Abstract
The small GTPases Cdc42 and Rac regulate a variety of biological processes, including actin polymerization, cell proliferation, and JNK/mitogen-activated protein kinase activation, conceivably via distinct effectors. Whereas the effector for mitogen-activated protein kinase activation appears to be p65PAK, the identity of effector(s) for actin polymerization remains unclear. We have found a putative effector for Drosophila Cdc42, Genghis Khan (Gek), which binds to Dcdc42 in a GTP-dependent and effector domain-dependent manner. Gek contains a predicted serine/threonine kinase catalytic domain that is 63% identical to human myotonic dystrophy protein kinase and has protein kinase activities. It also possesses a large coiled-coil domain, a putative phorbol ester binding domain, a pleckstrin homology domain, and a Cdc42 binding consensus sequence that is required for its binding to Dcdc42. To study the in vivo function of gek, we generated mutations in the Drosophila gek locus. Egg chambers homozygous for gek mutations exhibit abnormal accumulation of F-actin and are defective in producing fertilized eggs. These phenotypes can be rescued by a wild-type gek transgene. Our results suggest that this multidomain protein kinase is an effector for the regulation of actin polymerization by Cdc42.
Small GTPases of the Rho subfamily fulfill important cellular functions (1, 2). In mammalian fibroblasts, Rho, Rac, and Cdc42 regulate actin polymerization that underlies the formation of stress fiber, lamellipodia, and filopodia, respectively. In addition to the regulation of the cytoskeleton, the Rho family GTPases are involved in the activation of kinase cascades and regulation of cell proliferation (1, 2). Indeed, perturbations of the activity of these GTPases in yeast, flies, and mice result in disruptions of many biological processes, including yeast budding (3), axon and dendrite outgrowth (4, 5), myoblast fusion (4), epithelial cell shapes (6, 7), and tissue polarity establishment (8).
Different functions of the Rho family GTPases are likely to be mediated by different downstream effectors. Several structurally distinct effectors have been found for Rho (9). Rac and Cdc42 share more sequence similarities than they do with Rho, and they also share some common effectors. Recent studies using effector-domain mutants have shown that different effectors serve different functions. For example, point mutations in Rac and Cdc42 that eliminate their abilities to bind to Pak, a protein kinase effector for both GTPases, disrupt their functions in mitogen-activated protein kinase activation, but not in cytoskeleton regulation (10, 11), suggesting that Pak is the effector for Rac and Cdc42 in the activation of the mitogen-activated protein kinase cascades. The effectors of Rac and Cdc42 for the regulation of actin polymerization remain to be characterized.
To understand the mechanisms by which Cdc42 and Rac regulate neuronal morphogenesis (12), we set out to identify additional components in the Cdc42/Rac signaling pathways in Drosophila. In this paper, we describe the study of a multidomain protein kinase, Genghis Khan (Gek), which appears to be a downstream effector of Drosophila Cdc42 and a regulator of actin polymerization.
MATERIALS AND METHODS
Two-Hybrid Screen.
Detailed procedures for library screening and subsequent testing of isolated clones are described in ref. 13. 3-amino-triazole was added at 20 mM to triple selection plates to reduce false positives during the selection for His+. Of 6 million clones screened, two independent clones of gek were isolated (c17 twice, c12 once, see Fig. 2a).
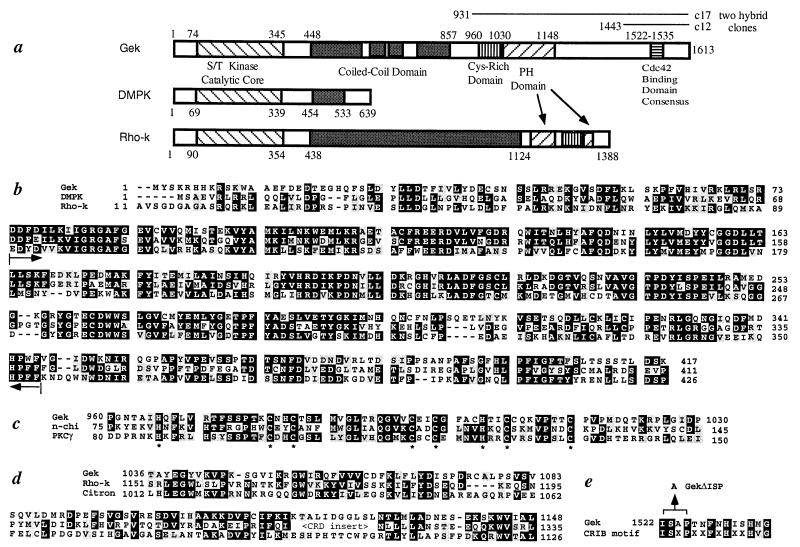
Figure 2.
Primary structure of Gek. (a) A schematic drawing of the Gek protein structure, with comparison to human DMPK (38) and bovine Rho-kinase (Rho-k) (25). Gek has four regions (448–589, 617–669, 685–738, and 784–857) that are predicted to form coiled-coil structures (score of 0.4 or above) (39). The lines above the Gek structure represent the original clones (c17 and c12) identified in the two-hybrid screen. (b) Sequence of the N terminus of Gek compared with DMPK and Rho-k. Brackets within the two arrows indicate the catalytic core of the kinase domain (20). Sequence similarities exist beyond the catalytic cores of these three proteins. (c) Sequence comparison of the Cys-rich domain of Gek with those of two phorbol ester binding proteins, n-chimerin (40), and protein kinase C γ (21). Conserved His and Cys residues are highlighted by ∗. (d) The pleckstrin homology (PH) domain of Gek aligned with those of Rho-k (25) and Citron (41). Citron also shares general structural similarity with Gek yet no kinase domain was reported (41). Note that the PH domain of Rho-k is split by a Cys-rich domain. (e) The Cdc42-binding domain of Gek aligned with a recently identified Cdc42/Rac interactive binding consensus sequence (23). The position of the three amino acids deleted in GekΔISP is indicated. In all parts, the numbers represent that of amino acids within their respective proteins, whereas black and gray shadows represent identity and conservative changes of amino acids among different proteins, respectively.
In Vitro Interaction.
35S-labeled full-length wild-type Gek and GekΔISP proteins were generated in vitro by using the TNT Coupled Reticulocyte Lysate System according to manufacturer’s specifications (Promega). Forty-five microliters of the reaction product was incubated with glutathione S-transferase fusion proteins of various GTPases purified from 400 μl of induced culture and 20 μl glutathione-Sepharose (Pharmacia) in 50 mM Tris⋅HCl, pH 7.5/50 mM NaCl/5 mM MgCl2/1 mM DTT/0.01% Nonidet P-40/33 μM GTPγS (total volume 300 μl) for 1 hr at 4°C. Proteins bound to the Sepharose beads were isolated by centrifugation, washed, and subjected to SDS/PAGE. The dried gel was exposed to x-ray film to reveal Gek proteins that were bound to the GTPases.
Molecular Biology.
A 5.1-kb cDNA clone that contains the entire ORF of Gek was isolated from the Brown’s embryonic library (14) by using digoxigenin-labeled (Boehringer) clone c17 (see Fig. 2a) as a probe. Sequence analysis revealed a single ORF from nucleotides 189–5020, encoding a protein of 1,613 amino acids (see Fig. 2a). The translation initiation codon was preceded by stop codons in all three reading frames. A polyadenylation signal AATAAA was identified at nucleotide 5086, followed by a poly(A) tail starting at nucleotide 5114. The complete cDNA and protein sequences are deposited in the GenBank. Homology search was performed by using the blast program. Sequence alignments (see Fig. 2 b–d) were performed by using the program clustal w, and identity and conservative changes were highlighted by using the boxshade program.
Point mutations in the kinase domain and in the Cdc42-binding domain were introduced via oligo-directed mutagenesis according to manufacturer’s specification (Amersham). Wild-type and A105K Gek (point mutations in the kinase domain) were fused in-frame with two copies of myc epitope at their N-termini and subcloned into a Schneider cell expression vector that contains the actin 5C promoter (15) for transfection.
A P-element insertion line obtained from the Drosophila Genome Project was found to have a P-element inserted between nucleotides 51 and 52 with respect to the numbering of the gek cDNA (see Fig. 4). Plasmid rescue recovered an XbaI fragment that contains 2 kb of sequences 5′ to the insert. The sequence from the rescued plasmid did not match the sequence from nucleotides 1–51 of the cDNA, or the sequence of the corresponding genomic DNA, and yet the clone maps to 60B by chromosome in situ hybridization, indicating that a deletion is associated with the P-element insertion (see Fig. 4). With primers from the plasmid rescue fragment and those of the gek coding region, we used PCR to identify deletions of gek generated from imprecise P-element excisions (see Fig. 4). The extent of deletion of gekD23 was determined by direct sequencing of the PCR products (the first 651 nucleotides of the cDNA and the coding sequences for the first 155 amino acids are deleted). Exon/intron structures were deduced from the comparison of genomic and cDNA sequences and from restriction enzyme analysis.
Figure 4.
Molecular map of the gek locus. 0 represents the 5′ end of a full-length cDNA. The transcription unit is shown below, with the introns in dotted lines and the ORF in bold. The P-element insertion site is indicated. Brackets mark the extent of deletions in the three gek alleles (D23, D31, and D134) used for the phenotypic analysis. The hatched bar represents the construct used to rescue the oogenesis phenotypes. Restriction enzymes: R, EcoRI; H, HindIII; S, SalI; and X, XbaI.
The rescue construct was made by piecing together genomic DNAs obtained from the gek region. The final construct contains DNA from −6 kb to +6.6 kb, with reference to the first nucleotide of the cDNA (see Fig. 4). Northern analysis was performed by using poly(A)+ mRNA derived from 250 μg of total RNA from ovary and hybridized with 32P-labeled DNA fragments corresponding to the entire rescue construct.
Transfection, Immunoprecipitation, and Kinase Assay.
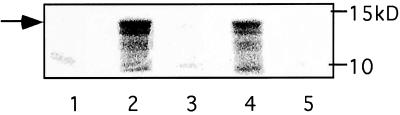
Schneider S2 cells were transfected with actin 5C promoter-myc-Gek (wild-type or A105K) expression constructs by using the calcium phosphate precipitation method as described (15). Thirty-six hours after transfection, cells from two 25-cm2 T flasks were collected and lysed for 20 min on ice in 400 μl of lysis buffer containing 50 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 25 mM NaF, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 0.2 mM orthovanadate, with the following protease inhibitor: 0.2 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 0.1 μg/ml aprotinin. The supernatant collected after a 10-min spin at 14,000 rpm at 4°C was mixed with equal volume of IP wash buffer (identical to lysis buffer except only 0.1% Triton X-100 was included) and precleared with 30 μl of protein G-Sepharose for 30 min at 4°C. The supernatant was incubated with 20 μl of mAb 9E10 against myc (Santa Cruz Biotechnology) for 4 hr at 4°C, with 30 μl of protein G-Sepharose for an additional 30 min. The immunoprecipitate was collected at 4,000 rpm for 30 sec, washed three times with IP wash buffer, and once with kinase buffer containing 2 mM DTT, 5 mM MnCl2, 10 mM MgCl2, 50 mM NaCl, 50 mM Hepes at pH 7.3, 0.03% Briji 35. The immunoprecipite was resuspended in a total volume of 30 μl containing 1 × kinase buffer, 5 μg of histone 2A (Sigma), and 44 μM (4 mCi) of [γ-32P]ATP. The kinase reaction was carried out at 30°C for 30 min. The reaction was stopped by the addition of equal volume of 2× SDS sample buffer and subjected to SDS/PAGE analysis. Phosphorylation of histone was visualized by PhosphorImager (see Fig. 3).
Figure 3.
Kinase activity of Gek. Histone phosphorylation by anti-myc immunoprecipitation complex is observed in S2 cells transfected with myc-Gek expression construct (lanes 2 and 4, duplicate experiments), but absent in mock transfected S2 cells (lane 1) as well as in S2 cells transfected with myc-Gek bearing the A105K mutation (lanes 3 and 5, duplicate experiments). The molecular mass marker is shown to the right, and the arrow represents the full-length histone 2A protein (14.5 kDa).
Analysis of Germ-Line Mosaic.
Third instar larval progeny of the cross yw,P[hsFLP]/Y;FRT,P[ovoD1]/CyO × yw; FRT,gek/CyO were subjected to two 30-min heat shock treatments at 37°C with 30 min rest at 25°C in between. Straight wing adult females (genotype: yw,P[hsFLP];FRT,P[ovoD1]/FRT,gek) were mated with sibling males and maintained in vials with fresh yeast for a few days, before egg collection for fertility analysis (see Fig. 5a) and ovary dissection for histological analysis. Ovaries were dissected in ice-cold 100 mM phosphate buffer pH 7.2 (PB), fixed in 4% formaldehyde in PB for 20 min at room temperature, and washed several times in PBT (PB + 0.1% Triton). Phalloidin and antibody staining was conducted in PBT with the following conditions: rhodamine-phalloidin (Molecular Probes), 4 units/ml; anti-Hts, 1:1; anti-Kelch, 1:1; anti-phosphotyrosine (4G10, Upstate Biotechnology, Lake Placid, NY), 1:200; dichlorotriazinly amio fluorescein-coupled secondary antibodies (The Jackson Laboratory), 1:200. Images were collected on a MRC 600 confocal microscope and processed with Adobe Photoshop and Adobe Illustrator.
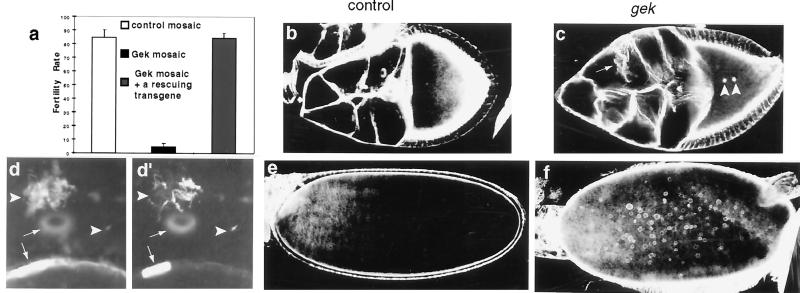
Figure 5.
gek mutants are defective in oogenesis and in actin cytoskeletal organization in egg chambers. (a) Fertility rate of eggs laid from control mosaic females (open bar, genotype: yw,P[hsFLP]/+;FRT,P[ovoD1]/FRT), gek mosaic females (black bar, genotype: yw,P[hsFLP]/+;FRT, P[ovoD1]/FRT,gekD23) and gek mosaic females in the presence of one copy of a rescuing transgene (gray bar, genotype: yw,P[hsFLP]/+;FRT,P[ovoD1]/FRT,gekD23;T35/+). (b and c) Phalloidin staining of stage 9 control (b) and gek (c) egg chambers. (d and d′) High-magnification images in nurse cells of a stage 9 gek mosaic egg chamber double-labeled with phalloidin (d) and anti-Hts (d′). In gek mutant clones, the cortical actin surrounding the nurse cells is disorganized (arrow in c) compared with the smooth structure in control mosaics (b). Ectopic actin polymerization occurs in oocytes (arrowheads in c) and in nurse cells (arrowheads in d). Such ectopic F-actin structures in nurse cells also are recognized by antibody to Hts (arrowheads in d′). Small arrows indicate the ring canals. (e and f) Phalloidin staining of stage 12 control (e) and gek (f) oocytes. In late-stage oocytes ectopic F-actin forms numerous spheres surrounding the yolk granules in gek mosaics (f). No such actin spheres are found in oocytes of control mosaics (e). Genotypes of control and gek mosaics in b–f are the same as in a.
Naming the Gene.
Prompted by the phenotype of ectopic F-actin accumulation, we named our protein after the powerful Mongolian emperor Genghis Khan. The shortened name, Gek, also stands for GTPase effector kinase.
RESULTS
Identification of Gek as a Drosophila Cdc42-Binding Protein.
To search for proteins that physically interact with the Drosophila small GTPases Drac1 and Dcdc42, we used the yeast two-hybrid method (16) and used as bait the LexA DNA-binding domain fused with Drac1 or Dcdc42 (4) bearing two modifications. First, Gly-12 was changed to Val-12 to disrupt the GTPase activity, so that the mutant proteins remain GTP-bound and thus are constitutively active (4, 17). Second, the C-terminal Cys-XXX motif responsible for lipid addition and membrane targeting (18) was changed to Ser-stop to facilitate nuclear targeting of the fusion proteins.
Two independent clones of gek (see Fig. 2a) were isolated from a yeast two-hybrid library containing cDNAs derived from 3- to 12-hr Drosophila embryos, based on their abilities to bind to Dcdc42V12 (Fig. 1a). Further tests revealed that Gek does not bind to Dcdc42N17 (Fig. 1a), a dominant negative mutant that preferentially stays in the GDP-bound state (17, 19). The ability of Gek to bind Dcdc42 in its GTP-bound, but not GDP-bound, form suggests that it is an effector of Dcdc42. This possibility is consistent with the finding that a mutation in the Dcdc42 effector domain (A35) important for signaling to downstream targets eliminated Gek binding. Gek does not bind to Drac1V12 (Fig. 1a).
Figure 1.
Gek binds specifically to Dcdc42. (a) Summary of two-hybrid interactions of Gek (clone c17, see Fig. 2a) with various GTPase baits. Interactions were assayed both for the ability to produce His+ colonies on triple selection plates, and for β-galactosidase activity, of yeast cells containing both plasmids (13). None of the GTPase fusion proteins interacted with the activation domain alone. (b) Binding of in vitro-translated full-length Gek wild-type (lanes 1–4) or GekΔISP bearing a three amino acid deletion (see Fig. 2e) in the Cdc42 binding domain (lanes 5–8) to various glutathione S-transferase-GTPases fusion proteins shown at the bottom of the figure. Only activated form of Dcdc42 (Dcdc42V12) can bind to wild-type Gek (lane 3). Mutations in the effector domain (A35) of Dcdc42 (lane 4) or in the Cdc42 binding domain of Gek (lanes 7 and 8) disrupt the binding. Drac1 does not bind to Gek (lanes 1, 2, 5, and 6). The arrow indicates the largest translation product of Gek, which matches the predicted molecular mass of 184 kDa.
To confirm the specific physical interaction of Dcdc42 and Gek, we incubated 35S-labeled in vitro-translated Gek with purified glutathione S-transferase fusion proteins of various forms of Dcdc42 and Drac1 and examined whether glutathione-Sepharose could coprecipitate Gek. We found that Dcdc42V12, but not Dcdc42V12A35, binds to wild-type Gek protein in this assay (Fig. 1b, lanes 3 and 4). Neither Drac1V12 nor Drac1V12A35 binds to Gek (Fig. 1b, lanes 1 and 2). Taken together with the data from the two-hybrid assay, these findings indicate that Gek is likely an effector specific for Dcdc42.
Primary Structure of Gek.
Sequence analysis of a full-length cDNA clone revealed that gek encodes a large protein of 1,613 amino acids (Fig. 2a). The N terminus of Gek contains a predicted Ser/Thr kinase catalytic domain (20) (Fig. 2b). It is followed by a large coiled-coil domain with sequence homology to myosin heavy chain (Fig. 2a), a Cys-rich domain similar to the phorbol ester/diacylglycerol binding domain of protein kinase C (21) (Fig. 2c), and a pleckstrin homology domain (Fig. 2d), which is found in many signaling molecules and used for protein-lipid interactions and the recruitment to the cell surface (22). Near the C terminus of Gek resides the sequence that resembles a Cdc42/Rac interactive binding (CRIB) domain (Fig. 2e) (23). Indeed, deletion of three residues in Gek (GekΔISP) (Fig. 2e) that correspond to three conserved residues of the CRIB domain disrupted its binding to Dcdc42 (Fig. 1b, compare lanes 3 and 7).
Gek exhibits strong sequence similarity with human myotonic dystrophy protein kinase (DMPK) (24). Gek and DMPK share 63% amino acid sequence identity within the 271-aa catalytic core (Fig. 2b), and the sequence similarity between these two proteins extends beyond the catalytic domain in both directions (Fig. 2 a and b). DMPK, however, is much smaller than Gek. Interestingly, a recently identified class of Rho-binding kinases (25–27), which may function as effectors of the small GTPase Rho, is similar to Gek in their domain structures (Fig. 2a). In addition, the kinase domain of the Rho-binding kinase is also similar to DMPK, although DMPK is more similar to Drosophila Gek than to mammalian Rho-kinase (63% vs. 49% identity in the catalytic core). Also the phorbol ester binding domain of protein kinase C bears stronger similarity to the Cys-rich domain of Gek (Fig. 2c) than that of the Rho-binding kinase (25–27).
Gek Is a Protein Kinase.
To test if Gek exhibits kinase activity, we transfected Drosophila Schneider cells (S2) with a myc-epitope-tagged wild-type Gek expression construct under the control of a ubiquitous actin promoter. We then used anti-myc antibody to immunoprecipitate the Gek protein. By using histone as a substrate, we detected kinase activity in the complex immunoprecipitated from cells transfected with myc-Gek but not from mock transfected cells (Fig. 3, compare lanes 2 and 4 with lane 1). To rule out the possibility that the kinase activity is caused by another kinase that is tightly associated with Gek during the immunoprecipitation procedure, we transfected S2 cell with a myc-tagged mutant Gek construct (A105K) bearing a mutation of the lysine residue in the kinase domain predicted to be essential for kinase activities (28). Immunoprecipitated myc-GekA105K did not exhibit any kinase activity above background (Fig. 3, lanes 3 and 5), although the GekA105K protein is expressed at a comparable level as wild-type Gek, indicated by Western blot probed with anti-myc antibody (data not shown). Because it is unlikely that a single point mutation in the kinase domain would disrupt the binding of Gek to other associated protein(s), we conclude that the Gek protein exhibits kinase activity.
gek Mutants Exhibit Abnormal Actin Polymerization.
To investigate the in vivo function of Gek, we initiated genetic analysis by generating loss-of-function mutations in the gek locus. Having mapped gek by chromosome in situ hybridization to region 60B (data not shown), we identified a Drosophila mutant with a P-element insertion in the 5′ untranslated region of the gek transcription unit (Fig. 3) along with a deletion of sequences upstream of the insertion site (see Materials and Methods). We then used a PCR-based screen and identified a number of imprecise P-element excisions that have additional deletions of the gek gene. We have used three such alleles for phenotypic analysis (Fig. 4). These alleles gave similar results in all of the studies described below. One of these alleles, gekD23, lacks the sequences between the P-element insertion site and the codon for amino acid 156 of the Gek protein, and hence has a deletion that removes the N-terminal third of the catalytic domain of the kinase (20).
Homozygous gek mutants die as larvae. Prompted by the high level of the gek transcript in precellularization stage embryos (data not shown), we investigated the possible function of gek in oogenesis by generating females that contained germ-line clones homozygous for gek by using the high-efficiency germ-line mosaic method (29). This method is designed to eliminate germ-line cells that are not homozygous for gek. Although the number of eggs produced from gek mosaic females was comparable to that from control mosaic females, only 5% of the eggs were fertilized (Fig. 5a). This suggests that Gek function is essential for proper oogenesis.
To test whether Gek is involved in the formation of the well characterized actin cytoskeleton in egg chambers (30), we compared the actin cytoskeleton of wild-type and gek mutant egg chambers by using phalloidin staining, which labels polymerized actin (F-actin). The F-actin-rich ring canals between nurse cells and between the oocyte and the nurse cells (30) were present in egg chambers homozygous for gekD23 (Fig. 5d, arrows). However, the cortical F-actin that surrounds the nurse cells appeared abnormal (compare Fig. 5 c and b), and ectopic F-actin blobs frequently were observed in the nurse cells (Fig. 5d, arrowheads) and oocytes (Fig. 5c). Interestingly, ectopic F-actin blobs in nurse cells (but not in oocytes) also contained proteins normally restricted to ring canals (31), including Hu-li tai shao (Hts) (Fig. 5d′), Kelch (not shown), and antigens recognized by antibodies to phosphotyrosine (not shown). Hts and Kelch are homologous to the actin binding proteins adducin and scruin, respectively (32, 33). These findings suggest that loss of Gek function in germ-line cells alters the distribution of F-actin and actin binding proteins. In late-stage oocytes, ectopic actin polymerization was manifested as numerous F-actin spheres surrounding the yolk granules (compare Fig. 5 e and f).
To confirm that the oogenesis defects were caused by loss of gek function and not a neighboring gene that might be deleted in these mutants (Fig. 4), we transformed flies with a transgene (T35) that includes a genomic DNA fragment containing the entire gek transcription unit and 6 kb of genomic sequences upstream from the beginning of the gek cDNA (Fig. 4). Although the lethality of homozygous gek mutants is not rescued by this transgene, possibly caused by the absence of some of the regulatory elements or another vital gene deleted in gek mutants, the fertility defect (Fig. 5a) and actin phenotypes in egg chambers (data not shown) are completely rescued in germ-line mosaic animals bearing such a transgene. Northern analysis revealed that in addition to a 5.1-kb transcript that represents gek, a 7.1-kb transcript was detected with probes upstream of the XbaI site located at −2.5 kb (Fig. 4). Even if no introns corresponded to this 7.1 transcript, the rescue construct could not have contained the entire transcription unit from this gene. Therefore we conclude that the oogenesis phenotypes are caused by loss of gek function.
DISCUSSION
Our studies suggest that Gek is an effector of Dcdc42 for its regulation of actin polymerization. The binding properties of Gek to Dcdc42 are consistent with its role as a downstream effector of Dcdc42. Moreover, the oogenesis phenotypes of gek mutants indicate that the function of Gek is required for the integrity of the actin cytoskeleton in vivo. Because the most prominent phenotype in gek mutants is ectopic actin polymerization, we infer that the normal function of Gek is to negatively regulate actin polymerization.
Expression of constitutively active and dominant negative Dcdc42 mutants in egg chambers results in qualitatively similar phenotypes (34). They include defective cortical F-actin around nurse cells and free floating ring canal components, similar to what we observed in gek mutants (Fig. 4). The phenotypes caused by overexpressing dominant mutants of Dcdc42, however, are more severe than those of gek mutants and are associated with a block of oogenesis at earlier stages as compared with loss of function gek mutants. The gek mutants we used are unlikely to have residual Gek activity given that part of the kinase domain is deleted in gekD23. Moreover, the mitotic recombinations that generate the germ-line clones happen several days before the maturation of egg chambers, rendering it unlikely that perdurance of the Gek protein should be a problem. This raises the possibility that Gek is not the only effector for Cdc42 in regulating actin polymerization. Potential candidates for additional Cdc42 effectors in regulating actin cytoskeleton include the recently identified WASP family of proteins (35, 36), and as-yet-unidentified effector(s) for filopodia formation that do not contain the Cdc42-Rac interactive binding domain, as inferred from studies of effector domain mutants (10).
It is intriguing that a class of recently identified mammalian Rho-binding kinases (25–27), which also was reported to bind to Rac (10, 11), share structural motifs similar to those of Gek (Fig. 2a). The Rho-binding kinases may be involved in regulating myosin phosphatase (37), stress fiber formation, and actin polymerization (27). It is thus likely that a family of these multidomain kinases exists in both Drosophila and mammals, and that each member of this family serves as an effector of one or a subset of the Rho family small GTP-binding proteins in regulating the actin cytoskeleton. Finally, the strong sequence similarities between fly Gek and human DMPK suggest a role for DMPK in regulating cellular morphology.
Acknowledgments
We thank C.-t. Chien for advice on the yeast two-hybrid screen; C.-t. Chien and S. Wang for the use of the two-hybrid library; the Berkeley Drosophila Genome project for P-element collections; L. Cooley for antibodies; and I. Clark, S. Younger, and R. Herbst for advice on oogenesis, mosaic analysis, and cell culture, respectively. We particularly thank M. Simon for lab space and reagents used to finish the study. This work was supported by the Howard Hughes Medical Institute at the University of California-San Francisco, where L.L. was an Associate and L.Y.J. and Y.N.J. are Investigators, and also by the start-up fund and National Institutes of Health Grant R01NS36623–01 (to L.L.) at Stanford University.
ABBREVIATIONS
- DMPK
myotonic dystrophy protein kinase
- PB
phosphate buffer
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF029395).
References
- 1.Hall A. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 2.Ridley A J. Curr Biol. 1996;6:1256–1264. doi: 10.1016/s0960-9822(02)70711-2. [DOI] [PubMed] [Google Scholar]
- 3.Chant J. Curr Opin Cell Biol. 1996;8:557–565. doi: 10.1016/s0955-0674(96)80035-4. [DOI] [PubMed] [Google Scholar]
- 4.Luo L, Liao Y J, Jan L Y, Jan Y N. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 5.Luo L, Hensch T K, Ackerman L, Barbel S, Jan L Y, Jan Y N. Nature (London) 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 6.Harden N, Loh H Y, Chia W, Lim L. Development (Cambridge, UK) 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- 7.Eaton S, Auvinen P, Luo L, Jan Y N, Simons K. J Cell Biol. 1995;131:151–164. doi: 10.1083/jcb.131.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strutt D I, Weber U, Mlodzik M. Nature (London) 1997;387:292–295. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- 9.Narumiya S. J Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- 10.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 11.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 12.Luo L, Jan L Y, Jan Y N. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- 13.Bartel P L, Chien C T, Sternglanz R, Fields S. In: Cellular Interactions in Development: A Practical Approach. Hartley D A, editor. Oxford: IRL; 1993. pp. 153–179. [Google Scholar]
- 14.Brown N H, Kafatos F C. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 15.Simon M A, Bowtell D D L, Rubin G M. Proc Natl Acad Sci USA. 1989;86:8333–8337. doi: 10.1073/pnas.86.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 18.Bourne H R, Sanders D A, McCormick F. Nature (London) 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 19.Kozma R, Ahmed S, Best A, Lim L. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor S S, Knighton D R, Zheng J, Ten Eyck L E, Sowadski J M. Annu Rev Cell Biol. 1992;8:429–462. doi: 10.1146/annurev.cb.08.110192.002241. [DOI] [PubMed] [Google Scholar]
- 21.Coussens L, Parker P J, Rhee L, Yang-Feng T L, Chen E, Waterfield M D, Francke U, Ullrich A. Science. 1986;233:859–866. doi: 10.1126/science.3755548. [DOI] [PubMed] [Google Scholar]
- 22.Lemmon M A, Ferguson K M, Schlessinger J. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 23.Burbelo P D, Drechsel D, Hall A. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 24.Brook J D, McCurrach M E, Harley H G, Buckler A J, Church D, et al. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 25.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 26.Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 27.Leung T, Chen X-Q, Manser E, Lim L. Mol Cell Biol. 1996;16:5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 29.Chou T B, Noll E, Perrimon N. Development (Cambridge, UK) 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 30.Cooley L, Theurkauf W E. Science. 1994;266:590–596. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- 31.Robinson D N, Cant K, Cooley L. Development (Cambridge, UK) 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- 32.Yue L, Spradling A. Genes Dev. 1992;6:2443–2454. doi: 10.1101/gad.6.12b.2443. [DOI] [PubMed] [Google Scholar]
- 33.Xue F, Cooley L. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 34.Murphy A M, Montell D J. J Cell Biol. 1996;133:617–630. doi: 10.1083/jcb.133.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Symons M, Derry J M, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 36.Miki H, Miura K, Takenawa T. EMBO J. 1996;19:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 38.Mahadevan M S, Amemiya C T, Jansen G, Sabourin L, Baird S, Neville C E, Wormskamp N, Segers B, Lamerdin J, de Jong P, Wieringa B, Korneluk R G. Hum Mol Genet. 1993;2:299–304. doi: 10.1093/hmg/2.3.299. [DOI] [PubMed] [Google Scholar]
- 39.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 40.Hall C, Monfries C, Smith P, Lim H H, Kozma R, Ahmed S, Vanniasingham V, Leung T, Lim L. J Mol Biol. 1990;211:11–16. doi: 10.1016/0022-2836(90)90006-8. [DOI] [PubMed] [Google Scholar]
- 41.Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]