Abstract
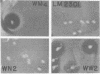
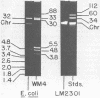
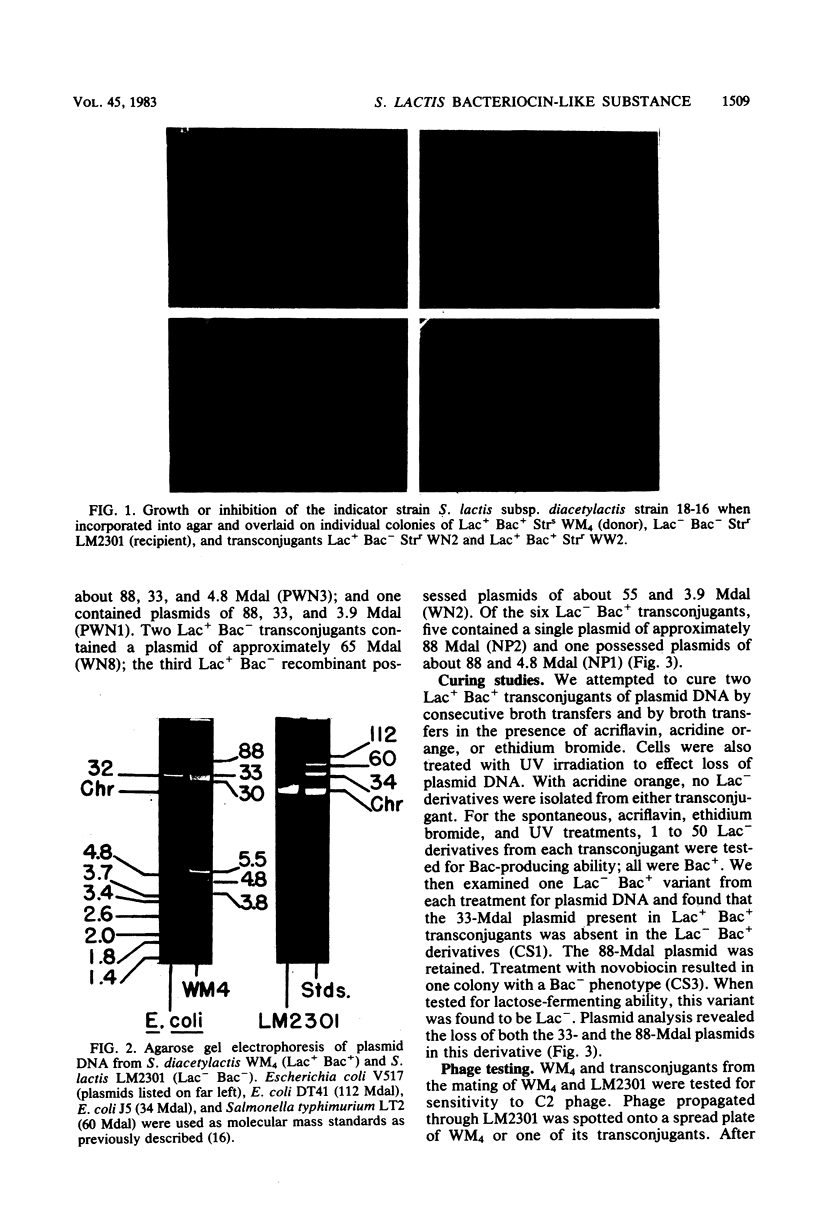
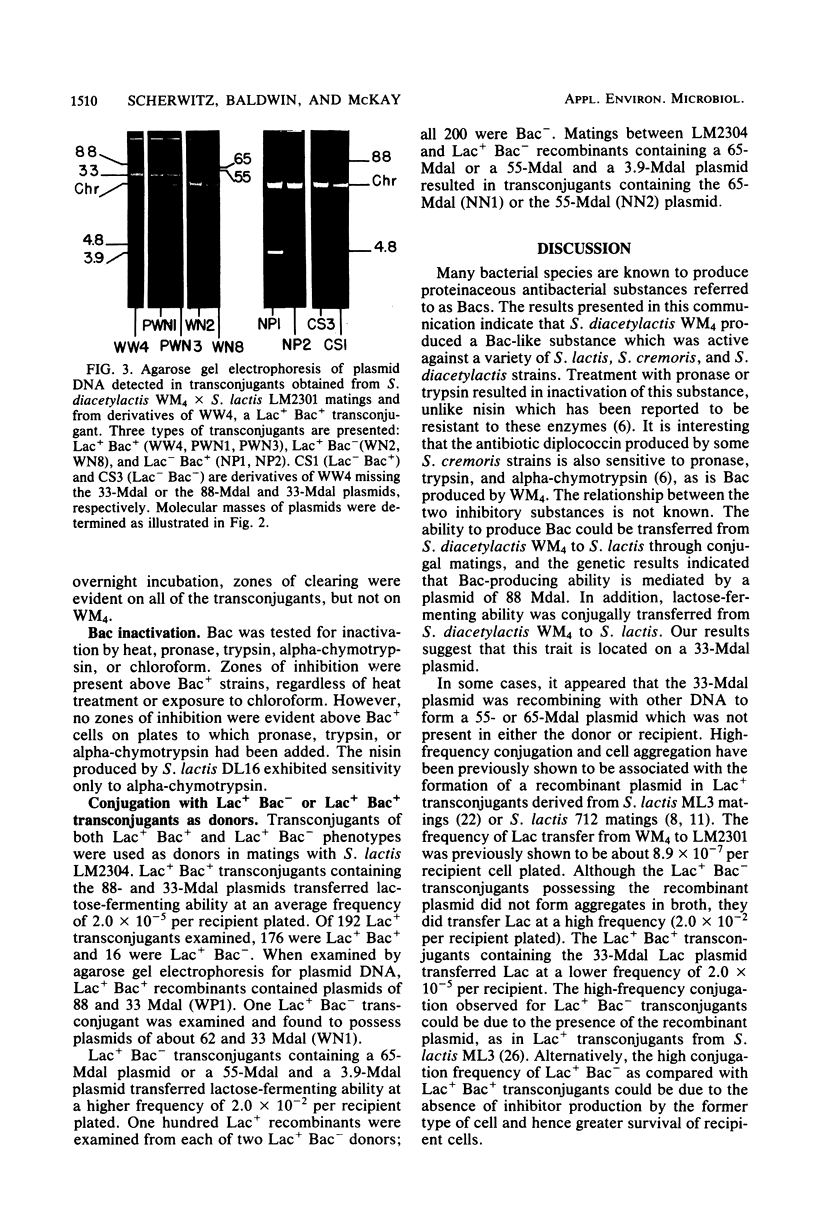
Streptococcus lactis subsp. diacetylactis strain WM4 transferred lactose-fermenting and bacteriocin-producing (Bac+) abilities to S. lactis LM2301, a lactose-negative, streptomycin-resistant (Lac- Strr), plasmid-cured derivative of S. lactis C2. Three types of transconjugants were obtained: Lac+ Bac+, Lac+ Bac-, and Lac-Bac+.S. diacetylactis WM4 possessed plasmids of 88, 33, 30, 5.5, 4.8, and 3.8 megadaltons (Mdal). In Lac+ Bac+ transconjugants, lactose-fermenting ability was linked to the 33-Mdal plasmid and bacteriocin-producing ability to the 88-Mdal plasmid. Curing the 33-Mdal plasmid from Lac+ Bac+ transconjugants resulted in loss of lactose-fermenting ability but not bacteriocin-producing ability (Lac- Bac+). These strains retained the 88-Mdal plasmid. Curing of both plasmids resulted in a Lac- Bac- phenotype. The Lac+ Bac- transconjugant phenotype was associated with a recombinant plasmid of 55 or 65 Mdal. When these transconjugants were used as donors in subsequent matings, the frequency of Lac transfer was about 2.0 X 10(-2) per recipient plated, whereas when Lac+ Bac+ transconjugants served as donors, the frequency of Lac transfer was about 2.0 X 10(-5) per recipient plated. Also, Lac- Bac+ transconjugants were found to contain the 88-Mdal plasmid. The data indicate that the ability of WM4 to produce bacteriocin is linked to an 88-Mdal conjugative plasmid and that lactose-fermenting ability resides on a 33-Mdal plasmid.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Davey G. P., Richardson B. C. Purification and Some Properties of Diplococcin from Streptococcus cremoris 346. Appl Environ Microbiol. 1981 Jan;41(1):84–89. doi: 10.1128/aem.41.1.84-89.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies F. L., Gasson M. J. Reviews of the progress of dairy science: genetics of lactic acid bacteria. J Dairy Res. 1981 Jun;48(2):363–376. doi: 10.1017/s0022029900021798. [DOI] [PubMed] [Google Scholar]
- Fuchs P. G., Zajdel J., Dobrzański W. T. Possible plasmid nature of the determinant for production of the antibiotic nisin in some strains of Streptococcus lactis. J Gen Microbiol. 1975 May;88(1):189–192. doi: 10.1099/00221287-88-1-189. [DOI] [PubMed] [Google Scholar]
- Gasson M. J., Davies F. L. High-frequency conjugation associated with Streptococcus lactis donor cell aggregation. J Bacteriol. 1980 Sep;143(3):1260–1264. doi: 10.1128/jb.143.3.1260-1264.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis A., Singh J., Teuber M. Potential of lactic streptococci to produce bacteriocin. Appl Environ Microbiol. 1983 Jan;45(1):205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst A. Nisin: its preservative effect and function in the growth cycle of the producer organism. Soc Appl Bacteriol Symp Ser. 1978;7:297–314. [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak W., Bardowski J., Dobrzański W. T. Lactostrepcins--acid bacteriocins produced by lactic streptococci. J Dairy Res. 1978 Jun;45(2):247–257. doi: 10.1017/s0022029900016423. [DOI] [PubMed] [Google Scholar]
- Kozar W., Rajchert-Trzpil M., Dobrzański W. T. The effect of proflavin, ethidium bromide and an elevated temperature on the appearance of nisin-negative clones in nisin-producing strains of Streptococcus lactis. J Gen Microbiol. 1974 Aug;83(2):295–302. doi: 10.1099/00221287-83-2-295. [DOI] [PubMed] [Google Scholar]
- Kékessy D. A., Piguet J. D. New method for detecting bacteriocin production. Appl Microbiol. 1970 Aug;20(2):282–283. doi: 10.1128/am.20.2.282-283.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh G. L., Swartz M. N. Elimination of plasmids from several bacterial species by novobiocin. Antimicrob Agents Chemother. 1977 Sep;12(3):423–426. doi: 10.1128/aac.12.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Induction of prophage in Streptococcus lactis C2 by ultraviolet irradiation. Appl Microbiol. 1973 Apr;25(4):682–684. doi: 10.1128/am.25.4.682-684.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Cords B. R., Baldwin K. A. Transduction of lactose metabolism in Streptococcus lactis C2. J Bacteriol. 1973 Sep;115(3):810–815. doi: 10.1128/jb.115.3.810-815.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P. M., McKay L. L. Recombinant plasmid associated cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. J Bacteriol. 1981 Jun;146(3):937–944. doi: 10.1128/jb.146.3.937-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]