Abstract
To gain more insight into the molecular mechanisms by which androgens stimulate lipogenesis and induce a marked accumulation of neutral lipids in the human prostate cancer cell line LNCaP, we studied their impact on the expression of lipogenic enzymes. Northern blot analysis of the steady-state mRNA levels of seven different lipogenic enzymes revealed that androgens coordinately stimulate the expression of enzymes belonging to the two major lipogenic pathways: fatty acid synthesis and cholesterol synthesis. In view of the important role of the recently characterized sterol regulatory element binding proteins (SREBPs) in the coordinate induction of lipogenic genes, we examined whether the observed effects of androgens on lipogenic gene expression are mediated by these transcription factors. Our findings indicate that androgens stimulate the expression of SREBP transcripts and precursor proteins and enhance the nuclear content of the mature active form of the transcription factor. Moreover, by using the fatty acid synthase gene as an experimental paradigm we demonstrate that the presence of an SREBP-binding site is essential for its regulation by androgens. These data support the hypothesis that SREBPs are involved in the coordinate regulation of lipogenic gene expression by androgens and provide evidence for the existence of a cascade mechanism of androgen-regulated gene expression.
By using the human prostatic adenocarcinoma cell line LNCaP as an experimental paradigm of androgen-sensitive prostate cancer cells, we recently demonstrated that androgens, in addition to their well-known effects on cell proliferation and on differentiated secretory function, also induce a remarkable accumulation of cytoplasmic lipid droplets (1). These lipid droplets mainly consist of triacylglycerols and cholesteryl esters (1), the intracellular storage products of fatty acids and cholesterol, respectively. Labeling studies with [14C]acetate revealed that the accumulating lipids are largely synthesized de novo (1), suggesting that LNCaP cells express all enzymes required for endogenous lipogenesis and that the expression and/or activity of one or more of these enzymes is regulated by androgens. One key lipogenic enzyme that we recently have shown to be regulated by androgens is fatty acid synthase (FAS) (2), a complex multifunctional enzyme that plays a central role in the de novo biosynthesis of fatty acids. Androgen treatment of LNCaP cells results in a 3- to 4-fold stimulation of the steady-state mRNA levels of FAS and leads to an up to 10-fold stimulation of FAS activity. Because FAS is not the only enzyme required for the biosynthesis of fatty acids de novo and because androgens also induce the synthesis and accumulation of cholesteryl esters, we investigated the possibility that androgens also affect the expression of other lipogenic enzymes.
Here we report that several other enzymes involved in fatty acid synthesis and several key enzymes of the cholesterol biosynthetic pathway are also controlled by androgens in LNCaP cells. These findings suggest that androgens might not affect the expression of all these enzymes individually, but may rather modulate the expression and/or activity of one or more common transcription factor(s) involved in the coordinate control of these lipogenic genes. Transcription factors that have been shown to play an important role in the coordinate control of lipogenic enzymes are the recently characterized sterol regulatory element binding proteins (SREBPs) (3). SREBPs are basic helix-loop-helix leucine zipper transcription factors that originally have been identified as transcription factors that bind to a 10-bp sterol regulatory element (SRE) (hence their name) of the low density lipoprotein receptor and HMG-CoA (3-hydroxy-3-methylglutaryl CoA) synthase promoters (4, 5), two key enzymes involved in the maintenance of cholesterol homeostasis. Two genes encoding functionally related SREBPs (SREBP-1 and -2) have been identified (5, 6). SREBPs are synthesized as 125-kDa precursor proteins that are anchored to intracellular membranes. When intracellular cholesterol levels drop below a threshold level a protease cleaves the precursor, the amino-terminal portion of the protein harboring the basic helix-loop-helix domain is released and translocates to the nucleus where it activates the transcription of SRE-containing genes (3). Consistent with the independent identification of a rat homologue of SREBP-1 as an adipocyte determination and differentiation factor (ADD-1) (7, 8), SREBP-regulated genes not only include the genes involved in the maintenance of cholesterol homeostasis (low density lipoprotein receptor, HMG-CoA synthase, HMG-CoA reductase, farnesyl diphosphate synthase, squalene synthase) (4–6, 9–13), but also several genes involved in the synthesis of fatty acids (acetyl-CoA carboxylase and FAS) (13–15). Based on these findings SREBPs have been proposed to play a central role in the coordinate control of lipogenic enzymes of the two major lipogenic pathways (14, 15). In support of this proposal, overexpression of a truncated mature SREBP in the liver of transgenic mice has been shown to lead to a massive accumulation of both triacylglycerols and cholesteryl esters (13).
Here we report that androgens stimulate the expression of SREBP transcripts and precursor proteins and enhance the nuclear content of the mature active form of the transcription factor in LNCaP cells. Together with our demonstration that SREBP-binding sites are essential for the androgen regulation of the FAS gene, these findings provide evidence for the involvement of SREBPs in a cascade mechanism for the coordinate regulation of lipogenic gene expression by androgens.
MATERIALS AND METHODS
Cell Culture.
The human prostatic adenocarcinoma cell line LNCaP was obtained from the American Type Culture Collection. Cells were maintained in a humidified atmosphere of 5% CO2 in air in RPMI 1640 medium, supplemented with 10% fetal calf serum (FCS), 3 mM l-glutamine, 100 μg/ml streptomycin, and 100 units/ml penicillin (Life Technologies, Paisley, Scotland). In experiments assessing the effects of steroids, FCS was pretreated with dextran-coated charcoal (CT-FCS) (16) to reduce the background levels of steroids. The synthetic androgen R1881 (methyltrienolone), purchased from DuPont/New England Nuclear, was dissolved in absolute ethanol and added to the cultures. Control cultures received similar amounts of ethanol only. Final ethanol concentrations did not exceed 0.1%. All experiments involving LNCaP cells were carried out with cells of passages 33–50.
cDNA Probes.
cDNA probes for acetyl-CoA carboxylase, ATP-citrate lyase, farnesyl diphosphate synthase, HMG-CoA synthase, and malic enzyme were prepared by PCR as follows. cDNA was generated by reverse transcription of RNA from LNCaP cells or from clinical human prostatic specimens by using Superscript II (Life Technologies) and oligo(dT) as primer. This cDNA was used as a template for PCR with the primer pairs listed in Table 1. PCR products were cloned into pGEM-T (Promega) and verified by nucleotide sequencing by using an ALF (automated laser fluorescence) automated sequencer (Pharmacia). A cDNA probe for FAS was prepared by PCR amplification of a 4.2-kb EcoRI insert of clone EST01325 (22) obtained from the American Type Culture Collection by using T7 and T3 primers (Promega). To produce single-stranded radiolabeled PCR probes, ≈5 ng of the PCR products were used in a radiolabeling reaction by using the 3′ primer and 50 μCi [α-32P]dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq) (Amersham) as described (23). Probes for HMG-CoA-reductase, SREBP-1, and SREBP-2 were generated by random primed labeling of the inserts of plasmids pHRed-102, pSREBP-1a, and pSREBP-2, all obtained from the American Type Culture Collection (5, 6, 24). A radiolabeled 18S rRNA probe was prepared as described (25). A mixture of random probes was generated by PCR with 5′ primer 5′-GATGGCATTG, 3′ primer 5′-(T)11GA, and LNCaP cDNA as template. The resulting PCR fragments were used to synthesize a mixture of single-stranded radiolabeled probes as described above.
Table 1.
Sequences of PCR primers for cloning human cDNA probes
| cDNA probe | Primer pair | Primer sequences | PCR product, bp | Ref. |
|---|---|---|---|---|
| Acetyl-CoA carboxylase-α | 5′ | 5′-TTCTCAGAGCTTCCGAACTTGCT | 840 | 17 |
| 3′ | 5′-CTAACCTGGCTCTACCAACCAC | |||
| ATP-citrate lyase | 5′ | 5′-TCCAGGAGTCAAAATGATTGTG | 267 | 18 |
| 3′ | 5′-ATCTCTCCAAGCTCATCAAAGC | |||
| Farnesyl diphosphate synthase | 5′ | 5′-TGACGGTGGTAGTAGCATTCC | 756 | 19 |
| 3′ | 5′-ACACTGCTGGCAGATCCAG | |||
| HMG-CoA synthase | 5′ | 5′-AATCGACACAACTAATGCATGC | 591 | 20 |
| 3′ | 5′-ATCCCCAAAGGCCTTCAG | |||
| Malic enzyme | 5′ | 5′-TGCTTCAGTTCTCAATGCATG | 400 | 21 |
| 3′ | 5′-GAATGTGCAATACTGGTTTCGA |
Northern Blot Analysis.
Cells were seeded in 150-mm dishes at 3 × 106 cells/plate in medium containing 10% FCS. The next day cells were washed with PBS (Life Technologies) and fresh medium with 5% CT-FCS was added. After 2 or 3 days, medium was replaced and cells were treated with steroids or ethanol vehicle. Three days later, plates were washed with PBS, quick-frozen in liquid nitrogen, and stored at −80°C. Total RNA was prepared by using a modified guanidinium/CsCl ultracentrifugation method as described (23). Equal aliquots of total RNA (20 μg) were denatured with formaldehyde and formamide and subjected to electrophoresis in a 1% agarose gel containing formaldehyde. The RNA was transferred to Biotrans+ membranes (ICN), prehybridized, hybridized, and washed as described (23). Blots were autoradiographed by exposure to Amersham hyperfilm-MP or to Kodak Biomax film (Amersham). Hybridization signals were quantitated with PhosphorImager screens (Molecular Dynamics).
Immunoblot Analysis of SREBPs.
To detect SREBP precursor proteins in total cell extracts, LNCaP cells were seeded in 5% CT-FCS in 6-cm plates at a density of 5 × 105 cells/plate. Three days later medium was replaced and cells were treated with 10−8 M R1881 or ethanol vehicle. At the indicated times after steroid addition cells were collected in SDS lysis buffer and boiled for 3 min. Protein concentrations were measured by the BCA procedure (Pierce) after trichloroacetic acid precipitation. To study the effects of androgens on the nuclear level of mature SREBPs, cells were seeded in 10-cm plates at a density of 2 × 106 cells/plate. After incubation for 3 days in 5% CT-FCS, cells were treated with 10−8 M R1881 or ethanol vehicle for 2 days and harvested. Nuclear fractions were prepared as described by Wang et al. (26) and Hua et al. (27) with minor modifications. Cells were resuspended in 10 mM Hepes-KOH (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM DTT. After incubation for 10 min at 4°C the cell suspensions were passed through a 22.5-gauge needle 15 times and centrifuged at 1,000 × g at 4°C for 5 min. The pellets were resuspended in 10 mM Hepes-KOH (pH 7.4), 0.42 M NaCl, 2.5% (vol/vol) glycerol, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 1 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. After rotation at 4°C for 30 min, the suspension was centrifuged at top speed in a microcentrifuge at 4°C. The supernatant was collected, the protein concentration was determined, and the proteins were denatured by boiling for 3 min in SDS-gel loading buffer. Equal amounts of protein were subjected to 7.5% SDS/PAGE. Proteins were blotted onto Hybond-C extra membranes (Amersham) and incubated with the indicated antibodies against SREBPs. Hybridoma cell lines producing mouse mAbs against the basic helix-loop-helix leucine zipper domain of human SREBP-1 (IgG-2A4) (28) and the carboxyl terminus of SREBP-2 (IgG-1C6) (29) were obtained from the American Type Culture Collection. The antibodies were purified from conditioned medium or from ascites fluid by protein A-Sepharose affinity chromatography. Immunoreactive proteins were visualized with horseradish peroxidase-conjugated goat anti-mouse IgG (Amersham) by using an Enhanced Chemiluminescence (ECL) Western Blotting Detection System (Amersham). Immunoreactive signals were quantitated by scanning with an LKB Bromma Utroscan XL Enhanced Laser Densitometer (Pharmacia LKB).
Promoter Reporter Gene Constructs.
A 178-bp promoter fragment of the human FAS gene was generated by Pfu DNA polymerase-mediated extension of two long complementary oligonucleotides with a central overlap of 24 bases. The 5′ primer (from base −167 to base −72) of the sequence reported by Hsu et al. (30) was provided with an XhoI restriction site at the 5′ end. The 3′ primer (from base +5 to base −95) was provided with a HindIII site. Fifty picomol of the two oligonucleotides was mixed in 1× Pfu DNA polymerase buffer (Stratagene) and supplemented with deoxynucleotides to a final concentration of 200 μM each and 5 units of Pfu DNA polymerase. After denaturation at 95°C for 1 min, the oligonucleotides were annealed at 64°C for 1 min and extended at 72°C for 5 min. Reaction products were purified on a QIA Quick PCR column (Qiagen, Hilden, Germany), digested with XhoI and HindIII, and ligated into the predigested pGL3-basic luciferase reporter vector (Promega). To generate a FAS promoter–reporter construct that lacks the major functional SREBP-binding site (15, 31), two complementary mutagenic primers containing an internal deletion of 18 bases (from base −63 to base −46) (30) were designed. These primers were annealed with the parental FAS promoter–reporter construct (FAS-luc) and extended by using Stratagene’s GC-PCR kit supplemented with Pfu DNA polymerase. Following temperature cycling the reaction product was digested with DpnI to remove the parental DNA and transformed into Escherichia coli. The sequences of both native and mutated FAS promoters were verified by ALF automated sequencing (Pharmacia).
Transfection and Luciferase Reporter Activity.
LNCaP cells were seeded in 6-cm dishes in DMEM containing 10% FCS at a density of 7 × 105 cells. The next day the medium was replaced with DMEM with 2% FCS. Cells were transfected with 5 μg of the indicated luciferase reporter constructs together with 0.5 μg of an androgen receptor-expressing vector (pSV-ARo) as described (32). After 4 h of exposure to DNA, cells were subjected to a glycerol shock, washed with PBS, and incubated with fresh medium containing 10% CT-FCS. The next day cells were incubated with 10−8 M R1881 or with ethanol vehicle. Twenty-four hours later cells were washed with PBS, harvested in 500 μl reporter lysis buffer (Promega), incubated on ice for 1 h, and vortex mixed. Aliquots of 10 μl of cleared lysate were assayed for luciferase activity by using a luciferase reporter assay kit from Promega and a Berthold Microlumat LB 96P luminometer.
RESULTS AND DISCUSSION
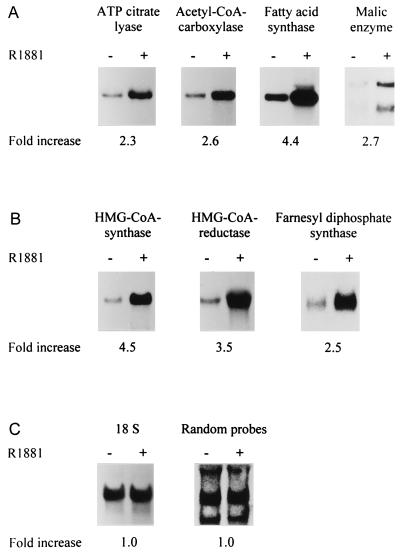
To explore the possibility that androgens, in addition to their effects on FAS (8) also affect the expression of other lipogenic enzymes, we incubated LNCaP cells for 3 days in the absence or presence of 10−8 M of the synthetic androgen R1881 [conditions that maximally stimulate the expression of FAS (2)]. Total RNA was prepared and subjected to Northern blot analysis with probes for four enzymes of the pathway of fatty acid synthesis: ATP-citrate lyase, acetyl-CoA-carboxylase, FAS, and malic enzyme. As Fig. 1A demonstrates, androgens stimulated the steady-state mRNA levels of all four enzymes tested. Because androgens also induce a marked accumulation of cholesteryl esters (1), which are the intracellular storage products of cholesterol, we also examined the impact of androgens on the expression of three key enzymes of the complex pathway leading to the synthesis of cholesterol: HMG-CoA synthase, HMG-CoA reductase, and farnesyl diphosphate synthase. Again, the expression of all three enzymes was affected by the androgen treatment (Fig. 1B). To ascertain that the observed effects were not caused by loading differences or to some general effect of androgens on the expression of polyadenylated mRNAs, two control hybridizations were performed: one with an 18S ribosomal RNA probe and one with a mixture of randomly generated probes of unknown identity. In both cases, none of the hybridization signals was affected by androgens (Fig. 1C). In agreement with the absence of a lipogenic response in the androgen receptor-negative cell lines PC-3 and DU-145, no effects of androgens on lipogenic gene expression were observed in these cell lines (data not shown).
Figure 1.
Androgen regulation of the mRNA expression of lipogenic enzymes. LNCaP cells were cultured in the absence (−) or presence (+) of 10−8 M of the synthetic androgen R1881 for 3 days. Total RNA was prepared, and 20 μg aliquots were subjected to Northern blot analysis with 32P-labeled probes for enzymes involved in fatty acid synthesis (A) or cholesterol synthesis (B). Two control hybridizations were performed (C): one with an 18S rRNA probe to demonstrate that similar amounts of RNA were present in both lanes, and one with a mixture of random probes of unidentified sequence (see Materials and Methods) to show that not all polyadenylated RNAs are affected by the androgen treatment. Blots were autoradiographed, and hybridization signals were quantified by using PhosphorImager screens. The fold-increase in mRNA expression, relative to that of the untreated condition, was calculated after correction for loading differences with 18S RNA and is shown below each blot.
Our finding that androgens stimulate the expression of multiple enzymes of the two major lipogenic pathways (fatty acid synthesis and cholesterol synthesis) are reminiscent of the coordinate control of lipogenic enzymes by SREBPs and suggest that androgens may not (only) affect the expression of all these genes individually, but may rather stimulate the expression and/or activity of these or other transcription factor(s) that are involved in the coordinate regulation of lipogenic gene expression.
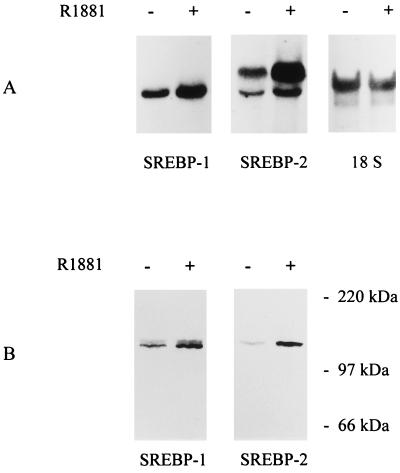
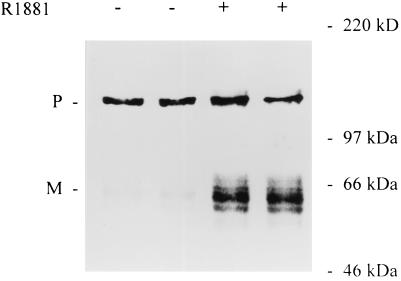
To assess whether SREBPs are involved in the androgen regulation of lipogenic gene expression in LNCaP cells we investigated whether androgens somehow affect the expression and/or the nuclear content of SREBPs. Northern blot analysis with radiolabeled probes for SREBP-1 and -2 revealed that LNCaP cells express both SREBP genes (Fig. 2A). Consistent with previous reports on SREBP mRNA expression in several other tissues and cell lines, one band corresponding to SREBP-1 was detected, whereas SREBP-2 transcripts were apparent as two discrete bands (5, 6). Interestingly, androgen treatment resulted in a 1.3- and 2.1-fold stimulation of the steady-state mRNA levels of SREBP-1 and -2, respectively. To test whether these increases in mRNA expression are reflected also at the protein level, total cellular extracts of LNCaP cells were prepared and subjected to Western blot analysis by using mAbs against SREBP-1 and SREBP-2. In both cases, one or two immunoreactive bands of ≈125 kDa were observed (Fig. 2B). The size of these proteins is consistent with the size of the immature precursor form of SREBP. In agreement with the effects of androgens on the steady-state mRNA levels, androgen treatment resulted in a 1.5- and 4-fold stimulation of SREBP precursor levels. Longer exposure times revealed the presence of immunoreactive bands of smaller size that may correspond to the mature nuclear form of SREBP. To study the impact of androgens on this mature nuclear form of SREBPs, nuclear extracts were prepared from LNCaP cells that had been cultured for 2 days in the absence or presence of R1881. Equal amounts of proteins were separated by SDS/PAGE and subjected to Western blot analysis with a monoclonal antiserum against the amino-terminal part of SREBP-1. As Fig. 3 shows, in addition to the 125-kDa precursor form that frequently contaminates these crude nuclear fractions (26), immunoreactive protein bands of 60–68 kDa were detected. The intensity of these bands was stimulated 2- to 6-fold after androgen treatment. The size of these bands is in the same range, or somewhat smaller, than the mature form found in nuclear extracts under sterol depleted conditions (26). Depending from experiment to experiment the immunoreactive bands appeared more or less heterogeneous. This heterogeneity may be explained not only by the existence of multiple transcripts and protein phosphorylation, but also by partial degradation caused by the instability of the protein, as mentioned in other reports (26). Although the effects of androgens on SREBP-2 precursor levels were more pronounced than those on SREBP-1, no antibody to the amino-terminal fragment of human SREBP-2 is available, limiting our studies on the effects of androgens on the nuclear content of SREBPs to SREBP-1.
Figure 2.
Effects of androgens on the steady-state mRNA and protein expression of SREBPs. LNCaP cells were incubated in the absence (−) or presence (+) of 10−8 M R1881. Three days later total RNA wa prepared and subjected to Northern blot analysis (A) with radiolabeled probes for SREBP-1 and -2 as described in the legend to Fig. 1. After removal of the probes, blots were hybridized with an 18S rRNA probe to demonstrate that similar amounts of RNA were present in both lanes. To assess the impact of androgens on SREBP expression at the protein level (B), LNCaP cells were lysed in an SDS gel loading buffer after 2 days of treatment. Proteins were separated on 7.5% reducing SDS/polyacrylamide gel and blotted onto a fortified nitrocellulose membrane. Blots were incubated with mAbs IgG-2A4 and IgG-1C6, directed against human SREBP-1 and -2, respectively. Immunoreactive signals were visualized by using an ECL Western blot analysis system and a peroxidase-coupled goat anti-mouse secondary antibody. Positions of molecular size markers are as indicated. The experiment shown is representative of four experiments.
Figure 3.
Effects of androgens on the nuclear content of mature SREBP-1. After incubation for 2 days in the absence (−) or in the presence (+) of 10−8 M R1881, LNCaP cells were harvested and a crude nuclear fraction was prepared. Equal amounts of proteins from the nuclear fractions of duplicate dishes were subjected to SDS/PAGE and blotted onto a fortified nitrocellulose membrane. The blot was incubated with mAb IgG-2A4, directed against SREBP-1. Immunoreactive bands were visualized by using an ELC Western blot analysis system and a peroxidase-coupled goat anti-mouse secondary antibody. Positions of molecular size markers are as indicated. The experiment shown is representative of three experiments performed. P, precursor; M, mature protein.
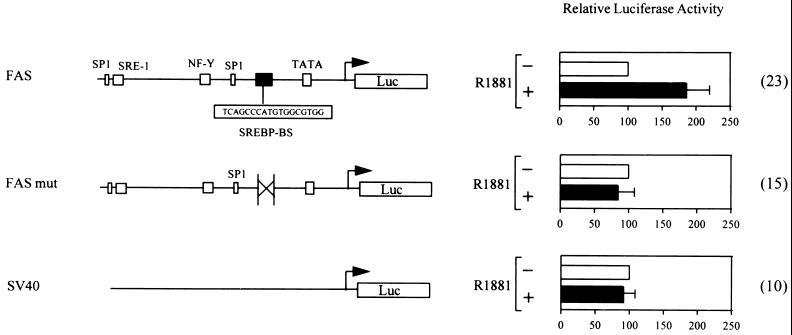
To demonstrate that the androgen-induced increase in the nuclear content of the mature form of SREBPs indeed results in increased transcription of lipogenic genes harboring SREBP-binding sites, a 178-bp promoter fragment of the human FAS gene was placed in front of a luciferase reporter gene (Fig. 4). This FAS promoter fragment harbors a complex SREBP-binding site that is involved in the stimulation of FAS transcription by cholesterol depletion (15, 31) and is essential for the increase in transcription in response to overexpression of a constitutively active amino-terminal fragment of SREBP-1 (ref. 31 and data not shown). Consistent with our finding that androgens stimulate the expression and nuclear content of SREBPs, transient transfection experiments with this FAS promoter–reporter construct (FAS-luc) revealed that androgens stimulate the transcriptional activity of this promoter fragment (Fig. 4). In further support of the involvement of SREBPs in the androgen regulation of the FAS promoter, the effects of androgens were completely abolished when the major SREBP-binding site was deleted (Fig. 4).
Figure 4.
Involvement of SREBP-binding sites in the androgen regulation of the FAS promoter. A luciferase reporter construct containing a 178-bp promoter fragment of the human FAS gene (FAS-luc) harboring a complex binding site for SREBPs (SREBP-BS) was generated. In the FASmut-luc construct this 18-bp binding site was deleted. LNCaP cells were transfected with 5 μg of these constructs or with 5 μg of a control plasmid containing the simian virus 40 (SV40) promoter (SV40-luc). The next day, the transfected cultures were treated with 10−8 M R1881 (+) or with ethanol vehicle (−). After 24 h cells were harvested and luciferase activity was determined. Luciferase activity was measured in triplicate dishes. Values are the mean ± SD of the indicated number of independent experiments (in parentheses) and are expressed relative to the activity of vehicle-treated cells.
Because most of the genes that we have analyzed in this study (with the exception of ATP-citrate lyase and malic enzyme) have been shown to harbor SREBP-binding sites in their regulatory sequences, these findings provide an explanation for the coordinate control of lipogenic gene expression by androgens. An important physiological implication of these observations is that SREBP genes may be critical downstream genes mediating a subset of androgen effects in the human prostate. The present findings do not exclude additional, more direct effects of androgens on lipogenic enzymes, which may allow fine tuning of individual activities.
One point that remains to be addressed is the mechanism by which androgens stimulate the expression and nuclear content of SREBPs. In view of the reports that SREBPs are subject to autoregulation (33), it is tempting to speculate that androgens stimulate or activate a protease that cleaves the SREBP precursors resulting in elevated nuclear levels of the mature transcription factor and in increased transcription of genes encoding lipogenic enzymes and SREBPs. Whether the androgen regulation of SREBP processing is independent of cell sterol levels or whether androgen action somehow results in a transient lowering of the regulatory pool of intracellular cholesterol will be exciting to explore. The observation that androgens increase rather than decrease total intracellular cholesterol (1) makes the latter hypothesis less attractive.
A final question that will be interesting to address is whether a similar mechanism involving SREBPs is involved in the induction of lipogenesis in other epithelial cell lines (e.g., T47 breast cancer cells in response to progesterone) and in prostate cancer cells in vivo (34–37).
Acknowledgments
We thank Joëlle Rosseels, Frank Vanderhoydonc, and Ludo Deboel for excellent technical assistance. This work was supported by a grant Geconcerteerde Onderzoeksactie van de Vlaamse Gemeenschap, by research grants from the Fund for Scientific Research-Flanders (Belgium) (F.W.O.) (to J.V.S. and to G.V.), by the Schenking Rimaux-Bartier (to J.V.S.), and by a grant Interuniversity Poles of Attraction Programme-Belgian State, Prime Minister’s Office, Federal Office for Scientific, Technical and Cultural Affairs. J.V.S. is a Senior Research Assistant of the Fund for Scientific Research-Flanders (Belgium) (F.W.O.).
ABBREVIATIONS
- FAS
fatty acid synthase
- FCS
fetal calf serum
- CT
charcoal-treated
- HMG-CoA
3-hydroxy-3-methylglutaryl CoA
- SRE
sterol regulatory element
- SREBP
sterol regulatory element binding protein
- ECL
enhanced chemiluminescence
References
- 1.Swinnen J V, Van Veldhoven P P, Esquenet M, Heyns W, Verhoeven G. Endocrinology. 1996;137:4468–4474. doi: 10.1210/endo.137.10.8828509. [DOI] [PubMed] [Google Scholar]
- 2.Swinnen J V, Esquenet M, Goossens K, Heyns W, Verhoeven G. Cancer Res. 1997;57:1086–1090. [PubMed] [Google Scholar]
- 3.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 4.Briggs M R, Yokoyama C, Wang X, Brown M S, Goldstein J L. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- 5.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 6.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J B, Spiegelman B M. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 9.Vallett S M, Sanchez H B, Rosenfeld J M, Osborne T F. J Biol Chem. 1996;271:12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- 10.Ericsson J, Jackson S M, Lee B C, Edwards P A. Proc Natl Acad Sci USA. 1996;93:945–950. doi: 10.1073/pnas.93.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson S M, Ericsson J, Metherall J E, Edwards P A. J Lipid Res. 1996;37:1712–1721. [PubMed] [Google Scholar]
- 12.Guan G, Jiang G, Koch R L, Shechter I. J Biol Chem. 1995;270:21958–21965. doi: 10.1074/jbc.270.37.21958. [DOI] [PubMed] [Google Scholar]
- 13.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez J M, Bennett M K, Sanchez H B, Rosenfeld J M, Osborne T F. Proc Natl Acad Sci USA. 1996;93:1049–1053. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennet M K, Lopez J M, Sanchez H B, Osborne T F. J Biol Chem. 1995;270:25578–25583. doi: 10.1074/jbc.270.43.25578. [DOI] [PubMed] [Google Scholar]
- 16.Leake R E, Freshney R I, Munir I. In: Steroid Hormones: A Practical Approach. Green B, Leake R E, editors. Washington, DC: IRL; 1987. p. 214. [Google Scholar]
- 17.Abu-Elheiga L, Jayakumar A, Baldini A, Chirala S S, Wakil S J. Proc Natl Acad Sci USA. 1995;92:4011–4015. doi: 10.1073/pnas.92.9.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes S A, Franklin M, Gloger I S. Eur J Biochem. 1992;204:491–499. doi: 10.1111/j.1432-1033.1992.tb16659.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilkin D J, Kutsunai S Y, Edwards P A. J Biol Chem. 1990;265:4607–4614. [PubMed] [Google Scholar]
- 20.Russ A P, Ruzicka V, Maerz W, Appelhans H, Gross W. Biochim Biophys Acta. 1992;1132:329–331. doi: 10.1016/0167-4781(92)90172-v. [DOI] [PubMed] [Google Scholar]
- 21.Loeber G, Dworkin M B, Infante A, Ahorn H. FEBS Lett. 1994;344:181–186. doi: 10.1016/0014-5793(94)00386-6. [DOI] [PubMed] [Google Scholar]
- 22.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 23.Swinnen J V, Esquenet M, Heyns W, Rombauts W, Verhoeven G. Mol Cell Endocrinol. 1994;104:153–162. doi: 10.1016/0303-7207(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 24.Luskey K L, Stevens B. J Biol Chem. 1985;260:10271–10277. [PubMed] [Google Scholar]
- 25.Verhoeven G, Deboel L, Swinnen J, Rombauts L, Vanderhoydonc F, Rosseels J, Hoeben E, Heyns W. Int J Androl. 1995;18:23–34. doi: 10.1111/j.1365-2605.1995.tb00931.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Sato R, Brown M, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 27.Hua X, Sakai J, Brown M S, Goldstein J L. J Biol Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 28.Sato R, Yang J, Wang X, Evans M J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- 29.Hua X, Sakai J, Ho Y K, Goldstein J L, Brown M. J Biol Chem. 1995;270:29422–29427. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 30.Hsu M H, Chirala S, Wakil S J. J Biol Chem. 1996;271:13584–13592. doi: 10.1074/jbc.271.23.13584. [DOI] [PubMed] [Google Scholar]
- 31.Magana M M, Osborne T F. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 32.Swinnen J V, Esquenet M, Rosseels J, Claessens F, Rombauts W, Heyns W, Verhoeven G. DNA Cell Biol. 1996;15:197–208. doi: 10.1089/dna.1996.15.197. [DOI] [PubMed] [Google Scholar]
- 33.Sato R, Inoue J, Kawabe Y, Komada T, Takano T, Maeda M. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 34.Chambon M, Rochefort H, Vial H, Chalbos D. J Steroid Biochem. 1989;33:915–922. doi: 10.1016/0022-4731(89)90240-9. [DOI] [PubMed] [Google Scholar]
- 35.Braunstein H. Am J Clin Pathol. 1964;41:44–48. doi: 10.1093/ajcp/41.1.44. [DOI] [PubMed] [Google Scholar]
- 36.Fisher E R, Jeffrey W. Am J Clin Pathol. 1965;44:119–134. doi: 10.1093/ajcp/44.2.119. [DOI] [PubMed] [Google Scholar]
- 37.Mao P, Nakao K, Angrist A. Cancer Res. 1966;26:955–973. [PubMed] [Google Scholar]