expressed in yeast cells, its activity varied at different temperatures (Fig. 1B). However, activity was similar in wild-type and hsp90G170D cells at 34–36°C. Thus, Hsp90 is not required for the de novo folding of this protein in vivo.
The expression of firefly luciferase was compromised by the Hsp90 mutation, but the effect was modest (Fig. 1C). At 36°C activity was reduced 3-fold. This reduction was matched by a similar reduction in firefly luciferase accumulation. Results with β-galactosidase (Fig. 1B) and bacterial luciferase (Fig. 1 D and E) indicate that the Hsp90 mutant causes no general defect in protein synthesis. Thus, the reduced accumulation of firefly luciferase at 36°C is most likely due to increased degradation. The firefly luciferase protein that does accumulate in Hsp90-deficient cells is active. Furthermore, at 34–35°C, temperatures at which the maturation of p60v-src and GR is greatly reduced in hsp90G170D cells, little or no reduction in firefly luciferase activity or accumulation was observed. If the maturation of firefly luciferase depends upon Hsp90 at all, it depends upon it much less than does the maturation of p60v-src and GR.
We also examined the effects of hsp90G170D on the de novo folding of bacterial luciferase fusion proteins. As with firefly luciferase, the activity of bacterial luciferase is measured by the production of light, but the two proteins employ different substrates and cofactors in their reaction cycles and are unrelated in sequence and structure. Thus, bacterial luciferase provides an independent test substrate for Hsp90 function.
When a bacterial luciferase fusion protein derived from V. harveyi MAV (bacterial luciferase-V.h.MAV) was synthesized in yeast, high levels of activity were observed in both wild-type and hsp90G170D cells at 25°C (Fig. 1D). However, when the protein was synthesized at 34°C very little activity was detected in either cell type, although the protein accumulated. As discussed below, once this protein has been matured at 25°C, its activity is stable at 34°C. Thus, the low levels of activity obtained when this enzyme is produced at 34°C are most likely due to the failure of S. cerevisiae chaperones to support its initial maturation at this temperature. The low level of activity in wild-type cells prevents testing the role of Hsp90 in the maturation of this substrate.
We therefore examined a closely related fusion protein constructed from the bacterial luciferase of the thermotolerant strain V. harveyi CTP5 (bacterial luciferase-V.h.CTP5). Twelve amino acid substitutions in this protein enhance its ability to fold in bacterial cells at high temperatures (31). In wild-type yeast cells, substantial activity was detected with this protein at 34°C (Fig. 1E). Note, however, that activity at 34°C was 1/4 of that at 25°C. Thus, this substrate will fold at higher temperatures in yeast, but it does so inefficiently. In hsp90G170D cells at 34°C, activity was reduced much more severely (1/25 of that in hsp90G170D cells at 25°C). Accumulation of bacterial luciferase-V.h.CTP5 was not compromised in either wild-type or hsp90G170D cells at 34°C. Thus, Hsp90 plays an important role in the de novo folding of this substrate to an active state in yeast cells.
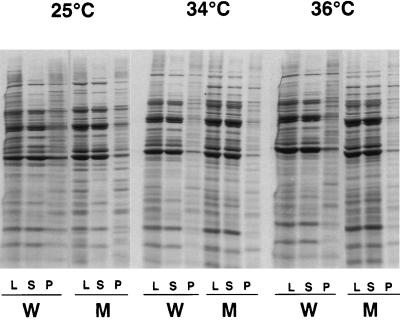
We next examined the general role of Hsp90 in the folding of yeast proteins. Because misfolded proteins have a strong tendency to aggregate, we reasoned that if Hsp90 was required for the de novo folding of a substantial fraction of yeast proteins, increased levels of aggregation would be found in the hsp90G170D mutant. In previous work with other chaperones, protein aggregation was readily detected by the pelleting of insoluble proteins at 340,000 × g (37). We used this method to examine the aggregation of newly synthesized cellular proteins in wild-type and hsp90G170D cells. Proteins were synthesized in the presence of [35S]methionine at 25°C, 34°C, or 36°C for 45 min, and cells were maintained at these temperatures for an additional hour in the presence of unlabeled methionine. No increase in aggregated protein was seen in hsp90G170D cells at any of the temperatures examined (Fig. 2). These results suggest that Hsp90 is not required for the folding of the majority of yeast proteins.
Figure 2.
The absence of Hsp90 function does not affect the solubility of most of newly synthesized yeast proteins. Cells grown to mid-logarithmic phase at 25°C were labeled with [35S]methionine at 25°C, 34°C, or 36°C for 45 min and maintained at the same temperatures for an additional hour in unlabeled medium. Solubility of proteins was assessed in wild-type (W) and hsp90G170D (M) cells by centrifugation of cell lysates at 340,000 × g, followed by SDS/PAGE of total lysates (L), supernatants (S), and pellets (P). Proteins were visualized by autoradiography.
Hsp90 Does Not Prevent the Thermal Inactivation of Proteins.
We next examined the role of Hsp90 in preventing the thermal inactivation of proteins in vivo. Wild-type and hsp90G170D cells were grown in galactose at 25°C to induce the expression of functional test proteins. They were then transferred to glucose to repress further expression and shifted to various temperatures to monitor enzyme inactivation.
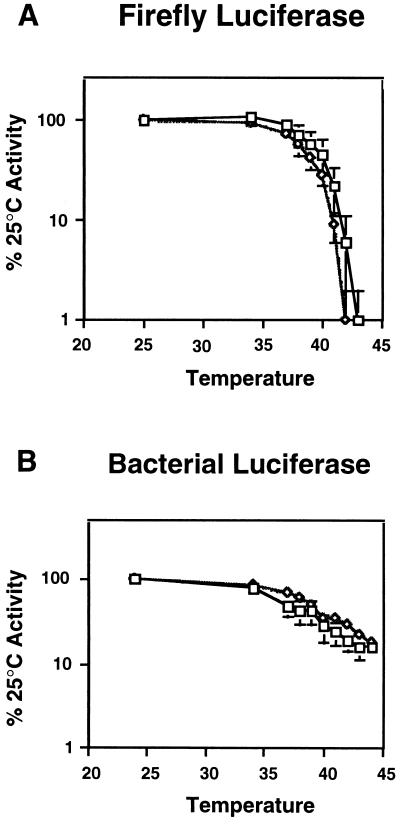
β-Galactosidase retained activity at all temperatures where cells remained viable (data not shown). Firefly luciferase precipitously lost activity at temperatures above 40°C (Fig. 3A). Surprisingly, bacterial luciferase-V.h.MAV was much more resistant to heat inactivation (Fig. 3B). Thus, this protein is temperature sensitive for folding (Fig. 1D), but once folded, is quite stable. With both luciferases, little difference was observed in the inactivation profiles of mutant and wild-type cells. Although conclusions drawn from only two substrates must be made with caution, the fact that these two substrates exhibit such different temperature-related folding problems implies that they are representative of a broader array of substrates. We suggest that Hsp90 is unlikely to play a role in protecting most proteins from heat inactivation.
Figure 3.
The absence of Hsp90 function has little effect on the heat-induced inactivation of test substrates. Wild-type (□) and hsp90G170D (⋄) cells expressing firefly luciferase (A) or bacterial luciferase-V.h.MAV (B) were grown to mid-logarithmic phase in galactose at 25°C to allow the accumulation of active test substrates. Cells were then shifted to the indicated temperatures for 15 min and the extent of enzyme inactivation was determined immediately. Enzyme activities were normalized to the activity present in each strain prior to heat inactivation. Data are the means and standard deviations of three independent experiments using three independent transformants.
We also examined the effect of heat on the aggregation of bulk yeast proteins labeled at 25°C. No significant difference was observed in the quantity of pelleted proteins in mutant and wild-type cells after exposure to 45°C for 15 min or to 44°C for 30 min (temperatures at which both cell types retained viability; data not shown).
Hsp90 Promotes the Recovery of Heat-Damaged Protein.
Finally, we examined the role of Hsp90 in the recovery of heat-damaged proteins in vivo by comparing recovery in the presence and absence of functional Hsp90. Test proteins were synthesized at 25°C and inactivated at 42°C, as before. The cells were then transferred to 34°C for recovery. At the same time, cycloheximide was added to block the synthesis of new protein. As expected, the bacterial luciferase-V.h.MAV protein, which was unable to fold at 34°C in either wild-type or hsp90G170D cells (Fig. 1D), was incapable of reactivation at 34°C (data not shown). More surprisingly, the bacterial luciferase-V.h.CTP5 protein, which could fold at 34°C in wild-type cells (Fig. 1E) was not reactivated at 34°C in either cell type. (Activity was restored at 25°C, but this regimen is not appropriate for examining the role of Hsp90 by this method because Hsp90G170D might regain activity at 25°C.)
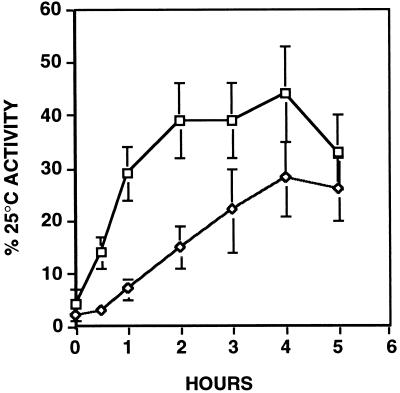
A role for Hsp90 in the reactivation of a heat-damaged protein was, however, readily demonstrated with firefly luciferase. Firefly luciferase recovered nearly 50% of its activity in wild-type cells at 34°C (Fig. 4). In hsp90G170D cells activity recovered to the same extent, but this recovery was achieved more slowly. Similar results were obtained in several other experiments, in which firefly luciferase was inactivated at different temperatures, to different levels (data not shown). Thus, the presence of Hsp90 speeds the recovery of the only heat-damaged substrate whose reactivation potential we could test by this method.
Figure 4.
Hsp90 speeds the recovery of heat-inactivated firefly luciferase. Firefly luciferase was inactivated at 42°C as described in the legend of Fig. 3. With the addition of a protein synthesis inhibitor, cells were returned to 34°C (a temperature at which most Hsp90 activity is lost in the hsp90G170D mutant). The recovery of previously synthesized, heat-inactivated firefly luciferase was monitored in wild-type (□) and hsp90G170D (⋄) cells as described in the text. Enzyme activities were normalized to the activity present in each strain prior to heat inactivation. Data are the means and standard deviations of three independent experiments using three independent transformants.
DISCUSSION
The Breadth of Hsp90 Function in Vivo.
We have taken advantage of a unique ts mutant of Hsp90, which rapidly and completely loses activity on shift to high temperatures (24), to investigate the role of Hsp90 in general protein folding in vivo. In vitro, Hsp90 can serve as a general chaperone, preventing the irreversible inactivation of several denatured substrates when present at high Hsp90-to-substrate ratios (11–18). Our data suggest, however, that the role of Hsp90 in living cells is more restricted: it is not required for the folding of most proteins, but plays an important role in the maturation of a select group of proteins whose folding is inherently problematic (38). It also facilitates, but is not required for, the reactivation of damaged protein in cells exposed to stress.
The Nature of Substrates That Depend on Hsp90 for de Novo Synthesis.
The role of Hsp90 in the maturation of total cellular proteins was assessed by determining their solubility. No increase in aggregation was observed among proteins synthesized in Hsp90-deficient cells. This assay is less direct than an enzymatic activity measurement. Nevertheless, if Hsp90 were required for the folding of the bulk of yeast proteins, we would have expected to see some difference in the state of aggregation in newly made proteins. Indeed, in similar experiments with a ts GroEL mutant in E. coli a dramatic increase in protein aggregation was observed (39).
Because chaperones solve folding problems common to many proteins, heterologous substrates that are easy to assay and manipulate experimentally provide valuable tools for deciphering chaperone functions. Accordingly, in addition to assaying the bulk of newly made yeast proteins, we employed heterologous test substrates that present different types of folding problems. The first test substrate we examined was β-galactosidase, a protein that presents several challenges to proper folding. Each subunit is large (116 kDa), composed of five structural domains, and must assemble into a tetramer to function (40). The maturation of this substrate was similar in the presence or absence of functional Hsp90. Taking our observations with total yeast proteins and β-galactosidase together, it seems highly unlikely that Hsp90 is required for the folding of most proteins in yeast.
Results with other test substrates, however, demonstrate that Hsp90 plays an important role in the maturation and maintenance of certain proteins and provide clues to the distinguishing features of those proteins. The test substrate whose folding was most dependent upon Hsp90 was a bacterial luciferase fusion protein. It accumulated at normal levels in Hsp90-deficient cells but was inactive. This was also the only test substrate that had difficulty achieving an active conformation even in wild-type cells at 34°C. Difficulty in acquiring a stable folded structure is a feature shared by the other known in vivo substrates of Hsp90—components of several signal transduction pathways illustrated by p60v-src and GR (22, 23). These proteins not only have difficulty achieving a stable conformation but also require continual association with Hsp90 to maintain their activity (24). Together, these data provide a compelling argument that the role of Hsp90 in de novo folding is generally restricted to proteins that have difficulty achieving stable structures. Notably, the folding properties of signal transducers are likely to be linked to their biological activities; their instability may facilitate the structural transitions by which they transduce signals. Thus, while the role of Hsp90 in bulk protein folding may be small, its role in governing the processes that control cell growth and differentiation may be large.
Effects of Hsp90 on Protein Degradation.
Hsp90 deficiencies exhibited a different effect on our third test substrate. Firefly luciferase was unaffected by the hsp90G170D mutation at 34°C and 35°C, where Hsp90 activity is very low. At 36°C, where Hsp90 activity is completely lost, accumulation of this luciferase was reduced 3-fold, but activity was normal for this quantity of protein. One explanation for these observations is that a fraction of firefly luciferase employs Hsp90 in its maturation, and does so efficiently even at 34°C when very little Hsp90 is available. Then, when Hsp90 function is completely lost, that fraction misfolds and is degraded. This, however, would distinguish firefly luciferase from all three of the other proteins we have studied that depend upon Hsp90 for de novo folding: bacterial luciferase, p60v-src, and GR accumulate in the hsp90G170D mutant as inactive proteins at near normal levels. Since 36°C is close to the denaturation temperature of firefly luciferase in yeast, it seems more likely that firefly luciferase matures in the absence of Hsp90 function, but some of molecules then unfold and are degraded.
Several laboratories have reported that Hsp90 substrates are degraded when cells or reticulocyte lysates are treated with the Hsp90 inhibitor geldanamycin (22). In vivo we have also observed a large reduction in the accumulation of Hsp90 substrates, with other Hsp90 mutants (24). These mutants, unlike hsp90G170D, have impaired Hsp90 function at normal temperatures and do not completely lose activity at elevated temperatures. Perhaps defects in Hsp90 activity lead to the degradation of its substrates only when a mutation or inhibitor causes Hsp90 to remain associated with the substrate in a futile or inefficient cycle (15).
A Useful Set of Proteins for Folding Studies.
Our results with bacterial and firefly luciferase establish a set of substrates that are easy to assay in yeast cells and have very different folding properties. The de novo folding of firefly luciferase is much less temperature sensitive than that of the bacterial luciferase fusion proteins. However, once these substrates have folded, firefly luciferase is much more sensitive to heat inactivation than bacterial luciferase is. [The ts-for-folding properties of bacterial luciferase are analogous to those of certain mutants of P22 tail-spike protein (41).] Once it is heat inactivated, firefly luciferase is more readily reactivated than is bacterial luciferase. Additionally, when the maturation of these substrates is compromised by the Hsp90 chaperone mutation, the accumulation of firefly luciferase is reduced, whereas bacterial luciferase accumulates as a nonfunctional but stable protein. The different properties of these substrates should prove of general utility for future in vivo protein folding and stress tolerance studies.
Role of Hsp90 Under Stressful Conditions.
Hsp90 deficiencies had no effect on the thermal inactivation profiles of either firefly luciferase or bacterial luciferase. They did, however, reduce the rate at which heat-inactivated firefly luciferase was reactivated. These results are in agreement with recent observations that geldanamycin inhibits the recovery of heat- or chemically inactivated firefly luciferase in SW620 colon carcinoma cells and reticulocyte lysates (15, 16, 42). If this ability of Hsp90 to promote the reactivation of heat-denatured firefly luciferase is indicative of a more general function in the recovery of heat-damaged proteins, the question arises: why do Hsp90 mutations have no notable effect in standard thermotolerance assays (8, 43), which measure survival after short exposures at extreme temperatures? Perhaps Hsp90 increases the rate at which damaged proteins refold but does not greatly affect the final yield.
Although Hsp90 does not appear to protect proteins from heat inactivation in vivo, it may interact with partially unfolded conformations, prevent further unfolding, and facilitate a more rapid recovery. Such an Hsp90 function might not have a dramatic effect on survival in a laboratory setting, but might play a vital role combating the selective pressures that operate on a species in evolution.
Although Hsp90 is an extremely abundant protein at normal temperatures, studies in yeast indicate that its levels can be reduced at least 10-fold without affecting cell viability. However, much higher concentrations are required for growth at higher temperatures (8). Our data provide a likely explanation for this observation. At normal temperatures, Hsp90 is required to facilitate the folding of a small subset of proteins that have difficulty reaching or maintaining their native conformations. At higher temperatures, proteins that have difficulty folding become even more problematic and some otherwise stable proteins encounter problems in folding and stability, thus increasing the requirement for Hsp90.
How Might General Protein Folding Proceed in Eukaryotes?
The question remains, how do eukaryotic cells ensure that the bulk of newly synthesized proteins acquire their native conformations at normal temperatures? The answer may lie in the suggestion by Netzer and Hartl (44) that protein folding is largely post-translational in prokaryotes and co-translational in eukaryotes. Post-translational folding would lead to the formation of unfolded aggregation-sensitive polypeptides, which require the GroEL/ES chaperone to reach their native conformation. In eukaryotes, on the other hand, the sequential co-translational folding of individual domains of polypeptide chains would reduce the formation of aggregation-sensitive intermediates. Thus, the Hsp70 chaperone system might be sufficient to mediate the folding of most cytosolic proteins, and only certain proteins with structurally unstable or discontiguous domains would require additional help from either TriC or Hsp90 (44). It seems likely that eukaryotic cells will prove to contain many other specialized chaperones that function in the context of specific substrates. However, the eukaryotic cytosol may not contain a chaperone with a function equivalent to that of GroEL/ES. Certainly, our results indicate Hsp90 is unlikely to play a major role in bulk protein folding.
Acknowledgments
We thank D. Morgan for the v-src plasmid, D. Gottschling for the β-galactosidase expression plasmid, A. Szalay for the bacterial luciferase plasmids, and J. Warner for the ribosomal protein L3 antibody. We acknowledge A. S. Kowal for his technical assistance. We are grateful to E. A. Craig, J. Glover, Y. Kimura, S. Rutherford, T. Serio, M. Singer, and N. Sondheimer for their thoughtful comments on the manuscript. D.F.N. was supported by National Institutes of Health Fellowship GM14745. This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health, Grant GM25843.
ABBREVIATIONS
- ts
temperature-sensitive
- bacterial luciferase-V.h.MAV
bacterial luciferase fusion protein from Vibrio harveyi strain MAV
- bacterial luciferase-V.h.CTP5
bacterial luciferase fusion protein from V. harveyi CTP5
- Hsp
heat shock protein
- GR
glucocorticoid receptor
References
- 1.Anfinsen C B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Buchner J. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 3.Hartl F U. Nature (London) 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 4.Johnson J L, Craig E A. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 5.Langer T, Lu C, Echols H, Flanagan J, Hayer M K, Hartl F U. Nature (London) 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 6.Buchberger A, Schroder H, Hesterkamp T, Schonfeld H J, Bukau B. J Mol Biol. 1996;261:328–333. doi: 10.1006/jmbi.1996.0465. [DOI] [PubMed] [Google Scholar]
- 7.Bose S, Weikl T, Bugl H, Buchner J. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 8.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardwell J C, Craig E A. J Bacteriol. 1988;170:2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutforth T, Rubin G M. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 11.Miyata Y, Yahara I. J Biol Chem. 1992;267:7042–7047. [PubMed] [Google Scholar]
- 12.Wiech H, Buchner J, Zimmermann R, Jakob U. Nature (London) 1992;358:169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]
- 13.Schumacher R J, Hurst R, Sullivan W P, McMahon N J, Toft D O, Matts R L. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- 14.Freeman B C, Morimoto R I. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider C, Sepp-Lorenzion L, Nimmesgem E, Querfelli O, Danishefsky S, Rosen N, Hartl F U. Proc Natl Acad Sci USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thulasiraman V, Matts R. Biochemistry. 1996;35:13443–13450. doi: 10.1021/bi9615396. [DOI] [PubMed] [Google Scholar]
- 17.Yonehara M, Minami Y, Kawata Y, Nagai J, Yahara I. J Biol Chem. 1996;271:2641–2645. doi: 10.1074/jbc.271.5.2641. [DOI] [PubMed] [Google Scholar]
- 18.Uma S, Hartson S D, Chen J, Matts R L. J Biol Chem. 1997;272:11648–11656. doi: 10.1074/jbc.272.17.11648. [DOI] [PubMed] [Google Scholar]
- 19.Freeman B C, Toft D O, Morimoto R I. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 20.Kimura Y, Rutherford S L, Miyata Y, Yahara I, Freeman B C, Yue L, Morimoto R I, Lindquist S. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 21.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 22.Pratt W B. Annu Rev Pharmacol Toxicol. 1997;37:297–326. doi: 10.1146/annurev.pharmtox.37.1.297. [DOI] [PubMed] [Google Scholar]
- 23.Pratt W B, Toft D O. Endocrinol Rev. 1997;18:319–360. [Google Scholar]
- 24.Nathan D F, Lindquist S. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith D F. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 26.Prodromou C, Roe S M, Piper P W, Pearl L H. Nat Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- 27.Stebbins C E, Russo A A, Schneider C, Rosen N, Hartl F U, Pavletich N P. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 28.Murphy S M, Bergman M, Morgan D O. Mol Cell Biol. 1993;13:5290–5300. doi: 10.1128/mcb.13.9.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottschling D. Proc Natl Acad Sci USA. 1982;79:7410–7414. [Google Scholar]
- 30.Escher A, O’Kane D J, Lee J, Szalay A A. Proc Natl Acad Sci USA. 1989;86:6528–6532. doi: 10.1073/pnas.86.17.6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escher A, Szalay A A. Mol Gen Genet. 1993;238:65–73. doi: 10.1007/BF00279532. [DOI] [PubMed] [Google Scholar]
- 32.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 33.Distel B, Gould S J, Voorn-Brouwer T, Van der Berg M, Tabak H F, Subramani S. New Biol. 1992;4:157–165. [PubMed] [Google Scholar]
- 34.Sikorski R S, Heiter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston M, Davis R W. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Lindquist S. Proc Natl Acad Sci USA. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 38.Smith D F, Whitesell L, Nair S C, Chen S, Prapapanich V, Rimerman R A. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horwich A L, Low K B, Fenton W A, Hirshfield I N, Furtak K. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson R H, Zhang X-J, DuBose R F, Matthews B W. Nature (London) 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 41.Mitraki A, Danner M, King J, Seckler R. J Biol Chem. 1993;268:20071–20075. [PubMed] [Google Scholar]
- 42.Schumacher R J, Hansen W J, Freeman B C, Alnemri E, Litwack G, Toft D O. Biochemistry. 1997;35:14889–14898. doi: 10.1021/bi961825h. [DOI] [PubMed] [Google Scholar]
- 43.Kimura Y, Matsumoto S, Yahara I. Mol Gen Genet. 1994;242:517–527. doi: 10.1007/BF00285275. [DOI] [PubMed] [Google Scholar]
- 44.Netzer W J, Hartl F U. Nature (London) 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]