Abstract
The insulin-like growth factor (IGF) binding proteins (IGFBPs) modulate the actions of the insulin-like growth factors in endocrine, paracrine, and autocrine settings. Additionally, some IGFBPs appear to exhibit biological effects that are IGF independent. The six high-affinity IGFBPs that have been characterized to date exhibit 40–60% amino acid sequence identity overall, with the most conserved sequences in their NH2 and COOH termini. We have recently demonstrated that the product of the mac25/IGFBP-7 gene, which shows significant conservation in the NH2 terminus, including an “IGFBP motif” (GCGCCXXC), exhibits low-affinity IGF binding. The closely related mammalian genes connective tissue growth factor (CTGF) gene, nov, and cyr61 encode secreted proteins that also contain the conserved sequences and IGFBP motifs in their NH2 termini. To ascertain if these genes, along with mac25/IGFBP-7, encode a family of low-affinity IGFBPs, we assessed the IGF binding characteristics of recombinant human CTGF (rhCTGF). The ability of baculovirus-synthesized rhCTGF to bind IGFs was demonstrated by Western ligand blotting, affinity cross-linking, and competitive affinity binding assays using 125I-labeled IGF-I or IGF-II and unlabeled IGFs. CTGF, like mac25/IGFBP-7, specifically binds IGFs, although with relatively low affinity. On the basis of these data, we propose that CTGF represents another member of the IGFBP family (IGFBP-8) and that the CTGF gene, mac25/IGFBP-7, nov, and cyr61 are members of a family of low-affinity IGFBP genes. These genes, along with those encoding the high-affinity IGFBPs 1–6, together constitute an IGFBP superfamily whose products function in IGF-dependent or IGF-independent modes to regulate normal and neoplastic cell growth.
Keywords: peptide hormones, serum inducible factors, transforming growth factor β
The insulin-like growth factor (IGF) binding proteins (IGFBPs) are a family of homologous proteins that regulate the biological activities of the IGFs and may also be capable of IGF-independent actions. Six distinct IGFBPs that bind IGFs with high affinity have been described (1–7). They share an overall protein sequence identity of 50% and contain 16–18 conserved cysteines in the NH2- and COOH-terminal regions (8). We have recently demonstrated that the protein product of the human mac25 cDNA, which is structurally related and which contains the “IGFBP motif” (GCGCCXXC) in its NH2 terminus, specifically binds IGFs, although with relatively low affinity, and constitutes another member of the IGFBP family, IGFBP-7 (9). This finding suggests that a family of low-affinity IGFBPs, distinct from the high-affinity members, may exist, and together these may constitute an IGFBP superfamily.
A closely related family of genes encoding connective tissue growth factor (CTGF) (10), the nov oncogene (11), and cyr61 (12) has been identified; the predicted proteins are products of “immediate-early genes” expressed after induction by serum, growth factors, or certain oncogenes (10–13). These proteins show an overall identity of 30–38% compared with IGFBPs 1–6. Although the similarity of the COOH-terminal sequences is low (<20%), the NH2-terminal region is well conserved among these new members and the IGFBPs. Moreover, these proteins also contain the conserved “IGFBP motif” (GCGCCXXC) in their NH2 terminus, and as many as 17 of the 18 cysteines are conserved in IGFBPs 1–6, suggesting that the CTGF/nov oncogene/cyr61 family shares significant structural homology with IGFBPs (14) and may potentially bind IGFs.
CTGF has been identified as a major chemotactic and mitogenic factor for connective tissue cells (10). It has platelet-derived growth factor (PDGF)-related biological and immunological activities, and it competes with PDGF for a cell-surface receptor (10). The CTGF gene, residing on chromosome 6q23.1, proximal to c-myb, was originally cloned from human umbilical vein endothelial cells (10, 11). CTGF gene expression is increased in human foreskin fibroblasts after activation with transforming growth factor β (TGF-β), but not other growth factors, including PDGF, epidermal growth factor, and basic fibroblast growth factor (15). The CTGF gene encodes a 38-kDa prepeptide of 349 amino acids (10).
In the present study, we report the expression of recombinant human CTGF (rhCTGF) in a baculovirus system, and we demonstrate that the 36-kDa CTGF protein specifically binds IGFs. CTGF thus meets criteria that define it as another member of the IGFBP superfamily, IGFBP-8. We further propose that the six high-affinity IGFBPs and the four potential low-affinity IGFBPs constitute a superfamily of proteins that regulate cell growth through both IGF-dependent and IGF-independent actions.
MATERIALS AND METHODS
Peptides and Proteins.
Recombinant human IGF-I was obtained from Bachem, and recombinant human IGF-II was provided by Eli Lilly. [Gln6,Ala7,Tyr18,Leu19,Leu27]IGF-II ([QAYLL]IGF-II), a synthetic IGF-II analog, was synthesized as described previously (16). Recombinant human IGFBP-3E. coli, a nonglycosylated 29-kDa core protein expressed in Escherichia coli, was a generous gift from Celtrix Laboratories (Santa Clara, CA). Recombinant human IGFBP-7 was expressed in a baculovirus system as described previously (9). Iodination was performed by a modification of the chloramine-T technique, to a specific activity of 350–500 μCi/μg for IGF-I and IGF-II (1 μCi = 37 kBq) (17). Iodinated prolactin was a gift from Diagnostic Systems Laboratories (Webster, TX). Endoglycosidase F (Endo F) was purchased from Boehringer Mannheim. Human multiple tissue Northern blots were obtained from CLONTECH.
Cloning of Human CTGF, nov Oncogene, and cyr61 cDNAs.
To characterize the family of low-affinity IGFBPs, partial cDNAs corresponding to all three members of this family were generated by using reverse transcription–PCR (RT-PCR) as described previously (18). In brief, degenerate primers were designed to conserved sequences in this family: 5′ primer GGNAT(A/C/T)(T/A)(C/G)NACN(A/C)GNGTNACNAA(C/T)GA(C/T)AA, corresponding to amino acids GISTRVTNDN, and 3′ primer GGNGT(A/G)CA(G/A)CANC(T/G)NCC(G/A)TC, corresponding to amino acids DGRCCTP (5′ amino acids 226–236 and 3′ amino acids 298–308 of GenBank sequence M92934). Five micrograms of total RNA isolated from the Hs578T human breast carcinoma cell line was used in RT-PCR as described previously (18). Fragments corresponding to all three cDNAs were identified by using gene-specific primers and by sequencing. Partial cDNAs were used to screen a cDNA library prepared from Hs578T cell line mRNA in the λZAP Express vector (Stratagene), and full-length cDNAs were isolated as described previously (19). The sequences of full-length CTGF and nov oncogene cDNAs were identical to the published sequences (10, 11), and the human cyr61 sequence (S.R.N., unpublished data) shares 90% identity to mouse cyr61 (12).
Expression of rhCTGF Protein.
To facilitate purification of recombinant CTGF, a FLAG epitope sequence (DYKDDDDK) was added at the COOH terminus by use of PCR. Primers 5′-GTCAGGCCTTGCGAAGCTGAC (845–868) and 5′-CGTGGTACCTTACTTGTCATCGTCGTCCTTGTAGTCTGCCATGTCTCCGTACAT (1160–1179), which was designed with the FLAG sequence followed by a stop codon and a restriction site for KpnI, were used. The resulting PCR product was digested with unique restriction enzymes StuI and KpnI and ligated into full-length cDNA digested with the same restriction enzymes, to replace the COOH terminus. After sequencing, the FLAG-tagged CTGF cDNA was subcloned into the baculovirus expression vector pFASTBAC1 (Life Technologies). The CTGF-pFASTBAC1 construct was transfected into Sf9 cells and positive viral recombinants were isolated by using the vender’s protocols. Western immunoblotting was performed with the FLAG sequence-specific anti-FLAG M2 antibody (Eastman Kodak) to confirm the expression of rhCTGF protein.
Protein Purification.
Large-scale protein purification was performed by infecting 108 HI-5 insect cells at a multiplicity of infection of 3 at 27°C for 2 days. The media from the infected cells were collected and centrifuged. The supernatant was bound to an anti-FLAG M2 antibody affinity column at 4°C for 2 hr. The column was washed three times with 5 ml of HBS (20 mM Hepes, pH 7.8/150 mM NaCl), and the protein was eluted with four 1-ml washes with HBS containing 0.5 mg/ml FLAG peptide. The purified protein was subjected to sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) in a 12% polyacrylamide gel and stained with Coomasie blue or transferred to nitrocellulose membrane for immunodetection.
Eluted fractions from an anti-FLAG M2 antibody affinity column were pooled and quantitated by using a densitometer and comparing with known amounts of IGFBP-3E. coli after silver staining.
Glycosylation Studies.
Proteins were deglycosylated with Endo F (9). As a positive control, acid-chromatographed normal human serum IGFBP fractions were prepared (20). Five hundred nanograms of FLAG-tagged-rhCTGF and 2 μl of acid-chromatographed normal human serum IGFBP fraction were treated with 100–600 milliunits of Endo F as described previously (9). Subsequent Western immunoblots or ligand blots were prepared as described below.
Western Ligand Blots (WLBs).
Unreduced samples of FLAG-tagged rhCTGF and IGFBP-3E. coli were subjected to SDS/PAGE in a 12% gel and electroblotted onto nitrocellulose membranes. The membranes were incubated overnight with 1.0 × 106 cpm of 125I-labeled IGF-I and IGF-II after blocking with normal saline containing 1% BSA at room temperature for 2 hr. The filters were then washed with normal saline containing 0.1% Tween 20 two times and normal saline three times for 15 min each time, dried, and exposed to film. Nondenaturing WLBs were prepared by PAGE without SDS, in stacking (pH 7.4) or resolving (pH 8.8) gels (21).
Affinity Cross-Linking.
FLAG-tagged rhCTGF and IGFBP-3E. coli were incubated overnight at 4°C with 125I-labeled IGF-I or IGF-II or prolactin (100,000 cpm) in the presence or absence of unlabeled peptides at the concentrations indicated in the text and figures. After cross-linking with disuccinimidyl suberate (0.6 mM), samples were subjected to SDS/PAGE and autoradiography (17). Bands were quantified by densitometry using the area under the curve, as calculated by an LKB densitometer.
Northern Blot Analysis.
Blots of 2 μg of poly(A)+ RNA from normal human tissues, which had been subjected to electrophoresis in a formaldehyde/1.5% agarose gel before transfer to nylon membranes, were purchased from CLONTECH. 32P-labeled antisense complementary RNA probes for CTGF, transcribed from the plasmid constructs, were used. Blots were hybridized and washed at high stringency as described previously (18).
RESULTS
Construction and Expression of FLAG Epitope-Tagged rhCTGF.
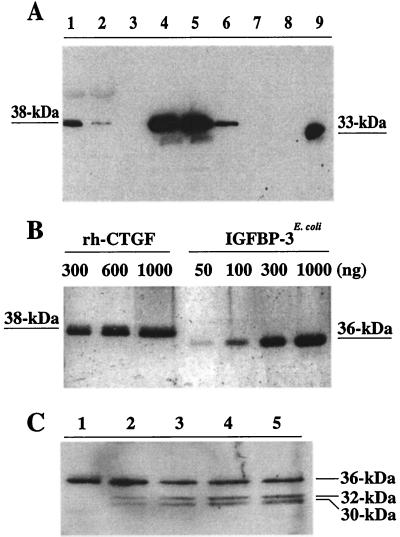
The immunoblot of the fractions collected during purification of FLAG-tagged rhCTGF by reducing SDS/12% PAGE (Fig. 1A) identifies an M2 antibody-specific protein of 38 kDa, which is compatible with the predicted size of CTGF as reported previously (10). Application of various amounts of the purified rhCTGF protein on a reducing SDS/12% polyacrylamide gel and subsequent silver staining (Fig. 1B) showed that the protein has approximately 99% purity and a molecular mass of approximately 36–38 kDa.
Figure 1.
Purification and characterization of FLAG epitope-tagged CTGF. (A) Purification and Western immunoblotting of FLAG-tagged rhCTGF using M2 monoclonal antibody under reducing conditions. Lanes represent 10 μl of samples: medium from HI-5 infected cells (lane 1), flow-through from the M2 affinity column (lane 2), wash from the same immunoaffinity column (lane 3), serial fractions of rhCTGF eluted by FLAG peptides (lanes 4–8), and 500 ng of FLAG-tagged rh-IGFBP-7 as a positive control (lane 9). (B) Silver staining of purified rhCTGF (lane 1, 300 ng; lane 2, 600 ng; lane 3, 1000 ng) and nonglycosylated IGFBP-3E. coli (lane 4, 50 ng; lane 5, 100 ng; lane 6, 300 ng; lane 7, 1000 ng) under reducing conditions. (C) Western immunoblot of rhCTGF with M2 monoclonal antibody after treatment with various concentrations of Endo F at 37°C for 3 hr under nonreducing conditions. Lanes represent 500 ng of rhCTGF alone (lane 1) or after Endo F treatment (100 milliunits of Endo F, lane 2; 200 milliunits, lane 3; 400 milliunits, lane 4; 600 milliunits, lane 5).
Because the predicted amino acid sequence analysis revealed that CTGF contains two potential N-glycosylation sites, located at amino acids 28 and 225 (Asn-Cys-Ser, Asn-Ala-Ser, respectively), we treated rhCTGF with various concentrations of Endo F (100–600 milliunits) to cleave the N-glycosylated carbohydrates. The size of rhCTGF (36 kDa under nonreducing conditions) was decreased to approximately 32 kDa and 30 kDa by treatment with 100–600 milliunits of Endo F, indicating that the secreted rhCTGF is a glycosylated protein with 2 kDa and 4 kDa of N-linked sugars and a 30-kDa core (Fig. 1C).
Characterization of rhCTGF as IGFBP-8.
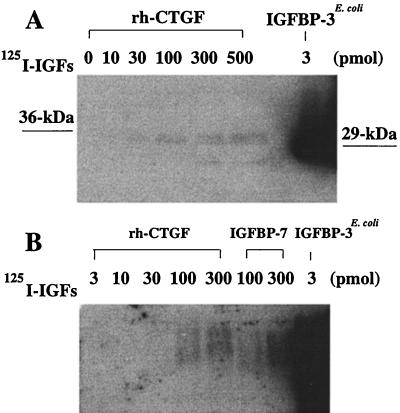
We prepared WLBs and performed IGF affinity cross-linking to characterize the affinity of rhCTGF for IGF peptides. In WLBs with 125I-labeled IGF-I and IGF-II under denaturing conditions, a 36-kDa band was detected under nonreducing conditions at amounts of rhCTGF ranging from 10 to 500 pmol, although with considerably lower binding ability than observed with IGFBP-3E. coli (Fig. 2A). A minor band at 26 kDa presumably represents a CTGF degradation product. In WLB with 125I-IGF-I and 125I-IGF-II after nondenaturing polyacrylamide gel electrophoresis, bands were also detected at CTGF amounts ranging from 30 to 300 pmol (Fig. 2B).
Figure 2.
(A) WLBs of various amounts of rhCTGF (0–500 pmol) and 3 pmol of IGFBP-3E. coli with 125I-IGFs after denaturing nonreducing SDS/PAGE. (B) WLBs of various amounts of rhCTGF, IGFBP-7 (100 and 300 pmol), and 3 pmol of IGFBP-3E. coli with 125I-IGFs after nondenaturing PAGE.
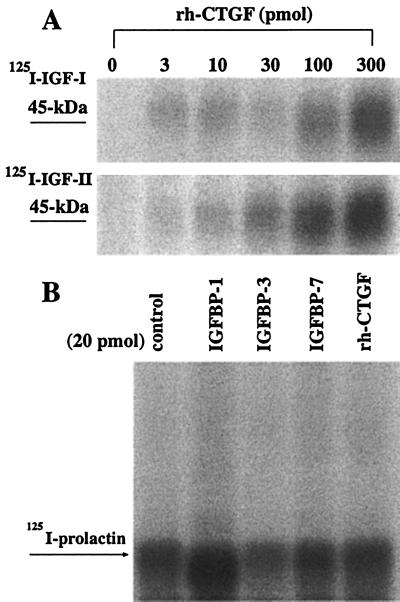
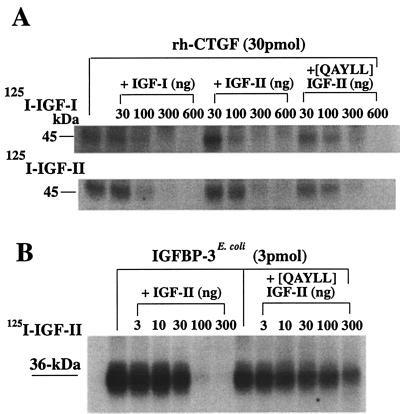
When we performed affinity cross-linking with 125I-IGF-I or -IGF-II and concentrations of rhCTGF similar to those used in WLB, a 45-kDa band was detected on a reducing SDS/12% polyacrylamide gel with concentrations of rhCTGF as low as 3 pmol (Fig. 3A). This 45-kDa band is consistent with the size of 38-kDa rhCTGF bound to 7-kDa 125I-IGF-I or 125I-IGF-II. Binding of 125I-IGF-I and 125I-IGF-II was linearly dependent on CTGF amount ranging from 3 to 300 pmol. To exclude the possibility that rhCTGF nonspecifically binds IGFs, we performed affinity cross-linking with 125I-prolactin and rhCTGF or other IGFBPs (IGFBPs-1, -3, -7). As shown in Fig. 3B, no band was detected for prolactin binding, indicating that the binding of IGF by CTGF is specific. Specificity of binding was further confirmed by competitive affinity binding assays using 125I-IGFs and unlabeled IGF-I and IGF-II (Fig. 4A). Displacement of 125I-IGF-I or -II from CTGF was observed at IGF-I and IGF-II amounts of 100–600 ng per lane. These data clearly demonstrate that the affinity of rhCTGF for IGF-I and IGF-II is significantly lower than that of rh-IGFBP-3E. coli. Interestingly, [QAYLL]IGF-II, which binds to IGFBPs 1–6 with 1/100 the affinity of IGF-II, appears to have an affinity for CTGF approximately equal to that of IGF-II (Fig. 4B).
Figure 3.
Affinity cross-linking of rhCTGF. (A) Autoradiogram of 125I-IGF-I (Upper) or 125I-IGF-II (Lower) cross-linked to rhCTGF. An approximately 45-kDa band can be seen that is compatible with the 38-kDa rhCTGF bound to 7-kDa 125I-IGF-I or 125I-IGF-II. (B) Autoradiogram of 125I-prolactin cross-linked to 20 pmol of rh-IGFBP-1 (lane 2), rh-IGFBP-3 (lane 3), rh-IGFBP-7 (lane 4) or rhCTGF (lane 5). No binding of prolactin to any of the IGFBPs was observed. The band seen at 33 kDa represents unbound 125I-prolactin.
Figure 4.
Competitive affinity cross-linking of rhCTGF or IGFBP-3E. coli. (A) Autoradiogram of 125I-IGF-I (Upper) or 125I-IGF-II (Lower) cross-linked to rhCTGF. Radiolabeled ligands (1.0 × 105 cpm) were incubated with 30 pmol of rhCTGF alone or in the presence of unlabeled IGF-I, IGF-II or [QAYLL]IGF-II, which has 1/100 the affinity for IGFBPs 1–6. (B) Autoradiogram of 125I-IGF-II cross-linked to 3 pmol of IGFBP-3E. coli in the absence or presence of different amounts of unlabeled IGF-II or [QAYLL]IGF-II.
Expression of CTGF mRNA in Normal Human Tissues.
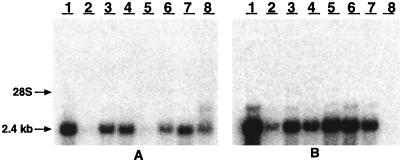
We performed Northern blot analysis to assess the expression of the CTGF gene in normal human tissues. The 2.4-kb CTGF mRNA was detected in a broad spectrum of normal human tissues (Fig. 5). In particular, CTGF mRNA was expressed at high levels in spleen, ovary, gastrointestinal tract, prostate, heart, and testis.
Figure 5.
Northern blot analysis of CTGF in normal human tissues. Human multiple tissue blots were probed with CTGF complementary RNA and exposed to Biomax film for 2 hr at −80°C. (A) Lanes 1–8 are as follows: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; and 8, pancreas. (B) Lanes 1–8: 1, spleen; 2, thymus; 3, prostate; 4, testis; 5, ovary; 6, small intestine; 7, colon; and 8, peripheral blood leukocyte.
DISCUSSION
CTGF is a cysteine-rich mitogen secreted by human umbilical vein endothelial (HUVE) cells. It was initially purified from conditioned media of HUVE cells subjected to PDGF-IgG affinity chromatography, but it was shown to not be composed of PDGF A or B chain peptides (10). Subsequent expression screening of an HUVE cell cDNA library with the anti-PDGF antibody led to the cloning and sequencing of a cDNA with an open reading frame encoding a 38-kDa protein (10). This cysteine-rich protein was shown to be the major PDGF-related mitogen and chemotactic factor secreted by HUVE cells and to compete with PDGF for binding to the PDGF cell-surface receptor on fibroblasts (10, 22). CTGF has, in fact, little peptide sequence homology with PDGF; it is believed that CTGF and PDGF monomers must share ternary structure, resulting in both common antigenic epitopes and competition for receptor binding. CTGF contains 39 cysteine residues, suggesting the presence of multiple intramolecular disulfide bonds and a complex ternary structure (14). The hydrophobic NH2-terminal sequence is consistent with its potential role as a signal peptide directing the secretion of processed CTGF.
Bork (14) has noted that CTGF is one of six different proteins, varying between 348 and 379 amino acids (including the signal peptides), that are the products of a group of “immediate-early genes” expressed after induction by growth factors or certain oncogenes. These genes are (i) human CTGF (10) and mouse CTGF (also known as fisp-12 or βIG-M2) (23), (ii) the chicken (24) and human (11) nov oncogenes, and (iii) the chicken gene cef10 (25) and the related mouse gene cyr61 (also known as βIG-M1) (12). Thirty-eight cysteines are conserved among the six proteins. It was further noted that these proteins are characterized by a modular architecture, with domains homologous to (i) IGFBPs, (ii) the von Willebrand factor type C repeat, (iii) thrombospondin type I repeat, and (iv) a COOH-terminal module. In addition to conferring the potential for binding IGF peptides, these modules may be involved in oligomerization, cell attachment through binding motifs for sulfated glycoconjugates, and dimerization (14).
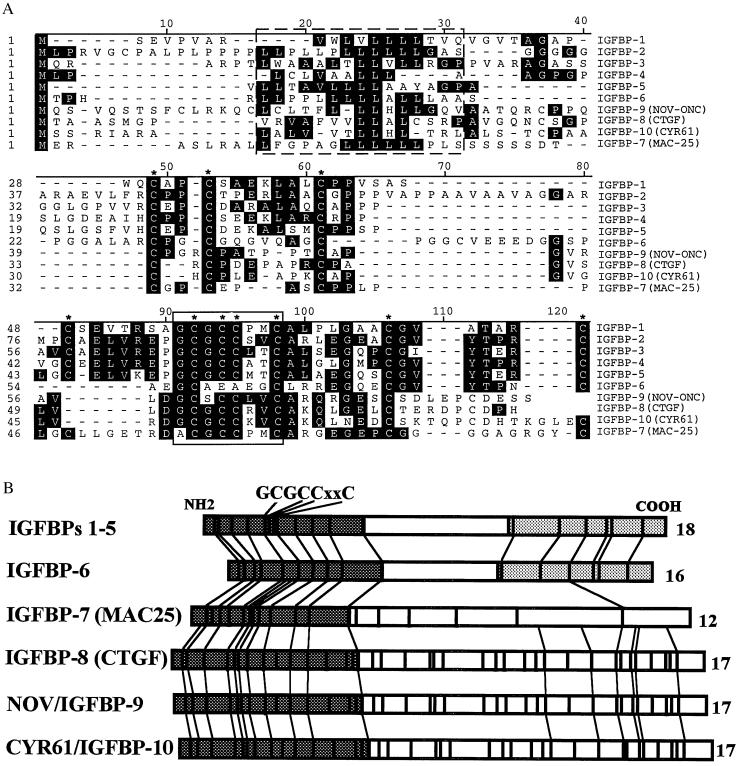
A number of characteristics of this family of proteins are of potential relevance to the IGFBPs: (i) as has been suggested for some of the IGFBPs, these proteins may be capable of IGF-independent regulation of cell growth (16, 17, 26); (ii) several of the proteins have been shown to interact with both cell surfaces and extracellular matrix and to be capable of binding to heparin, properties also shared by some members of the IGFBP family (27); and (iii) several genes from this family are induced by transforming growth factor β, as is the case for some IGFBPs (15, 28). Most importantly, these proteins share significant sequence homology with the IGFBPs, including preservation of as many as 17 of the 18 cysteines conserved in IGFBPs 1–6, and conservation of the “GCGCCXXC” motif found in the NH2 terminus of the IGFBPs (Fig. 6).
Figure 6.
Comparison of NH2-terminal amino acid sequences (A) and total cysteine residues (B) of IGFBP superfamily. (A) Sequences of IGFBPs 1–7 (1–7), CTGF (10), nov oncogene (22), and cyr61 (S.R.N., unpublished data) were aligned using the DNAstar MegAlign program (Madison, WI). Consensus signal sequences are boxed by a broken line. The characteristic IGFBP motif (GCGCCXXC) is boxed by a solid line. Conserved cysteine residues in the family are marked with asterisks. We have named mac-25 IGFBP-7 and have named CTGF IGFBP-8. We have tentatively named the protein products of the nov and cyr61 genes IGFBP-9 and IGFBP-10, respectively. (B) The conservation of the cysteine residues is shown by the vertical lines. The numbers on the right represent the total conserved cysteines among IGFBPs. The shaded regions represent the conserved domain of IGFBPs in their NH2 or COOH terminus.
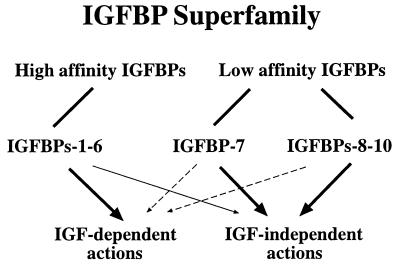
CTGF shares an overall 28–38% amino acid identity to IGFBPs 1–6. Both WLB, after either denaturing or nondenaturing PAGE, and affinity cross-linking demonstrate that CTGF specifically binds IGF-I and -II, although with relatively low affinity as compared with IGFBPs 1–6. Nevertheless, the specificity of IGF binding, coupled with the structural similarity to IGFBPs 1–7, indicates that CTGF meets the criteria necessary to define it as a member of the IGFBP family, IGFBP-8. Specificity of binding was demonstrated by the failure of CTGF (like IGFBPs 1, 3, and 7) to bind prolactin, as well as by the dose-dependent inhibition of binding by unlabeled IGF-I or -II. Interestingly, although the affinity of IGFBP-8 for IGFs is significantly reduced, the affinity of IGFBP-8 for [QAYLL]IGF-II appears to be approximately equivalent to its affinity for IGF-II, in contrast to the situation for IGFBPs 1–6, where [QAYLL]IGF-II binds with 1/100 the affinity. These results suggest that the IGF binding site in IGFBP-8 may differ somewhat from the binding site in IGFBPs 1–6. Whether this reflects the larger, more complex structure of IGFBP-8, or results from the less rigorous conservation of structures in the NH2 terminus or lack of conserved sequences in the COOH terminus is unclear. It is of note that the IGFBP-3 fragments derived from proteolysis appear to exhibit significantly reduced affinity, similar to that of IGFBP-8 (29). Furthermore, a baculovirus-expressed IGFBP-3 fragment corresponding to the NH2-terminal portion of IGFBP-3 (amino acids 1–97) was able to bind IGF-I and IGF-II with low affinity, which is comparable to those of low-affinity IGF binders (IGFBPs 7 and 8) (unpublished results). Nevertheless, it appears that the IGFBP superfamily, which now includes 10 potential members, can be divided into at least two subgroups: high-affinity IGF binders (IGFBPs 1–6) and low-affinity IGF binders (IGFBPs 7 and 8 and, potentially, the protein products of the nov and cyr61 genes) (Fig. 7). Although the latter two proteins have yet to be evaluated for IGF affinity, given their structural similarity with CTGF/IGFBP-8, it seems reasonable to consider them as potential members of the IGFBP superfamily, pending further studies.
Figure 7.
The IGFBP superfamily composed of high-affinity IGFBPs and low-affinity IGFBPs. The thick and thin arrows indicate proposed primary and secondary biological actions, respectively, and the dashed arrows represent potential actions of IGFBPs that have not been verified experimentally.
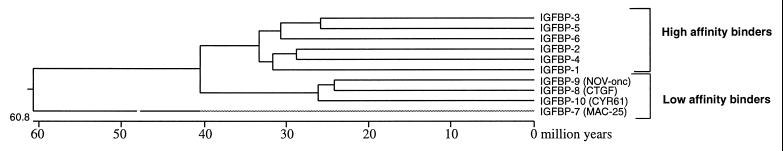
The dendrogram depicted in Fig. 8 indicates that, on the basis of structural similarities, all 10 members of the superfamily can be traced back to an ancestral gene approximately 60 million years ago (30). IGFBP-7 is the most divergent, followed by the CTGF (IGFBP-8), nov oncogene (provisionally IGFBP-9) and cyr61 (provisionally IGFBP-10). It is of note that IGFBPs or IGFBP-like activity have been identified in fish, amphibians, reptiles, and birds, indicating that significant conservation exists among vertebrates (31). The detection of an IGFBP in lamprey serum suggests that IGFBPs have been present since early vertebrate evolution (32). On the other hand, the fact that the genes for IGFBPs 1–8 are located on six different chromosomes demonstrates that some degree of divergence has also occurred.
Figure 8.
Phylogenetic tree of the IGFBP superfamily, derived from multiple comparisons of full-length sequences of known IGFBPs 1–7, CTGF, nov oncogene, and cyr61, using the method of Hein (30) as implemented by the DNAstar MegAlign program.
The existence of an IGFBP superfamily has important implications for our understanding of the biological roles of these proteins, including the conventional IGFBPs. While the lower affinity for IGFs exhibited by IGFBP-7 and -8 does not exclude a role for them as IGF transporters or modulators of IGF action, it is more likely that these proteins regulate cell proliferation and chemotaxis in an IGF-independent manner. It is noteworthy that a number of recent studies have indicated that several of the conventional IGFBPs, most notably IGFBP-3, are also capable of IGF-independent effects, and that these actions may be of importance in modulation of normal and malignant cell growth (16, 17, 26).
We speculate that the IGFBP superfamily is derived from an ancestral gene/protein that was critically involved in the regulation of cell growth and was capable of binding IGF peptides. Over the course of evolution, some members evolved into high-affinity IGF binders and others into low-affinity IGF binders, thereby conferring on the IGFBP superfamily the ability to influence cell growth by both IGF-dependent and IGF-independent mechanisms.
Acknowledgments
This work was supported in part by National Institutes of Health Grant CA58110 (to R.G.R.), National Institutes of Health Grant DK50810 (to C.T.R.), and U.S. Army Grant DAMD17-96-1–6204 (to Y.O.).
ABBREVIATIONS
- IGF
insulin-like growth factor
- IGFBP
IGF binding protein
- CTGF
connective tissue growth factor
- rhCTGF
recombinant human CTGF
- PDGF
platelet-derived growth factor
- [QAYLL]IGF-II
[Gln6,Ala7,Tyr18,Leu19,Leu27]IGF-II
- Endo F
endoglycosidase F
- WLB
Western ligand blot
References
- 1.Lee Y-L, Hintz R L, James P M, Lee P D K, Shively J E, Powell D R. Mol Endocrinol. 1988;2:404–411. doi: 10.1210/mend-2-5-404. [DOI] [PubMed] [Google Scholar]
- 2.Binkert C, Landwehr J, Mary J-L, Schwander J, Heinrich G. EMBO J. 1989;8:2497–2502. doi: 10.1002/j.1460-2075.1989.tb08386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood W I, Cachianes G, Henzel W J, Winslow G A, Spencer S A, Hellmiss R, Martin J L, Baxter R C. Mol Endocrinol. 1989;2:1176–1185. doi: 10.1210/mend-2-12-1176. [DOI] [PubMed] [Google Scholar]
- 4.LaTour D, Mohan S, Linkhart T A, Baylink D J, Strong D D. Mol Endocrinol. 1990;4:1806–1814. doi: 10.1210/mend-4-12-1806. [DOI] [PubMed] [Google Scholar]
- 5.Kiefer M C, Ioh R S, Bauer D M, Zapf J. Biochem Biophys Res Commun. 1991;176:219–225. doi: 10.1016/0006-291x(91)90912-q. [DOI] [PubMed] [Google Scholar]
- 6.Kiefer M C, Masiarz F R, Bauer D M, Zapf J. J Biol Chem. 1991;266:9043–9049. [PubMed] [Google Scholar]
- 7.Shimasaki S, Shimonaka M, Zhang H-P, Ling N. J Biol Chem. 1991;266:10646–10653. [PubMed] [Google Scholar]
- 8.Shimasaki S, Ling N. Prog Growth Factor Res. 1992;3:243–266. doi: 10.1016/0955-2235(91)90003-m. [DOI] [PubMed] [Google Scholar]
- 9.Oh Y, Nagalla S R, Yamanaka Y, Kim H S, Wilson E, Rosenfeld R G. J Biol Chem. 1996;271:30322–30325. doi: 10.1074/jbc.271.48.30322. [DOI] [PubMed] [Google Scholar]
- 10.Bradham D M, Igarashi A, Potter R L, Grotendorst G R. J Cell Biol. 1991;114:1285–1294. doi: 10.1083/jcb.114.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinerie C, Viegas-Pequignot E, Guenard I, Dutrillaux B, Nguyen V C, Bernheim A, Perbal B. Oncogene. 1992;7:2529–2534. [PubMed] [Google Scholar]
- 12.O’Brien T P, Yang G P, Sanders L, Lau L F. Mol Cell Biol. 1990;10:3569–3577. doi: 10.1128/mcb.10.7.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kireeva M L, Mo F-E, Yang G P, Lau L F. Mol Cell Biol. 1996;16:1326–1334. doi: 10.1128/mcb.16.4.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bork P. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi A, Okochi H, Bradham D M, Grotendorst G R. Mol Biol Cell. 1993;4:637–645. doi: 10.1091/mbc.4.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh Y, Muller H L, Zhang H, Ling N, Rosenfeld R G. Adv Exp Med Biol. 1994;343:41–54. doi: 10.1007/978-1-4615-2988-0_5. [DOI] [PubMed] [Google Scholar]
- 17.Oh Y, Muller H L, Pham H M, Rosenfeld R G. J Biol Chem. 1993;268:26045–26048. [PubMed] [Google Scholar]
- 18.Nagalla S R, Barry B J, Falick A M, Gibson B W, Taylor J E, Dong J Z, Spindel E R. J Biol Chem. 1996;271:7731–7737. doi: 10.1074/jbc.271.13.7731. [DOI] [PubMed] [Google Scholar]
- 19.Nagalla S R, Barry B J, Spindel E R. Mol Endocrinol. 1994;8:943–951. doi: 10.1210/mend.8.8.7997236. [DOI] [PubMed] [Google Scholar]
- 20.Gargosky S E, Pham H P, Wilson K F, Liu F, Giudice L C, Rosenfeld R G. Endocrinology. 1992;131:3051–3060. doi: 10.1210/endo.131.6.1280211. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.DiCorleto P E. Exp Cell Res. 1984;153:167–172. doi: 10.1016/0014-4827(84)90458-0. [DOI] [PubMed] [Google Scholar]
- 23.Ryseck R-P, Macdonald-Bravo H, Mattei M-G, Bravo R. Cell Growth Differ. 1991;2:225–233. [PubMed] [Google Scholar]
- 24.Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons D L, Levy D B, Yannoni Y, Erikson R L. Proc Natl Acad Sci USA. 1989;86:1178–1182. doi: 10.1073/pnas.86.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh Y, Muller H L, Lamson G, Rosenfeld R G. J Biol Chem. 1993;268:14964–14971. [PubMed] [Google Scholar]
- 27.Yang G P, Lau L F. Cell Growth Differ. 1991;2:351–357. [PubMed] [Google Scholar]
- 28.Oh Y, Muller H L, Ng L, Rosenfeld R G. J Biol Chem. 1995;270:13589–13592. doi: 10.1074/jbc.270.23.13589. [DOI] [PubMed] [Google Scholar]
- 29.Lalou C, Lassarre C, Binoux M. Endocrinology. 1996;137:3206–3212. doi: 10.1210/endo.137.8.8754741. [DOI] [PubMed] [Google Scholar]
- 30.Hein J. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- 31.Kelley K M, Oh Y, Gargosky S E, Gucev Z, Matsumoto T, Hwa V, Ng L, Simpson D M, Rosenfeld R G. Intl J Biochem Cell Biol. 1996;28:619–637. doi: 10.1016/1357-2725(96)00005-2. [DOI] [PubMed] [Google Scholar]
- 32.Upton A, Chan S J, Steiner D F, Wallace J C, Ballard F J. Growth Regul. 1992;3:29–32. [PubMed] [Google Scholar]